Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges
Abstract
:1. Introduction
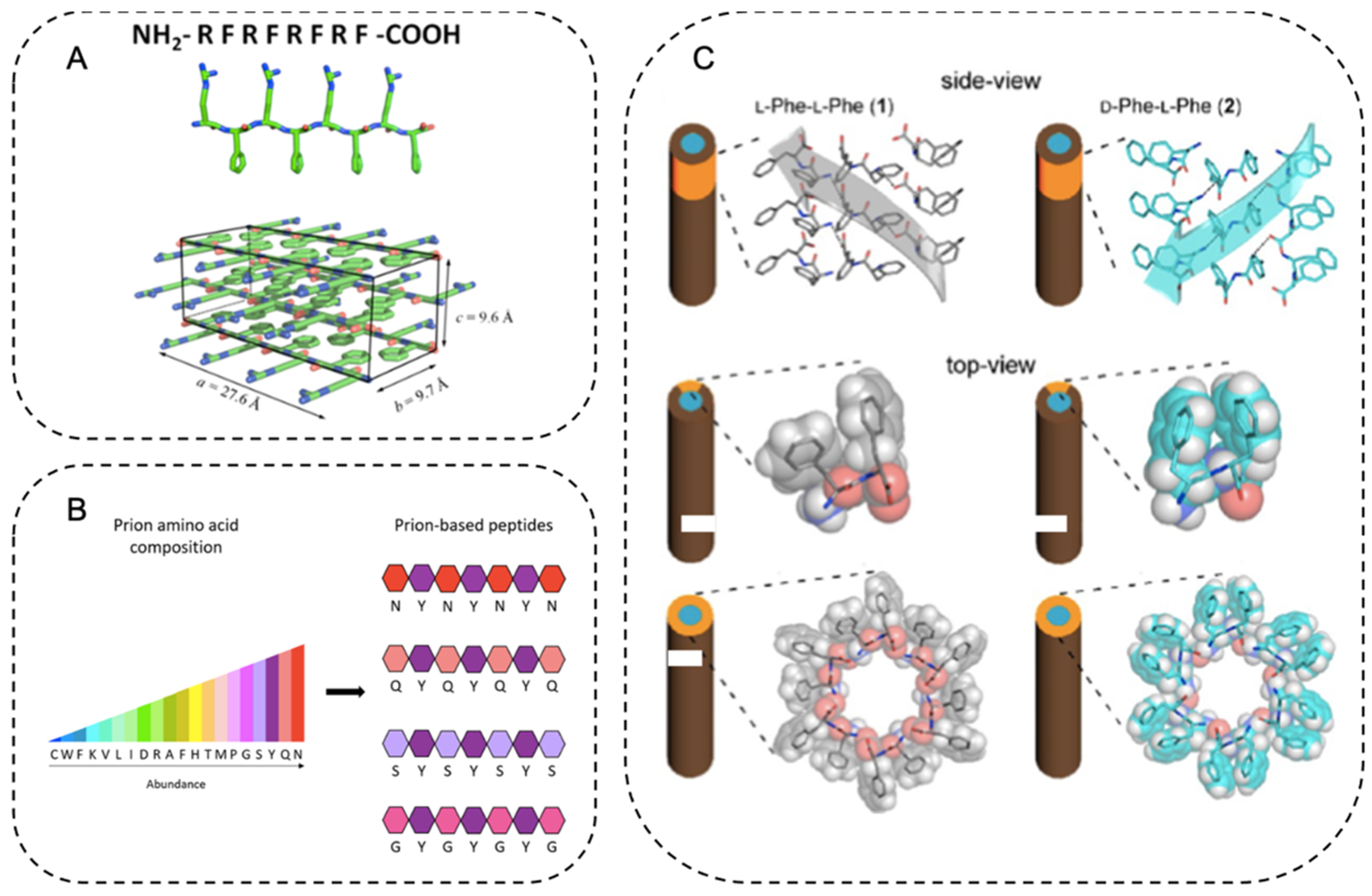
2. Structure and Property of Amyloid Fibrils
3. Macroscopic Functional Amyloid Materials
3.1. Functional Amyloids in Nature
3.2. Artificial Macroscopic Functional Amyloids
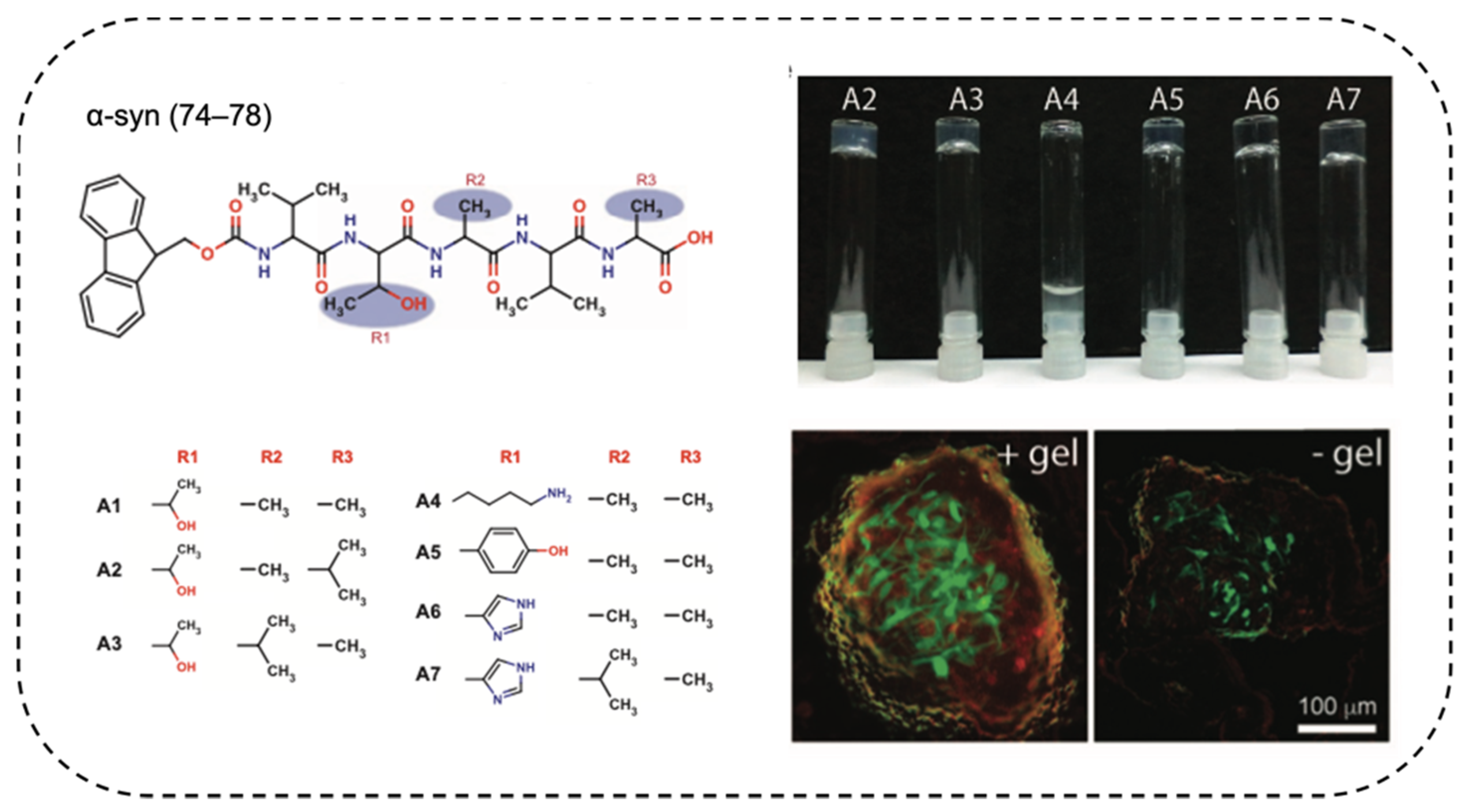
3.2.1. Amyloid-Based Hydrogels
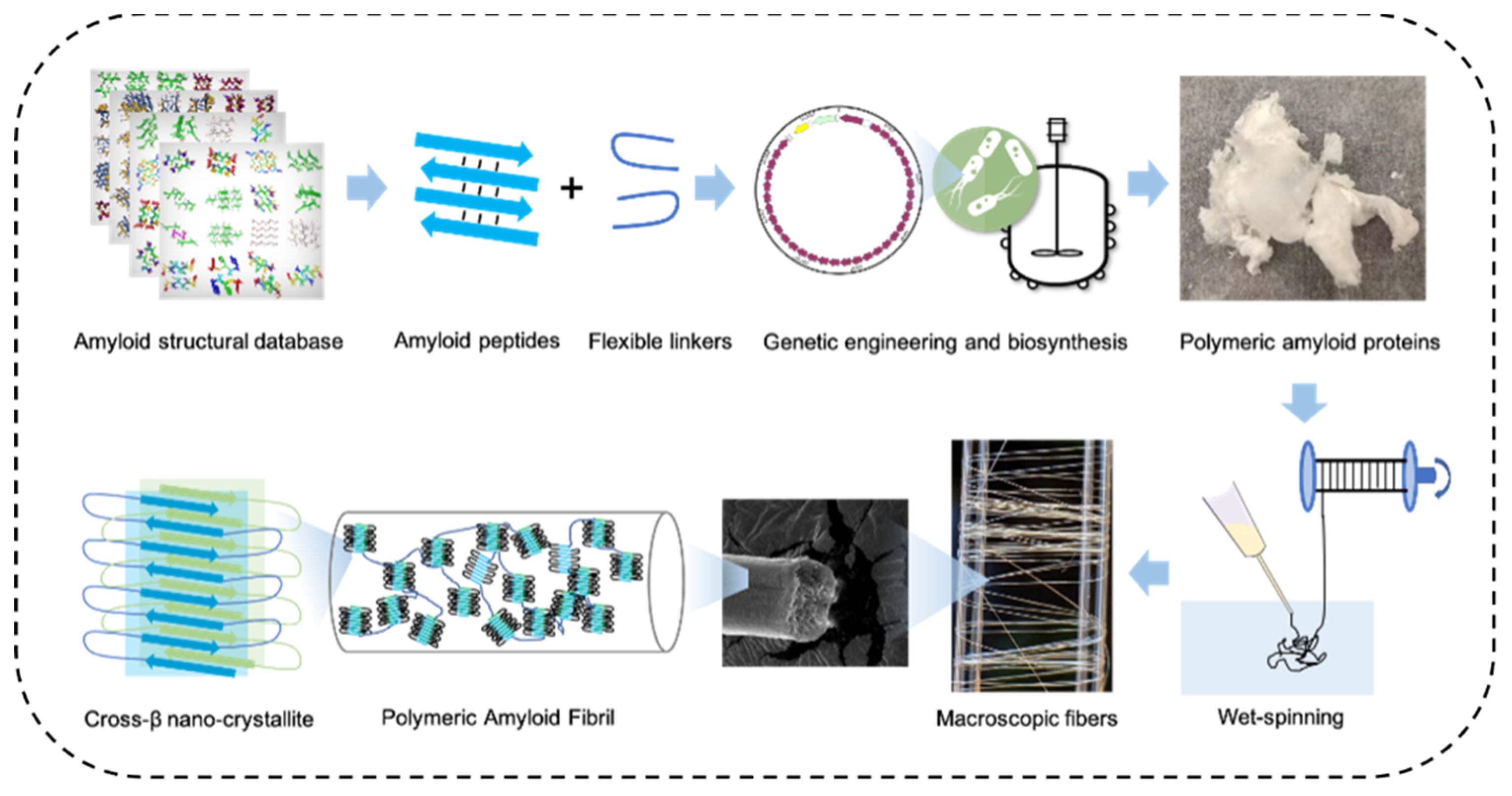
3.2.2. High-Strength Materials
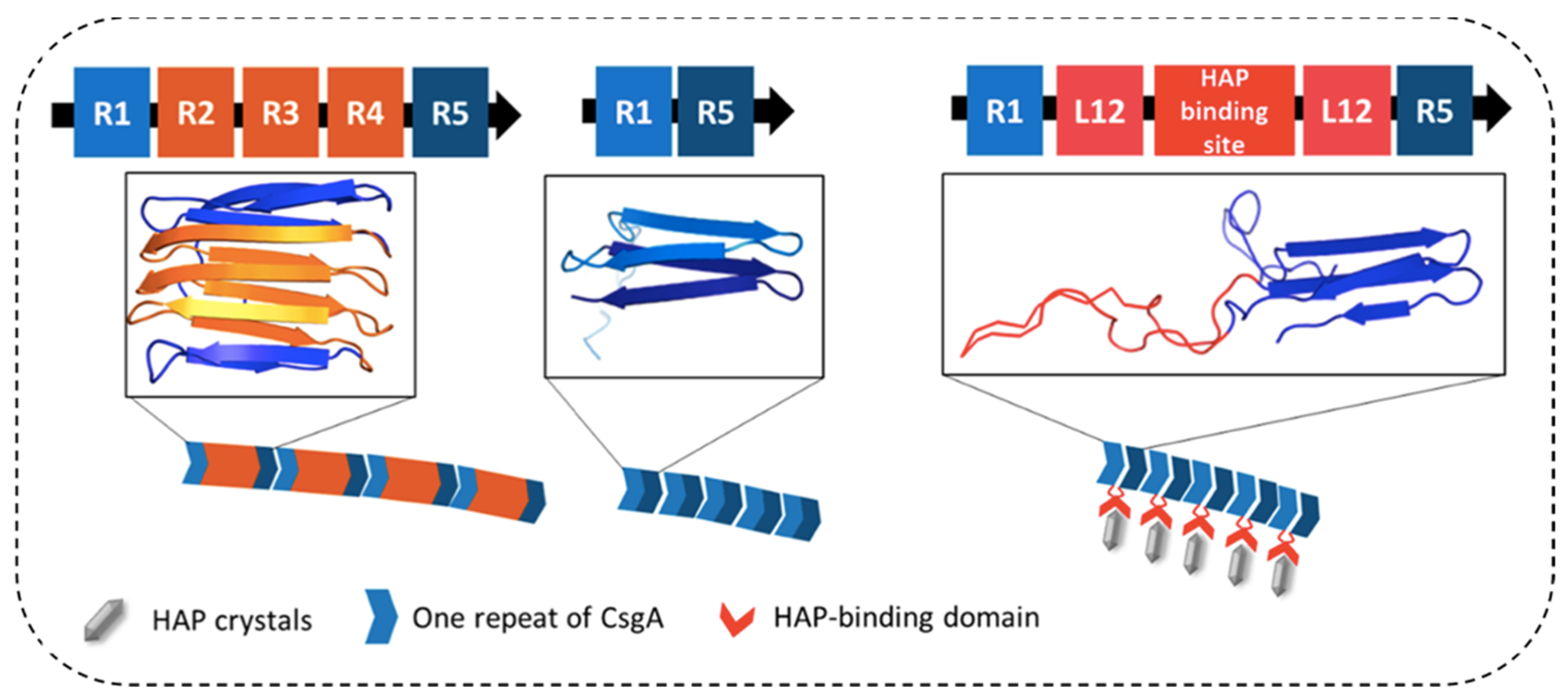
3.2.3. Amyloid-Inorganic Hybrid Composite Materials
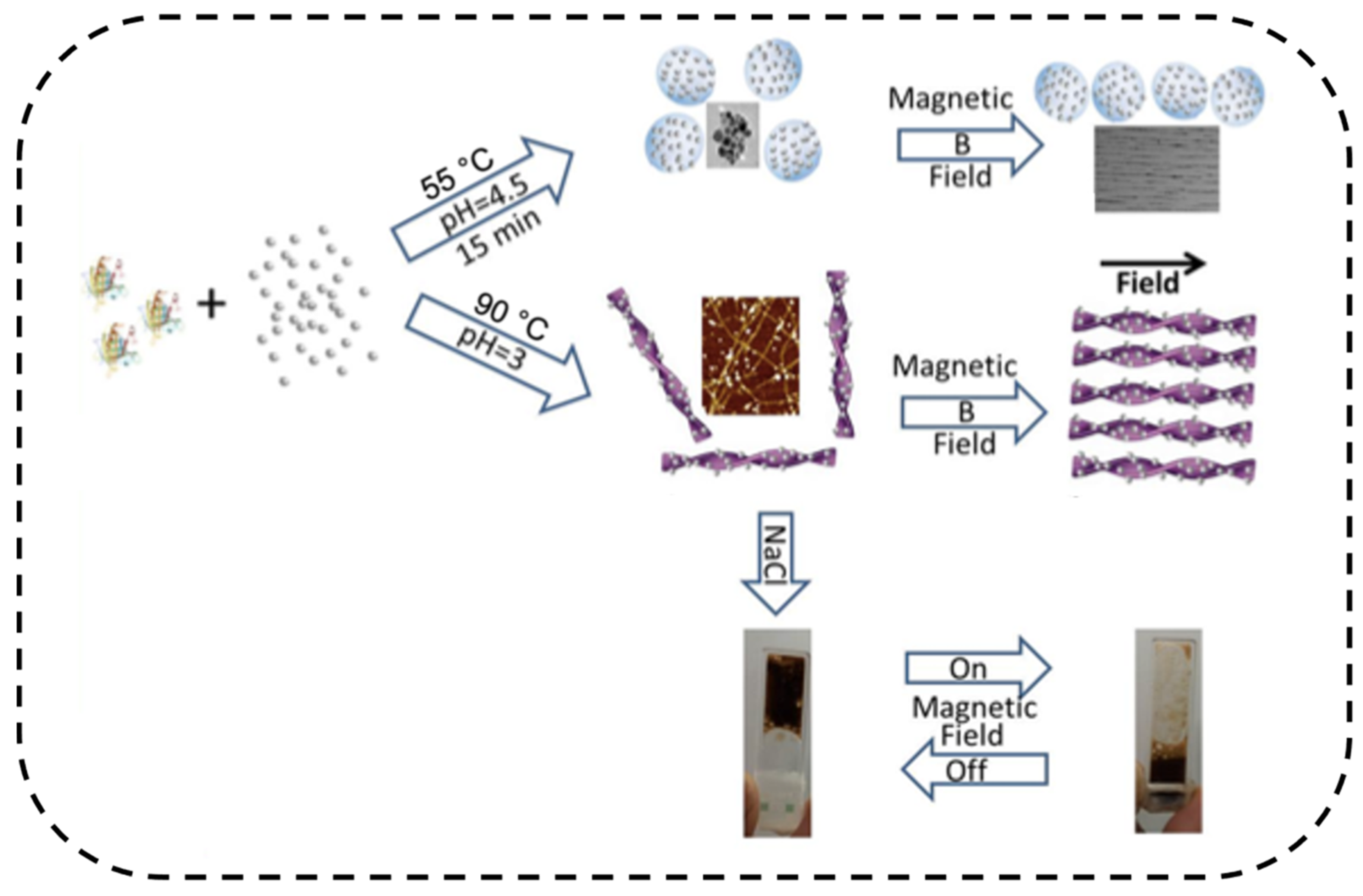
3.2.4. Responsive Materials for Sensing
3.2.5. Extracellular Matrix (ECM) to Sustain Viable Cells
3.2.6. Conductive Materials
3.2.7. Catalytic Materials
4. Perspectives and Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liang, P.; MacRae, T.H. Molecular chaperones and the cytoskeleton. J. Cell Sci. 1997, 110, 1431–1440. [Google Scholar] [CrossRef]
- Shao, Z.; Vollrath, F. Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef]
- Bowen, C.H.; Reed, T.J.; Sargent, C.J.; Mpamo, B.; Galazka, J.M.; Zhang, F. Seeded Chain-Growth Polymerization of Proteins in Living Bacterial Cells. ACS Synth. Biol. 2019, 8, 2651–2658. [Google Scholar] [CrossRef]
- Bai, W.; Sargent, C.J.; Choi, J.M.; Pappu, R.V.; Zhang, F. Covalently-assembled single-chain protein nanostructures with ultra-high stability. Nat. Commun. 2019, 10, 3317. [Google Scholar] [CrossRef]
- Kim, E.; Dai, B.; Qiao, J.B.; Li, W.; Fortner, J.D.; Zhang, F. Microbially Synthesized Repeats of Mussel Foot Protein Display Enhanced Underwater Adhesion. ACS Appl. Mater. Interfaces 2018, 10, 43003–43012. [Google Scholar] [CrossRef]
- Bowen, C.H.; Sargent, C.J.; Wang, A.; Zhu, Y.; Chang, X.; Li, J.; Mu, X.; Galazka, J.M.; Jun, Y.S.; Keten, S.; et al. Microbial production of megadalton titin yields fibers with advantageous mechanical properties. Nat. Commun. 2021, 12, 5182. [Google Scholar] [CrossRef]
- Sutherland, T.D.; Huson, M.G.; Rapson, T.D. Rational design of new materials using recombinant structural proteins: Current state and future challenges. J. Struct. Biol. 2018, 201, 76–83. [Google Scholar] [CrossRef]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [Green Version]
- Sipe, J.D.; Benson, M.D.; Buxbaum, J.N.; Ikeda, S.; Merlini, G.; Saraiva, M.J.; Westermark, P. Nomenclature 2014: Amyloid fibril proteins and clinical classification of the amyloidosis. Amyloid 2014, 21, 221–224. [Google Scholar] [CrossRef]
- Reiss, A.B.; Arain, H.A.; Stecker, M.M.; Siegart, N.M.; Kasselman, L.J. Amyloid toxicity in Alzheimer’s disease. Rev. Neurosci. 2018, 29, 613–627. [Google Scholar] [CrossRef]
- Kametani, F.; Hasegawa, M. Reconsideration of Amyloid Hypothesis and Tau Hypothesis in Alzheimer’s Disease. Front. Neurosci. 2018, 12, 25. [Google Scholar] [CrossRef] [Green Version]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Bondi, M.W.; Edmonds, E.C.; Salmon, D.P. Alzheimer’s Disease: Past, Present, and Future. J. Int. Neuropsychol. Soc. 2017, 23, 818–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irwin, D.J.; Lee, V.M.; Trojanowski, J.Q. Parkinson’s disease dementia: Convergence of alpha-synuclein, tau and amyloid-beta pathologies. Nat. Rev. Neurosci. 2013, 14, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Van Den Berge, N.; Ferreira, N.; Gram, H.; Mikkelsen, T.W.; Alstrup, A.K.O.; Casadei, N.; Tsung-Pin, P.; Riess, O.; Nyengaard, J.R.; Tamguney, G.; et al. Evidence for bidirectional and trans-synaptic parasympathetic and sympathetic propagation of alpha-synuclein in rats. Acta Neuropathol. 2019, 138, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, N.; Goncalves, N.P.; Jan, A.; Jensen, N.M.; van der Laan, A.; Mohseni, S.; Vaegter, C.B.; Jensen, P.H. Trans-synaptic spreading of alpha-synuclein pathology through sensory afferents leads to sensory nerve degeneration and neuropathic pain. Acta Neuropathol. Commun. 2021, 9, 31. [Google Scholar] [CrossRef]
- Ferreira, N.; Gram, H.; Sorrentino, Z.A.; Gregersen, E.; Schmidt, S.I.; Reimer, L.; Betzer, C.; Perez-Gozalbo, C.; Beltoja, M.; Nagaraj, M.; et al. Multiple system atrophy-associated oligodendroglial protein p25alpha stimulates formation of novel alpha-synuclein strain with enhanced neurodegenerative potential. Acta Neuropathol. 2021, 142, 87–115. [Google Scholar] [CrossRef]
- Jan, A.; Goncalves, N.P.; Vaegter, C.B.; Jensen, P.H.; Ferreira, N. The Prion-Like Spreading of Alpha-Synuclein in Parkinson’s Disease: Update on Models and Hypotheses. Int. J. Mol. Sci. 2021, 22, 8338. [Google Scholar] [CrossRef] [PubMed]
- Downing, K.H.; Nogales, E. Tubulin and microtubule structure. Curr. Opin. Cell Biol. 1998, 10, 16–22. [Google Scholar] [CrossRef]
- Knowles, T.P.; Mezzenga, R. Amyloid Fibrils as Building Blocks for Natural and Artificial Functional Materials. Adv. Mater. 2016, 28, 6546–6561. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid—From bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.W.; Balch, W.E. Amyloid as a natural product. J. Cell Biol. 2003, 161, 461–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006, 4, e6. [Google Scholar] [CrossRef]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [Green Version]
- Otzen, D.; Nielsen, P.H. We find them here, we find them there: Functional bacterial amyloid. Cell Mol. Life Sci. 2008, 65, 910–927. [Google Scholar] [CrossRef]
- Cao, Y.; Mezzenga, R. Food protein amyloid fibrils: Origin, structure, formation, characterization, applications and health implications. Adv. Colloid Interface Sci. 2019, 269, 334–356. [Google Scholar] [CrossRef]
- Goldschmidt, L.; Teng, P.K.; Riek, R.; Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2010, 107, 3487–3492. [Google Scholar] [CrossRef] [Green Version]
- Shimanovich, U.; Michaels, T.C.T.; De Genst, E.; Matak-Vinkovic, D.; Dobson, C.M.; Knowles, T.P.J. Sequential Release of Proteins from Structured Multishell Microcapsules. Biomacromolecules 2017, 18, 3052–3059. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.F.; Knowles, T.P.; Dobson, C.M.; Macphee, C.E.; Welland, M.E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA 2006, 103, 15806–15811. [Google Scholar] [CrossRef] [Green Version]
- Balbirnie, M.; Grothe, R.; Eisenberg, D.S. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc. Natl. Acad. Sci. USA 2001, 98, 2375–2380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasso, L.; Suei, S.; Domigan, L.; Healy, J.; Nock, V.; Williams, M.A.; Gerrard, J.A. Versatile multi-functionalization of protein nanofibrils for biosensor applications. Nanoscale 2014, 6, 1629–1634. [Google Scholar] [CrossRef]
- Dai, B.; Li, D.; Xi, W.; Luo, F.; Zhang, X.; Zou, M.; Cao, M.; Hu, J.; Wang, W.; Wei, G.; et al. Tunable assembly of amyloid-forming peptides into nanosheets as a retrovirus carrier. Proc. Natl. Acad. Sci. USA 2015, 112, 2996–3001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maji, S.K.; Schubert, D.; Rivier, C.; Lee, S.; Rivier, J.E.; Riek, R. Amyloid as a depot for the formulation of long-acting drugs. PLoS Biol. 2008, 6, e17. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, Y.; Yu, H.; Dai, B.; Jun, Y.S.; Zhang, F. Microbially Synthesized Polymeric Amyloid Fiber Promotes beta-Nanocrystal Formation and Displays Gigapascal Tensile Strength. ACS Nano 2021, 15, 11843–11853. [Google Scholar] [CrossRef]
- Bowen, C.H.; Dai, B.; Sargent, C.J.; Bai, W.; Ladiwala, P.; Feng, H.; Huang, W.; Kaplan, D.L.; Galazka, J.M.; Zhang, F. Recombinant Spidroins Fully Replicate Primary Mechanical Properties of Natural Spider Silk. Biomacromolecules 2018, 19, 3853–3860. [Google Scholar] [CrossRef] [Green Version]
- Gras, S.L.; Tickler, A.K.; Squires, A.M.; Devlin, G.L.; Horton, M.A.; Dobson, C.M.; MacPhee, C.E. Functionalised amyloid fibrils for roles in cell adhesion. Biomaterials 2008, 29, 1553–1562. [Google Scholar] [CrossRef]
- Li, C.; Born, A.K.; Schweizer, T.; Zenobi-Wong, M.; Cerruti, M.; Mezzenga, R. Amyloid-hydroxyapatite bone biomimetic composites. Adv. Mater. 2014, 26, 3207–3212. [Google Scholar] [CrossRef]
- Jacob, R.S.; Ghosh, D.; Singh, P.K.; Basu, S.K.; Jha, N.N.; Das, S.; Sukul, P.K.; Patil, S.; Sathaye, S.; Kumar, A.; et al. Self healing hydrogels composed of amyloid nano fibrils for cell culture and stem cell differentiation. Biomaterials 2015, 54, 97–105. [Google Scholar] [CrossRef]
- Li, C.; Qin, R.; Liu, R.; Miao, S.; Yang, P. Functional amyloid materials at surfaces/interfaces. Biomater. Sci. 2018, 6, 462–472. [Google Scholar] [CrossRef]
- Kim, S.; Kim, J.H.; Lee, J.S.; Park, C.B. Beta-Sheet-Forming, Self-Assembled Peptide Nanomaterials towards Optical, Energy, and Healthcare Applications. Small 2015, 11, 3623–3640. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Li, F.; Zou, Z.; Saeed, M.; Xu, Z.; Yu, H. Bio-inspired amyloid polypeptides: From self-assembly to nanostructure design and biotechnological applications. Appl. Mater. Today 2021, 22, 100966. [Google Scholar] [CrossRef]
- Balasco, N.; Diaferia, C.; Morelli, G.; Vitagliano, L.; Accardo, A. Amyloid-Like Aggregation in Diseases and Biomaterials: Osmosis of Structural Information. Front. Bioeng. Biotechnol. 2021, 9, 641372. [Google Scholar] [CrossRef]
- Hamley, I.W. Small Bioactive Peptides for Biomaterials Design and Therapeutics. Chem. Rev. 2017, 117, 14015–14041. [Google Scholar] [CrossRef] [Green Version]
- Mason, T.O.; Shimanovich, U. Fibrous Protein Self-Assembly in Biomimetic Materials. Adv. Mater. 2018, 30, e1706462. [Google Scholar] [CrossRef]
- Das, S.; Jacob, R.S.; Patel, K.; Singh, N.; Maji, S.K. Amyloid Fibrils: Versatile Biomaterials for Cell Adhesion and Tissue Engineering Applications. Biomacromolecules 2018, 19, 1826–1839. [Google Scholar] [CrossRef]
- Astbury, W.T.; Dickinson, S.; Bailey, K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 1935, 29, 2351–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fandrich, M.; Dobson, C.M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002, 21, 5682–5690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, R.; Sawaya, M.R.; Balbirnie, M.; Madsen, A.O.; Riekel, C.; Grothe, R.; Eisenberg, D. Structure of the cross-beta spine of amyloid-like fibrils. Nature 2005, 435, 773–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsemekhman, K.; Goldschmidt, L.; Eisenberg, D.; Baker, D. Cooperative hydrogen bonding in amyloid formation. Protein Sci. 2007, 16, 761–764. [Google Scholar] [CrossRef] [Green Version]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef]
- Chou, K.-C.; Pottle, M.; Némethy, G.; Ueda, y.; Scheraga, H.A. Structure of β-sheets: Origin of the right-handed twist and of the increased stability of antiparallel over parallel sheets. J. Mol. Biol. 1982, 162, 89–112. [Google Scholar] [CrossRef]
- Soriaga, A.B.; Sangwan, S.; Macdonald, R.; Sawaya, M.R.; Eisenberg, D. Crystal Structures of IAPP Amyloidogenic Segments Reveal a Novel Packing Motif of Out-of-Register Beta Sheets. J. Phys. Chem. B 2016, 120, 5810–5816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Sawaya, M.R.; Eisenberg, D. Beta(2)-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat. Struct. Mol. Biol. 2011, 18, 49–55. [Google Scholar] [CrossRef] [Green Version]
- Knowles, T.P.; Fitzpatrick, A.W.; Meehan, S.; Mott, H.R.; Vendruscolo, M.; Dobson, C.M.; Welland, M.E. Role of intermolecular forces in defining material properties of protein nanofibrils. Science 2007, 318, 1900–1903. [Google Scholar] [CrossRef]
- Toksoz, S.; Acar, H.; Guler, M.O. Self-assembled one-dimensional soft nanostructures. Soft Matter 2010, 6, 5839–5849. [Google Scholar] [CrossRef]
- Stankovic, I.M.; Niu, S.; Hall, M.B.; Zaric, S.D. Role of aromatic amino acids in amyloid self-assembly. Int. J. Biol. Macromol. 2020, 156, 949–959. [Google Scholar] [CrossRef]
- Berryman, J.T.; Radford, S.E.; Harris, S.A. Systematic examination of polymorphism in amyloid fibrils by molecular-dynamics simulation. Biophys. J. 2011, 100, 2234–2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.T.; Lin, Y.; Spencer, R.K.; Thomas, M.R.; Nguyen, A.I.; Amdursky, N.; Pashuck, E.T.; Skaalure, S.C.; Song, C.Y.; Parmar, P.A.; et al. Sequence-Dependent Self-Assembly and Structural Diversity of Islet Amyloid Polypeptide-Derived beta-Sheet Fibrils. ACS Nano 2017, 11, 8579–8589. [Google Scholar] [CrossRef]
- Naiki, H.; Hashimoto, N.; Suzuki, S.; Kimura, H.; Nakakuki, K.; Gejyo, F. Establishment of a kinetic model of dialysis-related amyloid fibril extensionin vitro. Amyloid 1997, 4, 223–232. [Google Scholar] [CrossRef]
- DeBenedictis, E.P.; Liu, J.; Keten, S. Adhesion mechanisms of curli subunit CsgA to abiotic surfaces. Sci. Adv. 2016, 2, e1600998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, S.J.; Kjellerup, B.V. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 2013, 97, 9909–9921. [Google Scholar] [CrossRef] [PubMed]
- Donato, V.; Ayala, F.R.; Cogliati, S.; Bauman, C.; Costa, J.G.; Lenini, C.; Grau, R. Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signalling pathway. Nat. Commun. 2017, 8, 14332. [Google Scholar] [CrossRef] [PubMed]
- Hobley, L.; Harkins, C.; MacPhee, C.E.; Stanley-Wall, N.R. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 2015, 39, 649–669. [Google Scholar] [CrossRef] [Green Version]
- Yates, M.D.; Strycharz-Glaven, S.M.; Golden, J.P.; Roy, J.; Tsoi, S.; Erickson, J.S.; El-Naggar, M.Y.; Barton, S.C.; Tender, L.M. Measuring conductivity of living Geobacter sulfurreducens biofilms. Nat. Nanotechnol. 2016, 11, 910–913. [Google Scholar] [CrossRef]
- Dueholm, M.S.; Nielsen, S.B.; Hein, K.L.; Nissen, P.; Chapman, M.; Christiansen, G.; Nielsen, P.H.; Otzen, D.E. Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 2011, 50, 8281–8290. [Google Scholar] [CrossRef] [Green Version]
- Hammer, N.D.; Schmidt, J.C.; Chapman, M.R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. USA 2007, 104, 12494–12499. [Google Scholar] [CrossRef] [Green Version]
- Dueholm, M.S.; Petersen, S.V.; Sonderkaer, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef] [Green Version]
- Zeng, G.; Vad, B.S.; Dueholm, M.S.; Christiansen, G.; Nilsson, M.; Tolker-Nielsen, T.; Nielsen, P.H.; Meyer, R.L.; Otzen, D.E. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front. Microbiol. 2015, 6, 1099. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, K.; Syed, A.K.; Stephenson, R.E.; Rickard, A.H.; Boles, B.R. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012, 8, e1002744. [Google Scholar] [CrossRef] [PubMed]
- Iconomidou, V.A.; Vriend, G.; Hamodrakas, S.J. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 2000, 479, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Si, K.; Lindquist, S.; Kandel, E.R. A Neuronal Isoform of the Aplysia CPEB Has Prion-Like Properties. Cell 2003, 115, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Banta, S.; Wheeldon, I.R.; Blenner, M. Protein engineering in the development of functional hydrogels. Annu. Rev. Biomed. Eng. 2010, 12, 167–186. [Google Scholar] [CrossRef] [Green Version]
- Rinaudo, M. Gelation of Polysaccharides. J. Intell. Mater. Syst. Struct. 1993, 4, 210–215. [Google Scholar] [CrossRef]
- Belwal, V.K.; Chaudhary, N. Amyloids and their untapped potential as hydrogelators. Soft Matter 2020, 16, 10013–10028. [Google Scholar] [CrossRef]
- Hu, B.; Shen, Y.; Adamcik, J.; Fischer, P.; Schneider, M.; Loessner, M.J.; Mezzenga, R. Polyphenol-Binding Amyloid Fibrils Self-Assemble into Reversible Hydrogels with Antibacterial Activity. ACS Nano 2018, 12, 3385–3396. [Google Scholar] [CrossRef]
- Gu, J.; Su, Y.; Liu, P.; Li, P.; Yang, P. An Environmentally Benign Antimicrobial Coating Based on a Protein Supramolecular Assembly. ACS Appl. Mater. Interfaces 2017, 9, 198–210. [Google Scholar] [CrossRef]
- Stephanopoulos, N.; Ortony, J.H.; Stupp, S.I. Self-Assembly for the Synthesis of Functional Biomaterials. Acta Mater. 2013, 61, 912–930. [Google Scholar] [CrossRef] [Green Version]
- Rehm, T.H.; Schmuck, C. Ion-pair induced self-assembly in aqueous solvents. Chem. Soc. Rev. 2010, 39, 3597–3611. [Google Scholar] [CrossRef] [PubMed]
- Medini, K.; Mansel, B.W.; Williams, M.A.K.; Brimble, M.A.; Williams, D.E.; Gerrard, J.A. Controlling gelation with sequence: Towards programmable peptide hydrogels. Acta Biomater. 2016, 43, 30–37. [Google Scholar] [CrossRef]
- Saini, A.; Chauhan, V.S. Self-assembling properties of peptides derived from TDP-43 C-terminal fragment. Langmuir 2014, 30, 3845–3856. [Google Scholar] [CrossRef]
- Decandio, C.C.; Silva, E.R.; Hamley, I.W.; Castelletto, V.; Liberato, M.S.; Oliveira, V.X., Jr.; Oliveira, C.L.; Alves, W.A. Self-Assembly of a Designed Alternating Arginine/Phenylalanine Oligopeptide. Langmuir 2015, 31, 4513–4523. [Google Scholar] [CrossRef]
- Measey, T.J.; Schweitzer-Stenner, R.; Sa, V.; Kornev, K. Anomalous Conformational Instability and Hydrogel Formation of a Cationic Class of Self-Assembling Oligopeptides. Macromolecules 2010, 43, 7800–7806. [Google Scholar] [CrossRef]
- Jain, R.; Khandelwal, G.; Roy, S. Unraveling the Design Rules in Ultrashort Amyloid-Based Peptide Assemblies toward Shape-Controlled Synthesis of Gold Nanoparticles. Langmuir 2019, 35, 5878–5889. [Google Scholar] [CrossRef]
- Goeden-Wood, N.L.; Keasling, J.D.; Muller, S.J. Self-Assembly of a Designed Protein Polymer into β-Sheet Fibrils and Responsive Gels. Macromolecules 2003, 36, 2932–2938. [Google Scholar] [CrossRef]
- Diaz-Caballero, M.; Navarro, S.; Fuentes, I.; Teixidor, F.; Ventura, S. Minimalist Prion-Inspired Polar Self-Assembling Peptides. ACS Nano 2018, 12, 5394–5407. [Google Scholar] [CrossRef]
- Kralj, S.; Bellotto, O.; Parisi, E.; Garcia, A.M.; Iglesias, D.; Semeraro, S.; Deganutti, C.; D’Andrea, P.; Vargiu, A.V.; Geremia, S.; et al. Heterochirality and Halogenation Control Phe-Phe Hierarchical Assembly. ACS Nano 2020, 14, 16951–16961. [Google Scholar] [CrossRef]
- Uptain, S.M.; Lindquist, S. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 2002, 56, 703–741. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Caballero, M.; Fernandez, M.R.; Navarro, S.; Ventura, S. Prion-based nanomaterials and their emerging applications. Prion 2018, 12, 266–272. [Google Scholar] [CrossRef]
- Marchesan, S.; Easton, C.D.; Kushkaki, F.; Waddington, L.; Hartley, P.G. Tripeptide self-assembled hydrogels: Unexpected twists of chirality. Chem. Commun. 2012, 48, 2195–2197. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, S.; Styan, K.E.; Easton, C.D.; Waddington, L.; Vargiu, A.V. Higher and lower supramolecular orders for the design of self-assembled heterochiral tripeptide hydrogel biomaterials. J. Mater. Chem. B 2015, 3, 8123–8132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bal, S.; Ghosh, C.; Ghosh, T.; Vijayaraghavan, R.K.; Das, D. Non-Equilibrium Polymerization of Cross-beta Amyloid Peptides for Temporal Control of Electronic Properties. Angew. Chem. Int. Ed. Engl. 2020, 59, 13506–13510. [Google Scholar] [CrossRef]
- Knowles, T.P.; Oppenheim, T.W.; Buell, A.K.; Chirgadze, D.Y.; Welland, M.E. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 2010, 5, 204–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kol, N.; Adler-Abramovich, L.; Barlam, D.; Shneck, R.Z.; Gazit, E.; Rousso, I. Self-assembled peptide nanotubes are uniquely rigid bioinspired supramolecular structures. Nano Lett. 2005, 5, 1343–1346. [Google Scholar] [CrossRef]
- Knowles, T.P.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 2011, 6, 469–479. [Google Scholar] [CrossRef]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef]
- Rising, A.; Johansson, J. Toward spinning artificial spider silk. Nat. Chem. Biol. 2015, 11, 309–315. [Google Scholar] [CrossRef]
- Dai, B.; Sargent, C.J.; Gui, X.; Liu, C.; Zhang, F. Fibril Self-Assembly of Amyloid-Spider Silk Block Polypeptides. Biomacromolecules 2019, 20, 2015–2023. [Google Scholar] [CrossRef]
- Xia, X.X.; Qian, Z.G.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef] [Green Version]
- Denny, M. The Physical Properties of Spider’s Silk and Their Role in the Design of Orb-Webs. J. Exp. Biol. 1976, 65, 483–506. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, P. Amyloid-Mediated Fabrication of Organic-Inorganic Hybrid Materials and Their Biomedical Applications. Adv. Mater. Interfaces 2020, 7, 2001060. [Google Scholar] [CrossRef]
- Nystrom, G.; Fernandez-Ronco, M.P.; Bolisetty, S.; Mazzotti, M.; Mezzenga, R. Amyloid Templated Gold Aerogels. Adv. Mater. 2016, 28, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bolisetty, S.; Cao, Y.; Handschin, S.; Adamcik, J.; Peng, Q.; Mezzenga, R. Selective and Efficient Removal of Fluoride from Water: In Situ Engineered Amyloid Fibril/ZrO2 Hybrid Membranes. Angew Chem. Int. Ed. Engl. 2019, 58, 6012–6016. [Google Scholar] [CrossRef]
- Bolisetty, S.; Arcari, M.; Adamcik, J.; Mezzenga, R. Hybrid Amyloid Membranes for Continuous Flow Catalysis. Langmuir 2015, 31, 13867–13873. [Google Scholar] [CrossRef]
- Bolisetty, S.; Coray, N.M.; Palika, A.; Prenosil, G.A.; Mezzenga, R. Amyloid hybrid membranes for removal of clinical and nuclear radioactive wastewater. Environ. Sci. Water Res. Technol. 2020, 6, 3249–3254. [Google Scholar] [CrossRef]
- Bolisetty, S.; Mezzenga, R. Amyloid-carbon hybrid membranes for universal water purification. Nat. Nanotechnol. 2016, 11, 365–371. [Google Scholar] [CrossRef]
- Bolisetty, S.; Reinhold, N.; Zeder, C.; Orozco, M.N.; Mezzenga, R. Efficient purification of arsenic-contaminated water using amyloid-carbon hybrid membranes. Chem. Commun. 2017, 53, 5714–5717. [Google Scholar] [CrossRef]
- Cao, Y.; Bolisetty, S.; Wolfisberg, G.; Adamcik, J.; Mezzenga, R. Amyloid fibril-directed synthesis of silica core-shell nanofilaments, gels, and aerogels. Proc. Natl. Acad. Sci. USA 2019, 116, 4012–4017. [Google Scholar] [CrossRef] [Green Version]
- Ha, Y.; Yang, J.; Tao, F.; Wu, Q.; Song, Y.J.; Wang, H.R.; Zhang, X.; Yang, P. Phase-Transited Lysozyme as a Universal Route to Bioactive Hydroxyapatite Crystalline Film. Adv. Funct. Mater. 2018, 28, 1704476. [Google Scholar] [CrossRef]
- Li, K.H.; Zhang, Z.F.; Li, D.P.; Zhang, W.S.; Yu, X.Q.; Liu, W.; Gong, C.C.; Wei, G.; Su, Z.Q. Biomimetic Ultralight, Highly Porous, Shape-Adjustable, and Biocompatible 3D Graphene Minerals via Incorporation of Self-Assembled Peptide Nanosheets. Adv. Funct Mater. 2018, 28, 1801056. [Google Scholar] [CrossRef]
- Yang, X.; Li, Z.H.; Xiao, H.; Wang, N.; Li, Y.P.; Xu, X.Y.; Chen, Z.J.; Tan, H.; Li, J.S. A Universal and Ultrastable Mineralization Coating Bioinspired from Biofilms. Adv. Funct. Mater. 2018, 28, 1802730. [Google Scholar] [CrossRef]
- Abdali, Z.; Aminzare, M.; Zhu, X.; DeBenedictis, E.; Xie, O.; Keten, S.; Dorval Courchesne, N.M. Curli-Mediated Self-Assembly of a Fibrous Protein Scaffold for Hydroxyapatite Mineralization. ACS Synth. Biol. 2020, 9, 3334–3343. [Google Scholar] [CrossRef]
- Saldanha, D.J.; Abdali, Z.; Modafferi, D.; Janfeshan, B.; Dorval Courchesne, N.M. Fabrication of fluorescent pH-responsive protein-textile composites. Sci. Rep. 2020, 10, 13052. [Google Scholar] [CrossRef]
- Li, C.; Mezzenga, R. Functionalization of multiwalled carbon nanotubes and their pH-responsive hydrogels with amyloid fibrils. Langmuir 2012, 28, 10142–10146. [Google Scholar] [CrossRef]
- Ozbas, B.; Kretsinger, J.; Rajagopal, K.; Schneider, J.P.; Pochan, D.J. Salt-Triggered Peptide Folding and Consequent Self-Assembly into Hydrogels with Tunable Modulus. Macromolecules 2004, 37, 7331–7337. [Google Scholar] [CrossRef]
- Bolisetty, S.; Vallooran, J.J.; Adamcik, J.; Mezzenga, R. Magnetic-responsive hybrids of Fe3O4 nanoparticles with beta-lactoglobulin amyloid fibrils and nanoclusters. ACS Nano 2013, 7, 6146–6155. [Google Scholar] [CrossRef]
- Lutz-Bueno, V.; Bolisetty, S.; Azzari, P.; Handschin, S.; Mezzenga, R. Self-Winding Gelatin-Amyloid Wires for Soft Actuators and Sensors. Adv. Mater. 2020, 32, e2004941. [Google Scholar] [CrossRef]
- Rabe, M.; Soragni, A.; Reynolds, N.P.; Verdes, D.; Liverani, E.; Riek, R.; Seeger, S. On-surface aggregation of alpha-synuclein at nanomolar concentrations results in two distinct growth mechanisms. ACS Chem. Neurosci. 2013, 4, 408–417. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, N.P.; Soragni, A.; Rabe, M.; Verdes, D.; Liverani, E.; Handschin, S.; Riek, R.; Seeger, S. Mechanism of membrane interaction and disruption by alpha-synuclein. J. Am. Chem. Soc. 2011, 133, 19366–19375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selkoe, D.J. The molecular pathology of Alzheimer’s disease. Neuron 1991, 6, 487–498. [Google Scholar] [CrossRef]
- Reynolds, N.P.; Charnley, M.; Mezzenga, R.; Hartley, P.G. Engineered lysozyme amyloid fibril networks support cellular growth and spreading. Biomacromolecules 2014, 15, 599–608. [Google Scholar] [CrossRef]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef]
- Pierschbacher, M.D.; Ruoslahti, E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 1984, 309, 30–33. [Google Scholar] [CrossRef]
- Huettner, N.; Dargaville, T.R.; Forget, A. Discovering Cell-Adhesion Peptides in Tissue Engineering: Beyond RGD. Trends Biotechnol. 2018, 36, 372–383. [Google Scholar] [CrossRef]
- Paiva Dos Santos, B.; Garbay, B.; Pasqua, M.; Chevron, E.; Chinoy, Z.S.; Cullin, C.; Bathany, K.; Lecommandoux, S.; Amedee, J.; Oliveira, H.; et al. Production, purification and characterization of an elastin-like polypeptide containing the Ile-Lys-Val-Ala-Val (IKVAV) peptide for tissue engineering applications. J. Biotechnol. 2019, 298, 35–44. [Google Scholar] [CrossRef]
- Deidda, G.; Jonnalagadda, S.V.R.; Spies, J.W.; Ranella, A.; Mossou, E.; Forsyth, V.T.; Mitchell, E.P.; Bowler, M.W.; Tamamis, P.; Mitraki, A. Self-Assembled Amyloid Peptides with Arg-Gly-Asp (RGD) Motifs As Scaffolds for Tissue Engineering. ACS Biomater. Sci. Eng. 2017, 3, 1404–1416. [Google Scholar] [CrossRef] [PubMed]
- Seras-Franzoso, J.; Peebo, K.; Luis Corchero, J.; Tsimbouri, P.M.; Unzueta, U.; Rinas, U.; Dalby, M.J.; Vazquez, E.; Garcia-Fruitos, E.; Villaverde, A. A nanostructured bacterial bioscaffold for the sustained bottom-up delivery of protein drugs. Nanomedicine 2013, 8, 1587–1599. [Google Scholar] [CrossRef]
- Cooke, M.J.; Zahir, T.; Phillips, S.R.; Shah, D.S.; Athey, D.; Lakey, J.H.; Shoichet, M.S.; Przyborski, S.A. Neural differentiation regulated by biomimetic surfaces presenting motifs of extracellular matrix proteins. J. Biomed. Mater. Res. A 2010, 93, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.D.; Mura, C.; Lampe, K.J. Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engineering. J. Am. Chem. Soc. 2019, 141, 4886–4899. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.P. Amyloid-like peptide nanofibrils as scaffolds for tissue engineering: Progress and challenges (Review). Biointerphases 2019, 14, 040801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Zhou, K.; Ghosh, D.; Jha, N.N.; Singh, P.K.; Jacob, R.S.; Bernard, C.C.; Finkelstein, D.I.; Forsythe, J.S.; Maji, S.K. Implantable amyloid hydrogels for promoting stem cell differentiation to neurons. NPG Asia Mater. 2016, 8, e304. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Cao, Y.; Bolisetty, S.; Tian, T.; Handschin, S.; Lu, C.; Mezzenga, R. Amyloid Fibril-Templated High-Performance Conductive Aerogels with Sensing Properties. Small 2020, 16, e2004932. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.J.; Adhikari, R.Y.; Holmes, D.E.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Electrically conductive pili from pilin genes of phylogenetically diverse microorganisms. ISME J. 2018, 12, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, P.Q.; Botyanszki, Z.; Tay, P.K.; Joshi, N.S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 2014, 5, 4945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorval Courchesne, N.M.; Duraj-Thatte, A.; Tay, P.K.R.; Nguyen, P.Q.; Joshi, N.S. Scalable Production of Genetically Engineered Nanofibrous Macroscopic Materials via Filtration. ACS Biomater. Sci. Eng. 2017, 3, 733–741. [Google Scholar] [CrossRef] [Green Version]
- Lovley, D.R. Live wires: Direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energy Environ. Sci. 2011, 4, 4896–4906. [Google Scholar] [CrossRef] [Green Version]
- Kalyoncu, E.; Ahan, R.E.; Olmez, T.T.; Seker, U.O.S. Genetically encoded conductive protein nanofibers secreted by engineered cells. RSC Adv. 2017, 7, 32543–32551. [Google Scholar] [CrossRef] [Green Version]
- Dorval Courchesne, N.M.; DeBenedictis, E.P.; Tresback, J.; Kim, J.J.; Duraj-Thatte, A.; Zanuy, D.; Keten, S.; Joshi, N.S. Biomimetic engineering of conductive curli protein films. Nanotechnology 2018, 29, 454002. [Google Scholar] [CrossRef]
- Duncan, K.L.; Ulijn, R.V. Short Peptides in Minimalistic Biocatalyst Design. Biocatalysis 2015, 1, 67–81. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Caballero, M.; Navarro, S.; Nuez-Martínez, M.; Peccati, F.; Rodríguez-Santiago, L.; Sodupe, M.; Teixidor, F.; Ventura, S. pH-Responsive Self-Assembly of Amyloid Fibrils for Dual Hydrolase-Oxidase Reactions. ACS Catal. 2020, 11, 595–607. [Google Scholar] [CrossRef]
- Zhang, C.; Shafi, R.; Lampel, A.; MacPherson, D.; Pappas, C.G.; Narang, V.; Wang, T.; Maldarelli, C.; Ulijn, R.V. Switchable Hydrolase Based on Reversible Formation of Supramolecular Catalytic Site Using a Self-Assembling Peptide. Angew Chem. Int. Ed. Engl. 2017, 56, 14511–14515. [Google Scholar] [CrossRef]
- Jang, H.S.; Lee, J.H.; Park, Y.S.; Kim, Y.O.; Park, J.; Yang, T.Y.; Jin, K.; Lee, J.; Park, S.; You, J.M.; et al. Tyrosine-mediated two-dimensional peptide assembly and its role as a bio-inspired catalytic scaffold. Nat. Commun. 2014, 5, 3665. [Google Scholar] [CrossRef] [Green Version]
- Garcia, A.M.; Kurbasic, M.; Kralj, S.; Melchionna, M.; Marchesan, S. A biocatalytic and thermoreversible hydrogel from a histidine-containing tripeptide. Chem. Commun. 2017, 53, 8110–8113. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, T.; Cringoli, M.C.; Kralj, S.; Kurbasic, M.; Fornasiero, P.; Pengo, P.; Marchesan, S. Biocatalysis of D,L-Peptide Nanofibrillar Hydrogel. Molecules 2020, 25, 2995. [Google Scholar] [CrossRef] [PubMed]
- Van’t Hag, L.; Handschin, S.; Gschwend, P.M.; Mezzenga, R. Light Gold: A Colloidal Approach Using Latex Templates. Adv. Funct Mater. 2020, 30, 1908458. [Google Scholar] [CrossRef]
- Pinheiro, F.; Santos, J.; Ventura, S. AlphaFold and the amyloid landscape. J. Mol. Biol. 2021, 167059. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhang, F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. Int. J. Mol. Sci. 2021, 22, 10698. https://doi.org/10.3390/ijms221910698
Li J, Zhang F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. International Journal of Molecular Sciences. 2021; 22(19):10698. https://doi.org/10.3390/ijms221910698
Chicago/Turabian StyleLi, Jingyao, and Fuzhong Zhang. 2021. "Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges" International Journal of Molecular Sciences 22, no. 19: 10698. https://doi.org/10.3390/ijms221910698
APA StyleLi, J., & Zhang, F. (2021). Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. International Journal of Molecular Sciences, 22(19), 10698. https://doi.org/10.3390/ijms221910698





