Exploration of the Hsa-miR-1587–Protein Interaction and the Inhibition to CASK
Abstract
:1. Introduction
2. Results and Discussion
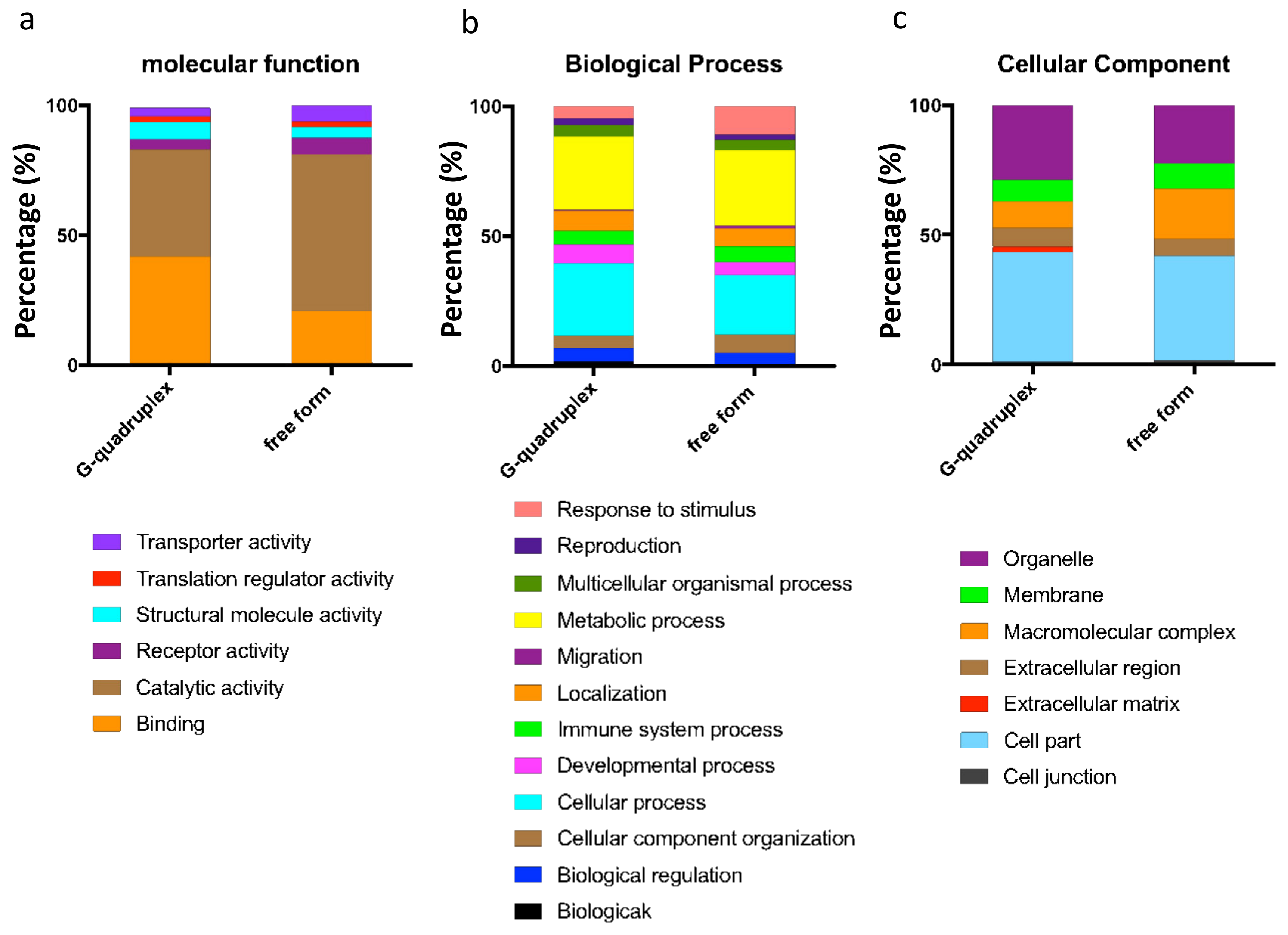
2.1. Different Structures of miR-1587 Bound to Different Proteins
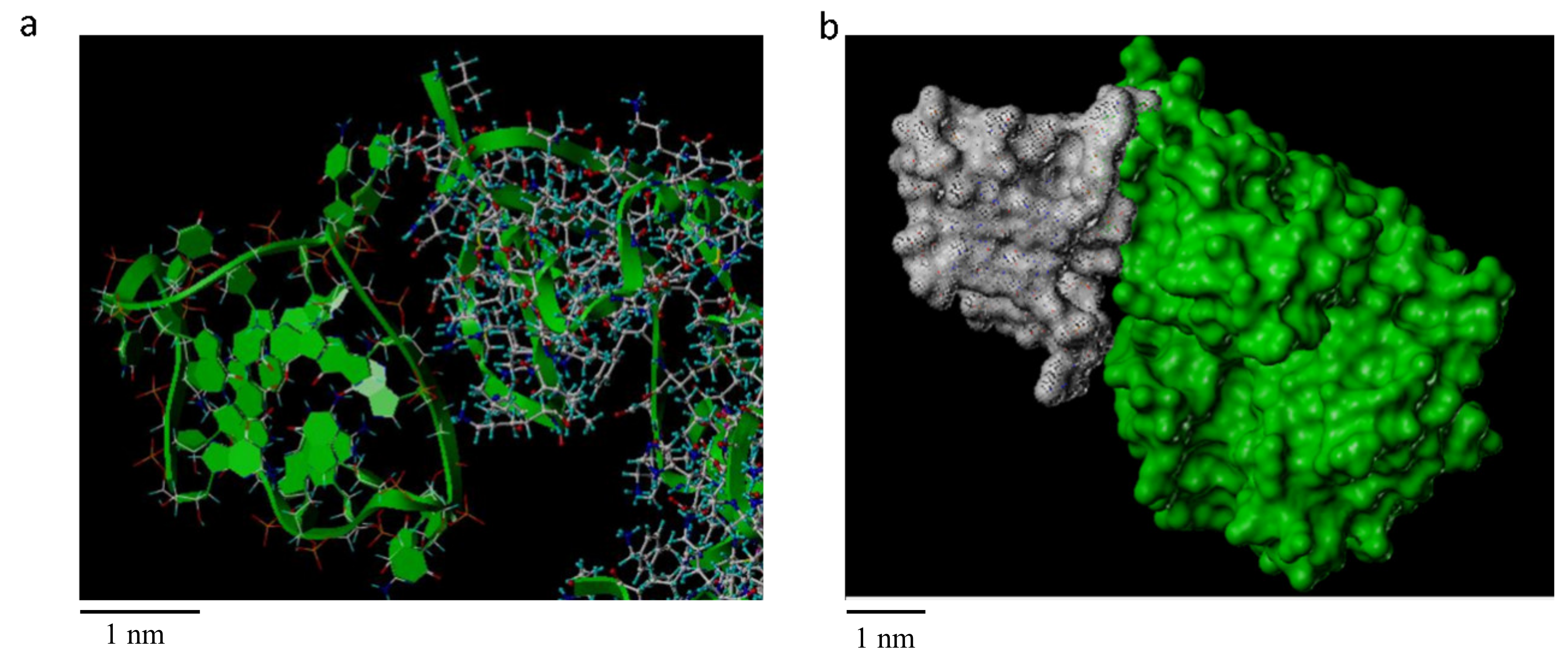
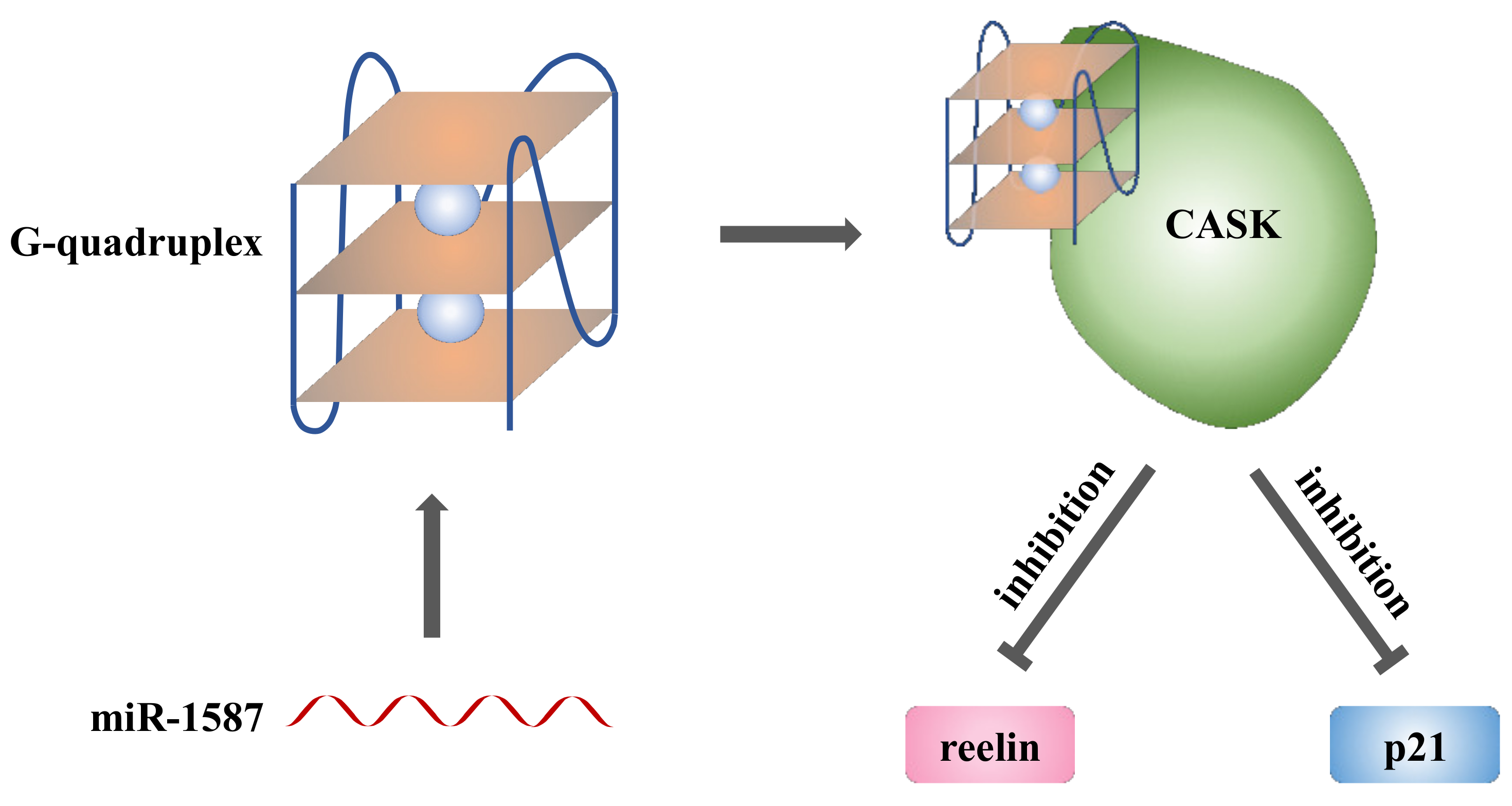
2.2. miR-1587 in G-Quadruplex Structure Bound to CASK
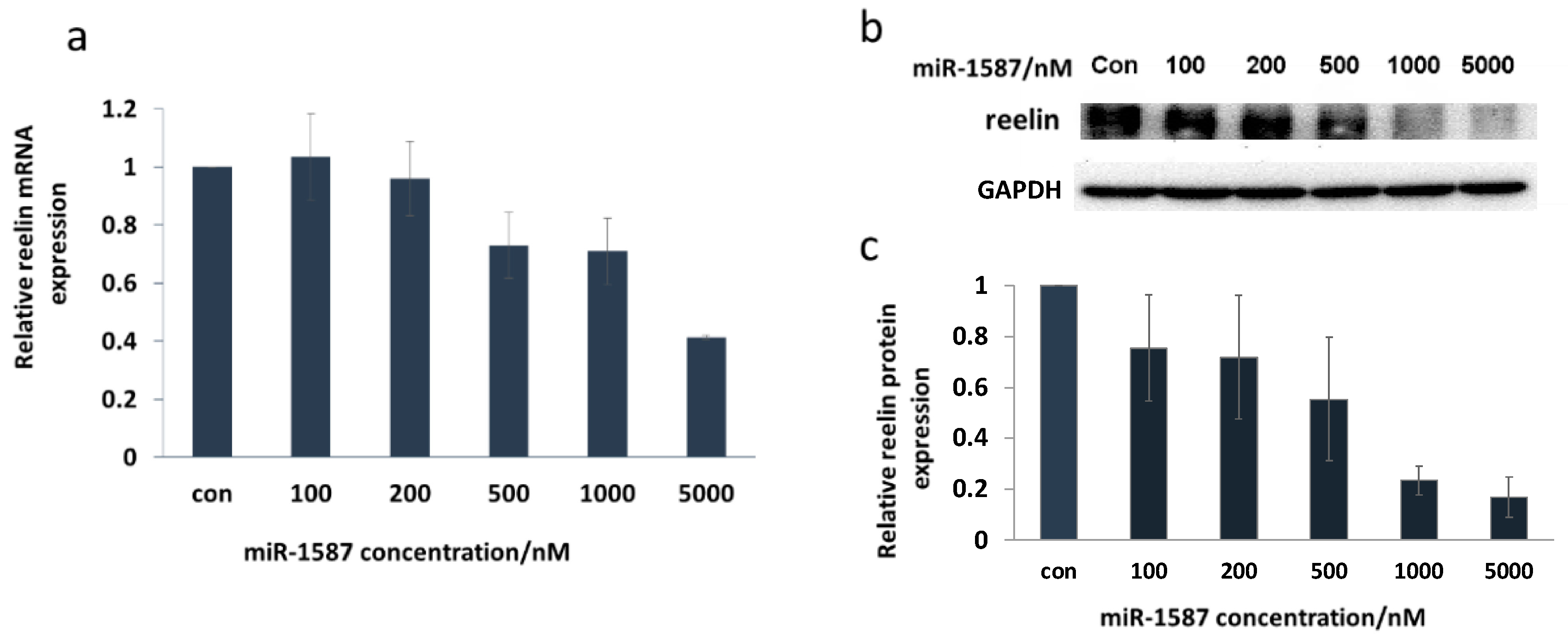
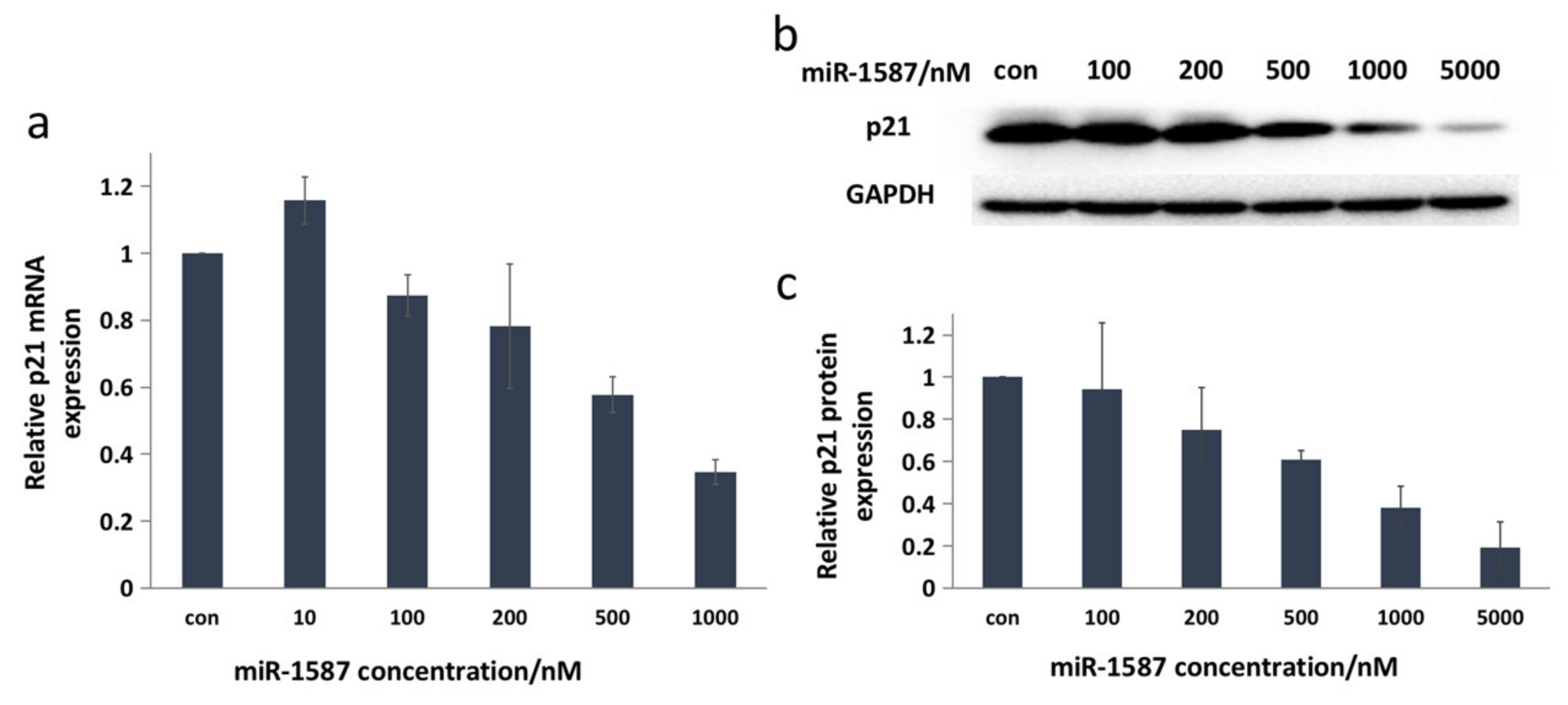
2.3. miR-1587 G-Quadruplex Influenced CASK Downstream Genes Reelin and p21
3. Materials and Methods
3.1. Materials
3.2. Human Proteome Microarray Assay
3.3. Computer Simulation
3.4. Quantitative Real-Time PCR
3.5. Western Blot
3.6. Bioinformatic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| miRNA/miR | MicroRNA |
| CASK | Calcium/Calmodulin-Dependent Serine Protein Kinase |
| CLL | Chronic Lymphocytic Leukemia |
| AGO | Archipelago |
| miRBP | MicroRNA Binding Protein |
| 3′-UTR | 3′-Untranslated Regions |
| GRSF | Guanine-Rich Sequence Factor |
| TUT4 | Terminal Uridylyltransferase 4 |
| PUP-2 | PAP-Associated Domain-Containing Protein |
| hnRNP | Heterogeneous Nuclear Ribonucleoprotein |
| CEBPA | CCAAT Enhancer Binding Protein Alpha |
| ESI-MS | Electrospray Ionization Mass Spectrometry |
| CD | Circular Dichroism |
| NMR | Nuclear Magnetic Resonance |
| NCOR1 | Nuclear Receptor Corepressor 1 |
| q-RT-PCR | Quantitative Real-Time Polymerase Chain Reaction |
| BSA | Bovine Serum Albumin |
| TBST | Tris Buffered Saline with Tween |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| FBS | Fetal Bovine Serum |
| SDS-PAGE | Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| PBST | Phosphate-Buffered Saline with Tween |
| SNR | Signal to Noise Ratio |
| NF-κB | Nuclear Factor kappa B |
| WNT6 | Wingless-Type MMTV Integration Site Family, member 6 |
| CCDC130 | Coiled-Coil Domain Containing 130 |
| NAA16 | N-Alpha-Acetyltransferase 16, NatA auxiliary subunit |
| BIRC5 | Baculoviral IAP Repeat Containing 5 |
References
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dan Dumitru, C.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.-F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8, R214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sempere, L.F.; Christensen, M.; Silahtaroglu, A.; Bak, M.; Heath, C.V.; Schwartz, G.; Wells, W.; Kauppinen, S.; Cole, C.N. Altered microRNA expression confined to specific epithelial cell Subpopulations in breast cancer. Cancer Res. 2007, 67, 11612–11620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, K.; Ghoshal, B.; Ghosh, S.; Chakrabarty, Y.; Shwetha, S.; Das, S.; Bhattacharyya, S.N. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 2016, 17, 1184–1203. [Google Scholar] [CrossRef] [PubMed]
- Poria, D.K.; Guha, A.; Nandi, I.; Ray, P.S. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 2016, 35, 1703–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA Stability Factor HuR Inhibits MicroRNA-16 Targeting of COX-2. Mol. Cancer Res. 2012, 10, 167–180. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Jo, M.H.; White, E.J.F.; De, S.; Hafner, M.; Zucconi, B.E.; Abdelmohsen, K.; Martindale, J.L.; Yang, X.L.; Wood, W.H.; et al. AUF1 promotes let-7b loading on Argonaute 2. Genes Dev. 2015, 29, 1599–1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Lv, J.; Liu, M.; Tang, H. miR-346 Up-regulates Argonaute 2 (AGO2) Protein Expression to Augment the Activity of Other MicroRNAs (miRNAs) and Contributes to Cervical Cancer Cell Malignancy. J. Biol. Chem. 2015, 290, 30342–30350. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, S.R.; Daley, G.Q. Lin28: A MicroRNA Regulator with a Macro Role. Cell 2010, 140, 445–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eiring, A.M.; Harb, J.G.; Neviani, P.; Garton, C.; Oaks, J.J.; Spizzo, R.; Liu, S.J.; Schwind, S.; Santhanam, R.; Hickey, C.J.; et al. miR-328 Functions as an RNA Decoy to Modulate hnRNP E2 Regulation of mRNA Translation in Leukemic Blasts. Cell 2010, 140, 652–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa, J.; Phillips, L.M.; Shahar, T.; Hossain, A.; Gumin, J.; Kim, H.; Bean, A.J.; Calin, G.A.; Fueyo, J.; Walters, E.T.; et al. Exosomes from Glioma-Associated Mesenchymal Stem Cells Increase the Tumorigenicity of Glioma Stem-like Cells via Transfer of miR-1587. Cancer Res. 2017, 77, 5808–5819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Long, H.; Cui, X.; Zhou, J.; Xu, M.; Yuan, G. Exploring the Formation and Recognition of an Important G-Quadruplex in a HIF1 alpha Promoter and Its Transcriptional Inhibition by a Benzo c phenanthridine Derivative. J. Am. Chem. Soc. 2014, 136, 2583–2591. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Zheng, H.; Dong, S.; Sun, R.; Qiao, S.; Sun, H.; Ma, X.; Zhang, T.; Sun, C. Facile detection of melamine by a FAM-aptamer-G-quadruplex construct. Anal. Bioanal. Chem. 2019, 411, 2521–2530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-N.; Yang, L.; Ling, J.-Y.; Czajkowsky, D.M.; Wang, J.-F.; Zhang, X.-W.; Zhou, Y.-M.; Ge, F.; Yang, M.-K.; Xiong, Q.; et al. Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc. Natl. Acad. Sci. USA 2015, 112, 15084–15089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.C.; Li, T.; Song, G.; He, Q.X.; Yin, Y.F.; Lu, J.Y.Y.; Bi, X.J.; Wang, K.L.; Luo, S.; Chen, Y.S.; et al. Insight into novel RNA-binding activities via large-scale analysis of lncRNA-bound proteome and IDH1-bound transcriptome. Nucleic Acids Res. 2019, 47, 2244–2262. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.L.; Qi, B.; Cai, Q.Q.; Fu, L.; Yang, Y.; Tang, C.; Zhu, P.; Chen, Q.W.; Pan, J.; Chen, M.H.; et al. LncRNA AY promotes hepatocellular carcinoma metastasis by stimulating ITGAV transcription. Theranostics 2019, 9, 4421–4436. [Google Scholar] [CrossRef]
- Hsueh, Y.P.; Wang, T.F.; Yang, F.C.; Sheng, M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature 2000, 404, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, J.Y.; Yang, C.H.; Wang, X.Q.; Cheng, L.; Hu, G.X.; Sun, Y.T.; Zhang, X.; Wu, M.; Liu, Z.H. CASK and its target gene Reelin were co-upregulated in human esophageal carcinoma. Cancer Lett. 2002, 179, 71–77. [Google Scholar] [CrossRef]
- Brugarolas, J.; Moberg, K.; Boyd, S.D.; Taya, Y.; Jacks, T.; Lees, J.A. Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G(1) arrest after gamma-irradiation. Proc. Natl. Acad. Sci. USA 1999, 96, 1002–1007. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.S.; Kuo, Y.H.; Kuo, H.C.; Hsieh, M.C.; Huang, C.Y.; Lee, K.C.; Lee, K.F.; Shen, C.H.; Tung, S.Y.; Teng, C.C. CIL-102-Induced Cell Cycle Arrest and Apoptosis in Colorectal Cancer Cells via Upregulation of p21 and GADD45. PLoS ONE 2017, 12, e0168989. [Google Scholar] [CrossRef]
- Ropponen, K.M.; Kellokoski, J.K.; Lipponen, P.K.; Pietilainen, T.; Eskelinen, M.J.; Alhava, E.M.; Kosma, V.M. P21/WAF1 expression in human colorectal carcinoma: Association with p53, transcription factor AP-2 and prognosis. Br. J. Cancer 1999, 81, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Sun, R.J.; Su, Y.Y.; Zhao, X.D.; Qi, J.; Luo, X.F.; Yang, Z.C.; Yao, Y.M.; Luo, X.D.; Xia, Z.F. Human calcium/calmodulin-dependent serine protein kinase regulates the expression of p21 via the E2A transcription factor. Biochem. J. 2009, 420, 493. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Kumar, K.; Hu, X.; Wallqvist, A.; Reifman, J. DOVIS 2.0: An efficient and easy to use parallel virtual screening tool based on AutoDock 4.0. Chem. Cent. J. 2008, 2, 18. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Zhou, J.; Xu, M.; Yuan, G. Exploration of the Hsa-miR-1587–Protein Interaction and the Inhibition to CASK. Int. J. Mol. Sci. 2021, 22, 10716. https://doi.org/10.3390/ijms221910716
Zhang L, Zhou J, Xu M, Yuan G. Exploration of the Hsa-miR-1587–Protein Interaction and the Inhibition to CASK. International Journal of Molecular Sciences. 2021; 22(19):10716. https://doi.org/10.3390/ijms221910716
Chicago/Turabian StyleZhang, Lulu, Jiang Zhou, Ming Xu, and Gu Yuan. 2021. "Exploration of the Hsa-miR-1587–Protein Interaction and the Inhibition to CASK" International Journal of Molecular Sciences 22, no. 19: 10716. https://doi.org/10.3390/ijms221910716
APA StyleZhang, L., Zhou, J., Xu, M., & Yuan, G. (2021). Exploration of the Hsa-miR-1587–Protein Interaction and the Inhibition to CASK. International Journal of Molecular Sciences, 22(19), 10716. https://doi.org/10.3390/ijms221910716






