Genetic Factors Affect the Survival and Behaviors of Selected Bacteria during Antimicrobial Blue Light Treatment
Abstract
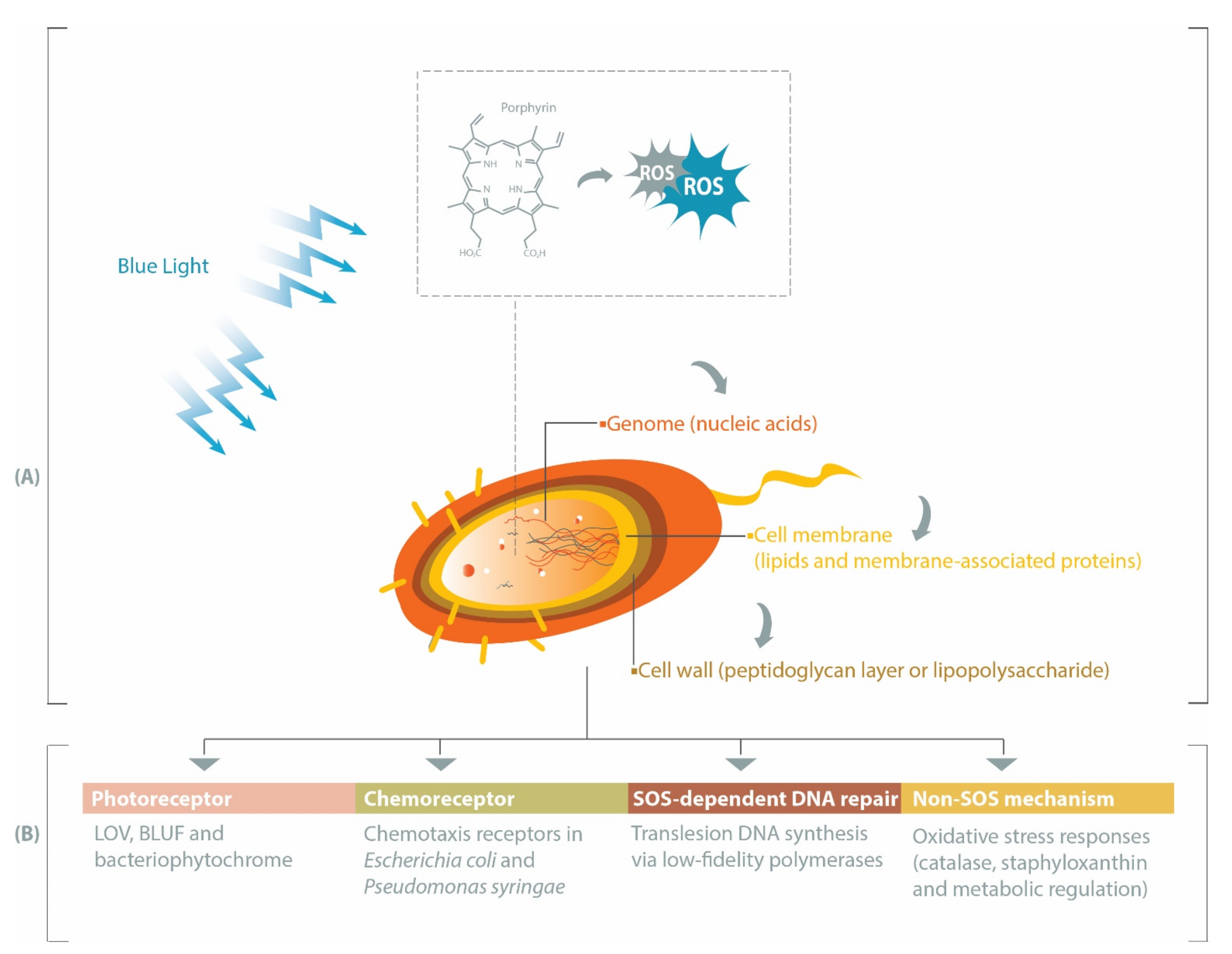
1. Introduction
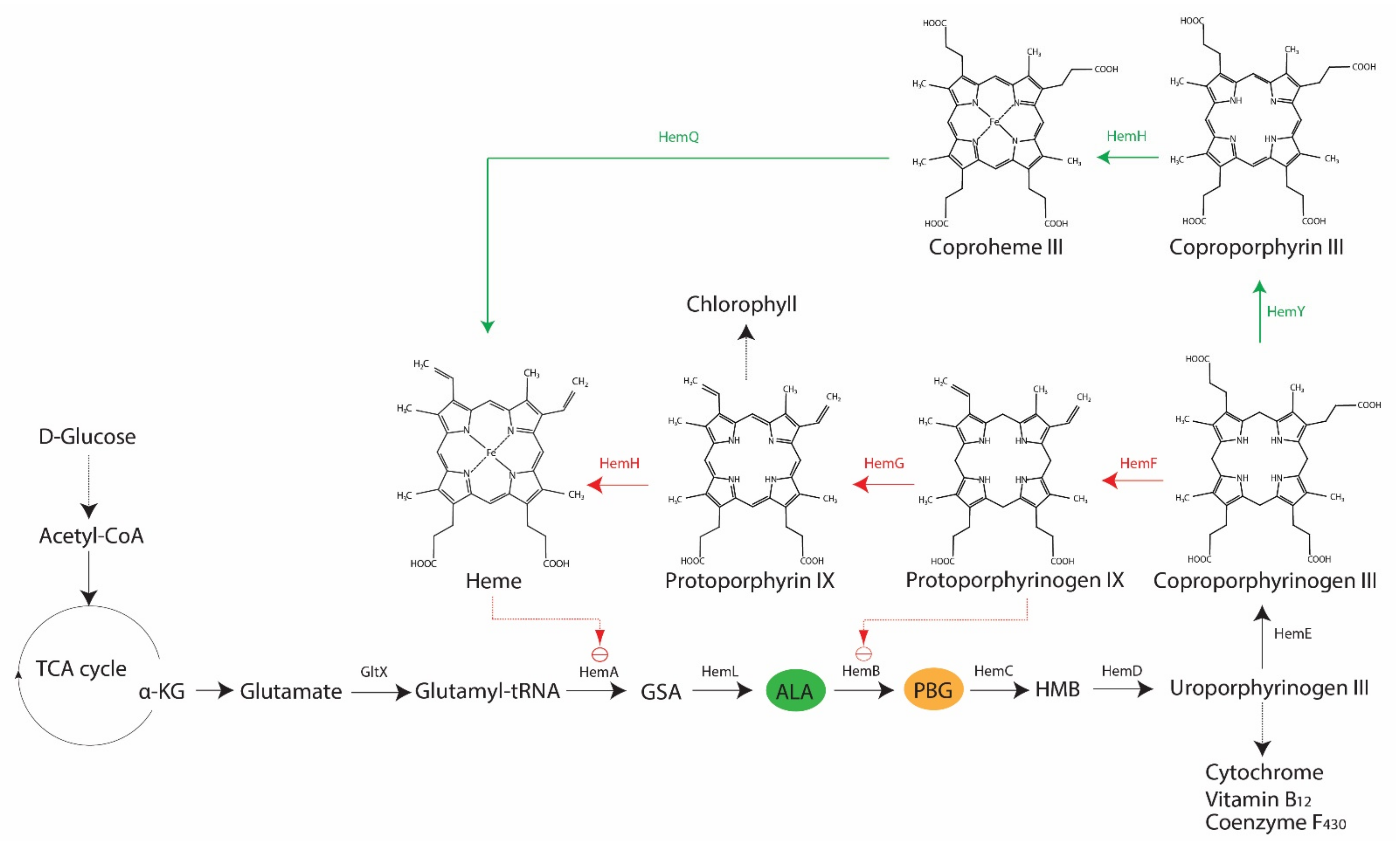
2. Porphyrin, Bacteria and Antimicrobial Blue Light
3. Heme Non-Producing Bacteria: Antimicrobial Blue Light or Photodynamic Therapy
4. Photoreceptors Facilitate Bacterial Responses to Blue Light
4.1. LOV-Mediated Response to Blue Light
4.1.1. Lmo0799 Photoreceptor and Transcription Factor Sig B (σB) in L. monocytogenes
4.1.2. YtvA Photoreceptor and Transcription Factor σB in Bacillus subtilis
4.1.3. LOV-Dependent Differential Physiological Behaviors of Pseudomonas spp. and Rhodobacter sphaeroides Containing Short LOV or LOV-Histidine Kinase
4.2. BLUF-Mediated Response to Blue Light
4.2.1. YcgF Photoreceptor in E. coli
4.2.2. BlsA Photoreceptor in A. baumannii
4.3. Blue Light-Sensing Bacteriophytochrome and PAS-Containing Photoreceptor in P. aeruginosa
5. Role of Chemoreceptors in Responses to Blue Light
6. Potential Development of Bacterial Tolerance to Blue Light via SOS-Dependent DNA Repair
7. Non-SOS Protective Mechanisms against Blue Light-Induced Oxidative Stress
8. Concluding Remarks and Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Gwynne, P.J.; Gallagher, M.P. Light as a broad-spectrum antimicrobial. Front. Microbiol. 2018, 9, 119. [Google Scholar] [CrossRef]
- Yin, R.; Dai, T.; Avci, P.; Jorge, A.E.S.; De Melo, W.C.M.A.; Vecchio, D.; Huang, Y.Y.; Gupta, A.; Hamblin, M.R. Light based anti-infectives: Ultraviolet C irradiation, photodynamic therapy, blue light, and beyond. Curr. Opin. Pharmacol. 2013, 13, 731–762. [Google Scholar] [CrossRef]
- D’Orazio, J.; Jarrett, S.; Amaro-Ortiz, A.; Scott, T. UV Radiation and the Skin. Int. J. Mol. Sci. 2013, 14, 12222–12248. [Google Scholar] [CrossRef] [PubMed]
- Kütting, B.; Drexler, H. UV-induced skin cancer at workplace and evidence-based prevention. Int. Arch. Occup. Environ. Health 2010, 83, 843–854. [Google Scholar] [CrossRef]
- Zaffina, S.; Camisa, V.; Lembo, M.; Vinci, M.R.; Tucci, M.G.; Borra, M.; Napolitano, A.; Cannatã, V. Accidental exposure to UV radiation produced by germicidal lamp: Case report and risk assessment. Photochem. Photobiol. 2012, 88, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- Kleinpenning, M.M.; Smits, T.; Frunt, M.H.A.; van Erp, P.E.J.; van de Kerkhof, P.C.M.; Gerritsen, R.M.J.P. Clinical and histological effects of blue light on normal skin. Photodermatol. Photoimmunol. Photomed. 2010, 26, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Angarano, V.; Smet, C.; Akkermans, S.; Watt, C.; Chieffi, A.; Van Impe, J.F.M. Visible light as an antimicrobial strategy for inactivation of Pseudomonas fluorescens and Staphylococcus epidermidis biofilms. Antibiotics 2020, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Dailey, H.A.; Dailey, T.A.; Gerdes, S.; Jahn, D.; Jahn, M.; O’Brian, M.R.; Warren, M.J. Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common Essential Product. Microbiol. Mol. Biol. Rev. 2017, 81, e00048-16. [Google Scholar] [CrossRef]
- Choby, J.E.; Skaar, E.P. Heme Synthesis and Acquisition in Bacterial Pathogens. J. Mol. Biol. 2016, 428, 3408–3428. [Google Scholar] [CrossRef]
- Hu, X.; Huang, Y.Y.; Wang, Y.; Wang, X.; Hamblin, M.R. Antimicrobial photodynamic therapy to control clinically relevant biofilm infections. Front. Microbiol. 2018, 9, 1299. [Google Scholar] [CrossRef]
- Kim, M.J.; Bang, W.S.; Yuk, H.G. 405 ± 5 nm light emitting diode illumination causes photodynamic inactivation of Salmonella spp. on fresh-cut papaya without deterioration. Food Microbiol. 2017, 62, 124–132. [Google Scholar] [CrossRef]
- Grinholc, M.; Rodziewicz, A.; Forys, K.; Rapacka-Zdonczyk, A.; Kawiak, A.; Domachowska, A.; Golunski, G.; Wolz, C.; Mesak, L.; Becker, K.; et al. Fine-tuning recA expression in Staphylococcus aureus for antimicrobial photoinactivation: Importance of photo-induced DNA damage in the photoinactivation mechanism. Appl. Microbiol. Biotechnol. 2015, 99, 9161–9176. [Google Scholar] [CrossRef]
- Wu, J.; Chu, Z.; Ruan, Z.; Wang, X.; Dai, T.; Hu, X. Changes of Intracellular Porphyrin, Reactive Oxygen Species, and Fatty Acids Profiles During Inactivation of Methicillin-Resistant Staphylococcus aureus by Antimicrobial Blue Light. Front. Physiol. 2018, 9, 1658. [Google Scholar] [CrossRef]
- Chu, Z.; Hu, X.; Wang, X.; Wu, J.; Dai, T.; Wang, X. Inactivation of Cronobacter sakazakii by blue light illumination and the resulting oxidative damage to fatty acids. Can. J. Microbiol. 2019, 65, 922–929. [Google Scholar] [CrossRef]
- Dai, T.; Gupta, A.; Huang, Y.Y.; Sherwood, M.E.; Murray, C.K.; Vrahas, M.S.; Kielian, T.; Hamblin, M.R. Blue light eliminates community-acquired methicillin-resistant Staphylococcus aureus in infected mouse skin abrasions. Photomed. Laser Surg. 2013, 31, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, Y.; Gupta, A.; Huang, Y.; Murray, C.K.; Vrahas, M.S.; Sherwood, M.E.; Baer, D.G.; Hamblin, M.R.; Dai, T. Antimicrobial blue light therapy for multidrug-resistant Acinetobacter baumannii infection in a mouse burn model: Implications for prophylaxis and treatment of combat-related wound infections. J. Infect. Dis. 2014, 209, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, M.; Landini, G.; Materassi, F.; Chellini, F.; Antonelli, A.; Tani, A.; Nosi, D.; Zecchi-Orlandini, S.; Rossolini, G.M.; Bani, D. Effects of photodynamic laser and violet-blue led irradiation on Staphylococcus aureus biofilm and Escherichia coli lipopolysaccharide attached to moderately rough titanium surface: In vitro study. Lasers Med. Sci. 2017, 32, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Fila, G.; Krychowiak, M.; Rychlowski, M.; Bielawski, K.P.; Grinholc, M. Antimicrobial blue light photoinactivation of Pseudomonas aeruginosa: Quorum sensing signaling molecules, biofilm formation and pathogenicity. J. Biophotonics 2018, 11, e201800079. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, D.L.; Lima, R.A.; Zanin, I.C.; Klein, M.I.; Janal, M.N.; Duarte, S. Effect of twice-daily blue light treatment on matrix-rich biofilm development. PLoS ONE 2015, 10, e0131941. [Google Scholar] [CrossRef]
- Ferrer-Espada, R.; Wang, Y.; Goh, X.S.; Dai, T. Antimicrobial Blue Light Inactivation of Microbial Isolates in Biofilms. Lasers Surg. Med. 2020, 52, 472–478. [Google Scholar] [CrossRef]
- Beirão, S.; Fernandes, S.; Coelho, J.; Faustino, M.A.F.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Tomé, A.C.; Almeida, A.; Cunha, A. Photodynamic inactivation of bacterial and yeast biofilms with a cationic porphyrin. Photochem. Photobiol. 2014, 90, 1387–1396. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Wang, Y.; Murray, C.K.; Hamblin, M.R.; Hooper, D.C.; Dai, T. Antimicrobial blue light inactivation of pathogenic microbes: State of the art. Drug Resist. Updates 2017, 33–35, 1–22. [Google Scholar] [CrossRef]
- Hyun, J.E.; Lee, S.Y. Blue light-emitting diodes as eco-friendly non-thermal technology in food preservation. Trends Food Sci. Technol. 2020, 105, 284–295. [Google Scholar] [CrossRef]
- Halstead, F.D.; Thwaite, J.E.; Burt, R.; Laws, T.R.; Raguse, M.; Moeller, R.; Webber, M.A.; Oppenheim, B.A. Antibacterial activity of blue light against nosocomial wound pathogens growing planktonically and as mature biofilms. Appl. Environ. Microbiol. 2016, 82, 4006–4016. [Google Scholar] [CrossRef]
- dos Anjos, C.; Sabino, C.P.; Bueris, V.; Fernandes, M.R.; Pogliani, F.C.; Lincopan, N.; Sellera, F.P. Antimicrobial blue light inactivation of international clones of multidrug-resistant Escherichia coli ST10, ST131 and ST648. Photodiagn. Photodyn. Ther. 2019, 27, 51–53. [Google Scholar] [CrossRef]
- Kim, M.J.; Adeline Ng, B.X.; Zwe, Y.H.; Yuk, H.G. Photodynamic inactivation of Salmonella enterica Enteritidis by 405 ± 5-nm light-emitting diode and its application to control salmonellosis on cooked chicken. Food Control 2017, 82, 305–315. [Google Scholar] [CrossRef]
- Rapacka-Zdonczyk, A.; Wozniak, A.; Nakonieczna, J.; Grinholc, M. Development of antimicrobial phototreatment tolerance: Why the methodology matters. Int. J. Mol. Sci. 2021, 22, 2224. [Google Scholar] [CrossRef]
- Nitzan, Y.; Salmon-Divon, M.; Shporen, E.; Malik, Z. ALA induced photodynamic effects on Gram positive and negative bacteria. Photochem. Photobiol. Sci. 2004, 3, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.M.; Bhayana, B.; Hamblin, M.R.; Dai, T. Antimicrobial blue light inactivation of Pseudomonas aeruginosa by photo-excitation of endogenous porphyrins: In vitro and in vivo studies. Lasers Surg. Med. 2016, 48, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, X.; Chen, J.; Amin, R.; Lu, M.; Bhayana, B.; Zhao, J.; Murray, C.K.; Hamblin, M.R.; Hooper, D.C.; et al. Antimicrobial Blue Light Inactivation of Gram-Negative Pathogens in Biofilms: In Vitro and in Vivo Studies. J. Infect. Dis. 2016, 213, 1380–1387. [Google Scholar] [CrossRef]
- Battisti, A.; Morici, P.; Ghetti, F.; Sgarbossa, A. Spectroscopic characterization and fluorescence imaging of Helicobacter pylori endogenous porphyrins. Biophys. Chem. 2017, 229, 19–24. [Google Scholar] [CrossRef]
- Zhang, J.; Kang, Z.; Chen, J.; Du, G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef]
- Fotinos, N.; Convert, M.; Piffaretti, J.C.; Gurny, R.; Lange, N. Effects on gram-negative and gram-positive bacteria mediated by 5-aminolevulinic acid and 5-aminolevulinic acid derivatives. Antimicrob. Agents Chemother. 2008, 52, 1366–1373. [Google Scholar] [CrossRef]
- Morimoto, K.; Ozawa, T.; Awazu, K.; Ito, N.; Honda, N.; Matsumoto, S.; Tsuruta, D. Photodynamic therapy using systemic administration of 5-aminolevulinic acid and a 410-nm wavelength light-emitting diode for methicillin-resistant Staphylococcus aureus-infected ulcers in mice. PLoS ONE 2014, 9, e105173. [Google Scholar] [CrossRef]
- Fyrestam, J.; Bjurshammar, N.; Paulsson, E.; Mansouri, N.; Johannsen, A.; Östman, C. Influence of culture conditions on porphyrin production in Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis. Photodiagn. Photodyn. Ther. 2017, 17, 115–123. [Google Scholar] [CrossRef]
- Biener, G.; Masson-Meyers, D.S.; Bumah, V.V.; Hussey, G.; Stoneman, M.R.; Enwemeka, C.S.; Raicu, V. Blue/violet laser inactivates methicillin-resistant Staphylococcus aureus by altering its transmembrane potential. J. Photochem. Photobiol. B Biol. 2017, 170, 118–124. [Google Scholar] [CrossRef]
- Abana, C.M.; Brannon, J.R.; Ebbott, R.A.; Dunigan, T.L.; Guckes, K.R.; Fuseini, H.; Powers, J.; Rogers, B.R.; Hadjifrangiskou, M. Characterization of blue light irradiation effects on pathogenic and nonpathogenic Escherichia coli. Microbiologyopen 2017, 6, e00466. [Google Scholar] [CrossRef]
- Ramstad, S.; Le Anh-Vu, N.; Johnsson, A. The temperature dependence of porphyrin production in Propionibacterium acnes after incubation with 5-aminolevulinic acid (ALA) and its methyl ester (m-ALA). Photochem. Photobiol. Sci. 2006, 5, 66–72. [Google Scholar] [CrossRef]
- Hessling, M.; Spellerberg, B.; Hoenes, K. Photoinactivation of bacteria by endogenous photosensitizers and exposure to visible light of different wavelengths—A review on existing data. FEMS Microbiol. Lett. 2017, 364, fnw270. [Google Scholar] [CrossRef]
- Baureder, M.; Hederstedt, L. Heme Proteins in Lactic Acid Bacteria. Adv. Microb. Physiol. 2013, 62, 1–43. [Google Scholar] [CrossRef]
- Kang, S.M.; Jung, H.I.; Kim, B. Il Susceptibility of oral bacteria to antibacterial photodynamic therapy. J. Oral Microbiol. 2019, 11, 1644111. [Google Scholar] [CrossRef] [PubMed]
- Hoenes, K.; Bauer, R.; Meurle, T.; Spellerberg, B.; Hessling, M. Inactivation Effect of Violet and Blue Light on ESKAPE Pathogens and Closely Related Non-pathogenic Bacterial Species—A Promising Tool Against Antibiotic-Sensitive and Antibiotic-Resistant Microorganisms. Front. Microbiol. 2021, 11, 3429. [Google Scholar] [CrossRef] [PubMed]
- Cieplik, F.; Späth, A.; Leibl, C.; Gollmer, A.; Regensburger, J.; Tabenski, L.; Hiller, K.A.; Maisch, T.; Schmalz, G. Blue light kills Aggregatibacter actinomycetemcomitans due to its endogenous photosensitizers. Clin. Oral Investig. 2014, 18, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Plavskii, V.Y.; Mikulich, A.V.; Tretyakova, A.I.; Leusenka, I.A.; Plavskaya, L.G.; Kazyuchits, O.A.; Dobysh, I.I.; Krasnenkova, T.P. Porphyrins and flavins as endogenous acceptors of optical radiation of blue spectral region determining photoinactivation of microbial cells. J. Photochem. Photobiol. B Biol. 2018, 183, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Cieplik, F.; Deng, D.; Crielaard, W.; Buchalla, W.; Hellwig, E.; Al-Ahmad, A.; Maisch, T. Antimicrobial photodynamic therapy–what we know and what we don’t. Crit. Rev. Microbiol. 2018, 44, 571–589. [Google Scholar] [CrossRef]
- Bumah, V.V.; Cortez, P.M.; Morrow, B.N.; Rojas, P.; Bowman, C.R.; Masson-Meyers, D.S.; Enwemeka, C.S. Blue light absorbing pigment in Streptococcus agalactiae does not potentiate the antimicrobial effect of pulsed 450 nm light. J. Photochem. Photobiol. B Biol. 2021, 216, 112149. [Google Scholar] [CrossRef]
- Manoil, D.; Filieri, A.; Gameiro, C.; Lange, N.; Schrenzel, J.; Wataha, J.C.; Bouillaguet, S. Flow cytometric assessment of Streptococcus mutans viability after exposure to blue light-activated curcumin. Photodiagn. Photodyn. Ther. 2014, 11, 372–379. [Google Scholar] [CrossRef]
- Luke-Marshall, N.R.; Hansen, L.A.; Shafirstein, G.; Campagnari, A.A. Antimicrobial Photodynamic Therapy with Chlorin e6 Is Bactericidal against Biofilms of the Primary Human Otopathogens. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Pileggi, G.; Wataha, J.C.; Girard, M.; Grad, I.; Schrenzel, J.; Lange, N.; Bouillaguet, S. Blue light-mediated inactivation of Enterococcus faecalis in vitro. Photodiagn. Photodyn. Ther. 2013, 10, 134–140. [Google Scholar] [CrossRef]
- van der Horst, M.A.; Key, J.; Hellingwerf, K.J. Photosensing in chemotrophic, non-phototrophic bacteria: Let there be light sensing too. Trends Microbiol. 2007, 15, 554–562. [Google Scholar] [CrossRef]
- Losi, A.; Gärtner, W. Bacterial bilin- and flavin-binding photoreceptors. Photochem. Photobiol. Sci. 2008, 7, 1168–1178. [Google Scholar] [CrossRef]
- Gomelsky, M.; Hoff, W.D. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 2011, 19, 441–448. [Google Scholar] [CrossRef]
- Glantz, S.T.; Carpenter, E.J.; Melkonian, M.; Gardner, K.H.; Boyden, E.S.; Wong, G.K.S.; Chow, B.Y. Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. USA 2016, 113, E1442–E1451. [Google Scholar] [CrossRef]
- Krauss, U.; Minh, B.Q.; Losi, A.; Gärtner, W.; Eggert, T.; Von Haeseler, A.; Jaeger, K.E. Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. J. Bacteriol. 2009, 191, 7234–7242. [Google Scholar] [CrossRef]
- Conrad, K.S.; Manahan, C.C.; Crane, B.R. Photochemistry of flavoprotein light sensors. Nat. Chem. Biol. 2014, 10, 801–809. [Google Scholar] [CrossRef]
- Losi, A.; Gärtner, W. Old chromophores, new photoactivation paradigms, trendy applications: Flavins in blue light-sensing photoreceptors. Photochem. Photobiol. 2011, 87, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Zoltowski, B.D.; Gardner, K.H. Tripping the light fantastic: Blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry 2011, 50, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Losi, A.; Gärtner, W. Solving Blue Light Riddles: New Lessons from Flavin-binding LOV Photoreceptors. Photochem. Photobiol. 2017, 93, 141–158. [Google Scholar] [CrossRef]
- Huala, E.; Oeller, P.W.; Liscum, E.; Han, I.S.; Larsen, E.; Briggs, W.R. Arabidopsis NPH1: A Protein Kinase with a Putative Redox-Sensing Domain. Science 1997, 278, 2120–2123. [Google Scholar] [CrossRef]
- Christie, J.M.; Salomon, M.; Nozue, K.; Wada, M.; Briggs, W.R. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 1999, 96, 8779–8783. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, M.T.A.; Domratcheva, T.; Bonetti, C.; Van Wilderen, L.J.G.W.; Van Grondelle, R.; Groot, M.L.; Hellingwerf, K.J.; Kennis, J.T.M. Primary reactions of the LOV2 domain of phototropin studied with ultrafast mid-infrared spectroscopy and quantum chemistry. Biophys. J. 2009, 97, 227–237. [Google Scholar] [CrossRef]
- Herrou, J.; Crosson, S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 2011, 9, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Losi, A.; Mandalari, C.; Gärtner, W. The Evolution and Functional Role of Flavin-based Prokaryotic Photoreceptors. Photochem. Photobiol. 2015, 91, 1021–1031. [Google Scholar] [CrossRef]
- Mandalari, C.; Losi, A.; Gärtner, W. Distance-tree analysis, distribution and co-presence of bilin- and flavin-binding prokaryotic photoreceptors for visible light. Photochem. Photobiol. Sci. 2013, 12, 1144–1157. [Google Scholar] [CrossRef]
- Rani, R.; Jentzsch, K.; Lecher, J.; Hartmann, R.; Willbold, D.; Jaeger, K.E.; Krauss, U. Conservation of dark recovery kinetic parameters and structural features in the pseudomonadaceae “short” light, oxygen, voltage (lov) protein family: Implications for the design of lov-based optogenetic tools. Biochemistry 2013, 52, 4460–4473. [Google Scholar] [CrossRef]
- Dikiy, I.; Edupuganti, U.R.; Abzalimov, R.R.; Borbat, P.P.; Srivastava, M.; Freed, J.H.; Gardner, K.H. Insights into histidine kinase activation mechanisms from the monomeric blue light sensor EL346. Proc. Natl. Acad. Sci. USA 2019, 116, 4963–4972. [Google Scholar] [CrossRef]
- Sankhe, G.D.; Dixit, N.M.; Saini, D.K. Activation of Bacterial Histidine Kinases: Insights into the Kinetics of the cis Autophosphorylation Mechanism. mSphere 2018, 3, e00111-18. [Google Scholar] [CrossRef]
- Campbell, E.A.; Westblade, L.F.; Darst, S.A. Regulation of bacterial RNA polymerase σ factor activity: A structural perspective. Curr. Opin. Microbiol. 2008, 11, 121–127. [Google Scholar] [CrossRef]
- Moy, B.E.; Seshu, J. STAS Domain Only Proteins in Bacterial Gene Regulation. Front. Cell. Infect. Microbiol. 2021, 11, 564. [Google Scholar] [CrossRef]
- Ryjenkov, D.A.; Tarutina, M.; Moskvin, O.V.; Gomelsky, M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005, 187, 1792–1798. [Google Scholar] [CrossRef]
- Williams, A.H.; Redzej, A.; Rolhion, N.; Costa, T.R.D.; Rifflet, A.; Waksman, G.; Cossart, P. The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Guerreiro, D.N.; Arcari, T.; O’Byrne, C.P. The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen. Front. Microbiol. 2020, 11, 1505. [Google Scholar] [CrossRef]
- Impens, F.; Rolhion, N.; Radoshevich, L.; Bécavin, C.; Duval, M.; Mellin, J.; Garciá Del Portillo, F.; Pucciarelli, M.G.; Williams, A.H.; Cossart, P. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat. Microbiol. 2017, 2, 1–12. [Google Scholar] [CrossRef]
- Ondrusch, N.; Kreft, J. Blue and Red Light Modulates SigB-Dependent Gene Transcription, Swimming Motility and Invasiveness in Listeria monocytogenes. PLoS ONE 2011, 6, e16151. [Google Scholar] [CrossRef]
- Tiensuu, T.; Andersson, C.; Rydén, P.; Johansson, J. Cycles of light and dark co-ordinate reversible colony differentiation in Listeria monocytogenes. Mol. Microbiol. 2013, 87, 909–924. [Google Scholar] [CrossRef]
- O’Donoghue, B.; NicAogáin, K.; Bennett, C.; Conneely, A.; Tiensuu, T.; Johansson, J.; O’Byrne, C. Blue-light inhibition of Listeria monocytogenes growth is mediated by reactive oxygen species and is influenced by σB and the blue-light sensor Lmo0799. Appl. Environ. Microbiol. 2016, 82, 4017–4027. [Google Scholar] [CrossRef]
- Dorey, A.L.; Lee, B.H.; Rotter, B.; O’Byrne, C.P. Blue Light Sensing in Listeria monocytogenes Is Temperature-Dependent and the Transcriptional Response to It Is Predominantly SigB-Dependent. Front. Microbiol. 2019, 10, 2497. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef]
- Chan, R.H.; Lewis, J.W.; Bogomolni, R.A. Photocycle of the LOV-STAS protein from the pathogen Listeria monocytogenes. Photochem. Photobiol. 2013, 89, 361–369. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Ogawa, M.; Hain, T.; Yoshida, M.; Fukumatsu, M.; Kim, M.; Mimuro, H.; Nakagawa, I.; Yanagawa, T.; Ishii, T.; et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 2009, 11, 1233–1240. [Google Scholar] [CrossRef]
- Mitchell, G.; Ge, L.; Huang, Q.; Chen, C.; Kianian, S.; Roberts, M.F.; Schekman, R.; Portnoy, D.A. Avoidance of autophagy mediated by PlcA or ActA is required for listeria monocytogenes growth in macrophages. Infect. Immun. 2015, 83, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Tamburro, M.; Sammarco, M.L.; Ammendolia, M.G.; Fanelli, I.; Minelli, F.; Ripabelli, G. Evaluation of transcription levels of inlA, inlB, hly, bsh and prfA genes in Listeria monocytogenes strains using quantitative reverse-transcription PCR and ability of invasion into human CaCo-2 cells. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef] [PubMed]
- Phelps, C.C.; Vadia, S.; Arnett, E.; Tan, Y.; Zhang, X.; Pathak-Sharma, S.; Gavrilin, M.A.; Seveau, S. Relative roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 2018, 86, e00555-18. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Marquis, H.; Boor, K.J. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 2005, 151, 3215–3222. [Google Scholar] [CrossRef] [PubMed]
- Vadia, S.; Seveau, S. Fluxes of Ca2+ and K+ are required for the listeriolysin O-dependent internalization pathway of Listeria monocytogenes. Infect. Immun. 2014, 82, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Marles-Wright, J.; Grant, T.; Delumeau, O.; van Duinen, G.; Firbank, S.J.; Lewis, P.J.; Murray, J.W.; Newman, J.A.; Quin, M.B.; Race, P.R.; et al. Molecular architecture of the “stressosome”, a signal integration and transduction hub. Science 2008, 322, 92–96. [Google Scholar] [CrossRef]
- Marles-Wright, J.; Lewis, R.J. The stressosome: Molecular architecture of a signalling hub. Biochem. Soc. Trans. 2010, 38, 928–933. [Google Scholar] [CrossRef]
- Losi, A.; Polverini, E.; Quest, B.; Gärtner, W. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 2002, 82, 2627–2634. [Google Scholar] [CrossRef]
- Ávila-Pérez, M.; Hellingwerf, K.J.; Kort, R. Blue light activates the σB-dependent stress response of Bacillus subtilis via YtvA. J. Bacteriol. 2006, 188, 6411–6414. [Google Scholar] [CrossRef]
- Gaidenko, T.A.; Kim, T.J.; Weigel, A.L.; Brody, M.S.; Price, C.W. The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J. Bacteriol. 2006, 188, 6387–6395. [Google Scholar] [CrossRef]
- Jurk, M.; Schramm, P.; Schmieder, P. The blue-light receptor YtvA from Bacillus subtilis is permanently incorporated into the stressosome independent of the illumination state. Biochem. Biophys. Res. Commun. 2013, 432, 499–503. [Google Scholar] [CrossRef]
- Suzuki, N.; Takaya, N.; Hoshino, T.; Nakamura, A. Enhancement of a σB-dependent stress response in Bacillus subtilis by ligh via YtvA photoreceptor. J. Gen. Appl. Microbiol. 2007, 53, 81–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van der Steen, J.B.; Ávila-Pérez, M.; Knippert, D.; Vreugdenhil, A.; van Alphen, P.; Hellingwerf, K.J. Differentiation of function among the RsbR paralogs in the general stress response of Bacillus subtilis with regard to light perception. J. Bacteriol. 2012, 194, 1708–1716. [Google Scholar] [CrossRef]
- Choi, S.; Nakasone, Y.; Hellingwerf, K.J.; Terazima, M. Photoreaction Dynamics of a Full-Length Protein YtvA and Intermolecular Interaction with RsbRA. Biochemistry 2020, 59, 4703–4710. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Ayala, F.; Bartolini, M.; Grau, R. The Stress-Responsive Alternative Sigma Factor SigB of Bacillus subtilis and Its Relatives: An Old Friend With New Functions. Front. Microbiol. 2020, 11, 1761. [Google Scholar] [CrossRef] [PubMed]
- Sumi, S.; Mutaguchi, N.; Ebuchi, T.; Tsuchida, H.; Yamamoto, T.; Suzuki, M.; Natsuka, C.; Shiratori-Takano, H.; Shintani, M.; Nojiri, H.; et al. Light response of Pseudomonas putida KT2440 mediated by class II LitR, a photosensor homolog. J. Bacteriol. 2020, 202, e00146-20. [Google Scholar] [CrossRef] [PubMed]
- Krauss, U.; Losi, A.; Gärtner, W.; Jaeger, K.E.; Eggert, T. Initial characterization of a blue-light sensing, phototropin-related protein from Pseudomonas putida: A paradigm for an extended LOV construct. Phys. Chem. Chem. Phys. 2005, 2804–2811. [Google Scholar] [CrossRef]
- Jentzsch, K.; Wirtz, A.; Circolone, F.; Drepper, T.; Losi, A.; Gärtner, W.; Jaeger, K.E.; Krauss, U. Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry 2009, 48, 10321–10333. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Buttani, V.; Losi, A.; Gärtner, W. A blue light inducible two-component signal transduction system in the plant pathogen Pseudomonas syringae pv. tomato. Biophys. J. 2008, 94, 897–905. [Google Scholar] [CrossRef]
- Moriconi, V.; Sellaro, R.; Ayub, N.; Soto, G.; Rugnone, M.; Shah, R.; Pathak, G.P.; Gärtner, W.; Casal, J.J. LOV-domain photoreceptor, encoded in a genomic island, attenuates the virulence of Pseudomonas syringae in light-exposed Arabidopsis leaves. Plant J. 2013, 76, 322–331. [Google Scholar] [CrossRef]
- Río-Álvarez, I.; Rodríguez-Herva, J.J.; Martínez, P.M.; González-Melendi, P.; García-Casado, G.; Rodríguez-Palenzuela, P.; López-Solanilla, E. Light regulates motility, attachment and virulence in the plant pathogen Pseudomonas syringae pv tomato DC3000. Environ. Microbiol. 2014, 16, 2072–2085. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; McGrane, R.S.; Beattie, G.A. Light regulation of swarming motility in Pseudomonas syringae integrates signaling pathways mediated by a bacteriophytochrome and a LOV protein. MBio 2013, 4, e00334-13. [Google Scholar] [CrossRef] [PubMed]
- Magerl, K.; Dick, B. Dimerization of LOV domains of: Rhodobacter sphaeroides (RsLOV) studied with FRET and stopped-flow experiments. Photochem. Photobiol. Sci. 2020, 19, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Hendrischk, A.K.; Moldt, J.; Frühwirth, S.W.; Klug, G. Characterization of an unusual LOV domain protein in the α-proteobacterium Rhodobacter sphaeroides. Photochem. Photobiol. 2009, 85, 1254–1259. [Google Scholar] [CrossRef]
- Conrad, K.S.; Bilwes, A.M.; Crane, B.R. Light-induced subunit dissociation by a light-oxygen-voltage domain photoreceptor from Rhodobacter sphaeroides. Biochemistry 2013, 52, 378–391. [Google Scholar] [CrossRef]
- Metz, S.; Jäger, A.; Klug, G. Role of a short light, oxygen, voltage (LOV) domain protein in blue light-and singlet oxygen-dependent gene regulation in Rhodobacter sphaeroides. Microbiology 2012, 158, 368–379. [Google Scholar] [CrossRef]
- Braatsch, S.; Gomelsky, M.; Kuphal, S.; Klug, G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 2002, 45, 827–836. [Google Scholar] [CrossRef]
- Masuda, S.; Bauer, C.E. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 2002, 110, 613–623. [Google Scholar] [CrossRef]
- Iseki, M.; Matsunaga, S.; Murakami, A.; Ohno, K.; Shiga, K.; Yoshida, K.; Sugai, M.; Takahashi, T.; Hori, T.; Watanabe, M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 2002, 415, 1047–1051. [Google Scholar] [CrossRef]
- Masuda, S. Light detection and signal transduction in the BLUF photoreceptors. Plant Cell Physiol. 2013, 54, 171–179. [Google Scholar] [CrossRef]
- Park, S.Y.; Tame, J.R.H. Seeing the light with BLUF proteins. Biophys. Rev. 2017, 9, 169–176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jung, A.; Domratcheva, T.; Tarutina, M.; Wu, Q.; Ko, W.H.; Shoeman, R.L.; Gomelsky, M.; Gardner, K.H.; Schlichting, I. Structure of a bacterial BLUF photoreceptor: Insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 2005, 102, 12350–12355. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, M.S.; Sharma, R.; Veetil, S.K.; Srivastava, S.K.; Kateriya, S. Modular diversity of the bluf proteins and their potential for the development of diverse optogenetic tools. Appl. Sci. 2019, 9, 3924. [Google Scholar] [CrossRef]
- Schroeder, C.; Werner, K.; Otten, H.; Krätzig, S.; Schwalbe, H.; Essen, L.O. Influence of a joining helix on the BLUF domain of the YcgF photoreceptor from Escherichia coli. ChemBioChem 2008, 9, 2463–2473. [Google Scholar] [CrossRef]
- Tschowri, N.; Busse, S.; Hengge, R. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 2009, 23, 522–534. [Google Scholar] [CrossRef]
- Tschowri, N.; Lindenberg, S.; Hengge, R. Molecular function and potential evolution of the biofilm-modulating blue light-signalling pathway of Escherichia coli. Mol. Microbiol. 2012, 85, 893–906. [Google Scholar] [CrossRef]
- Nakasone, Y.; Ono, T.A.; Ishii, A.; Masuda, S.; Terazima, M. Temperature-sensitive reaction of a photosensor protein YcgF: Possibility of a role of temperature sensor. Biochemistry 2010, 49, 2288–2296. [Google Scholar] [CrossRef]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef]
- Chitrakar, I.; Iuliano, J.N.; He, Y.L.; Woroniecka, H.A.; Tolentino Collado, J.; Wint, J.M.; Walker, S.G.; Tonge, P.J.; French, J.B. Structural Basis for the Regulation of Biofilm Formation and Iron Uptake in A. baumannii by the Blue-Light-Using Photoreceptor, BlsA. ACS Infect. Dis. 2020, 6, 2592–2603. [Google Scholar] [CrossRef]
- Mussi, M.A.; Gaddy, J.A.; Cabruja, M.; Arivett, B.A.; Viale, A.M.; Rasia, R.; Actis, L.A. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 2010, 192, 6336–6345. [Google Scholar] [CrossRef]
- Wood, C.R.; Squire, M.S.; Finley, N.L.; Page, R.C.; Actis, L.A. Structural and functional analysis of the Acinetobacter baumannii BlsA photoreceptor and regulatory protein. PLoS ONE 2019, 14, e220918. [Google Scholar] [CrossRef]
- Tuttobene, M.R.; Cribb, P.; Mussi, M.A. BlsA integrates light and temperature signals into iron metabolism through fur in the human pathogen Acinetobacter baumannii. Sci. Rep. 2018, 8, 7728. [Google Scholar] [CrossRef]
- Tuttobene, M.R.; Fernández-García, L.; Blasco, L.; Cribb, P.; Ambroa, A.; Müller, G.L.; Fernández-Cuenca, F.; Bleriot, I.; Rodríguez, R.E.; Barbosa, B.G.V.; et al. Quorum and light signals modulate acetoin/Butanediol catabolism in Acinetobacter spp. Front. Microbiol. 2019, 10, 1376. [Google Scholar] [CrossRef]
- Golic, A.E.; Valle, L.; Jaime, P.C.; Álvarez, C.E.; Parodi, C.; Borsarelli, C.D.; Abatedaga, I.; Mussi, M.A. BlsA is a low to moderate temperature blue light photoreceptor in the human pathogen Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1925. [Google Scholar] [CrossRef] [PubMed]
- Abatedaga, I.; Valle, L.; Golic, A.E.; Müller, G.L.; Cabruja, M.; Morán Vieyra, F.E.; Jaime, P.C.; Mussi, M.A.; Borsarelli, C.D. Integration of Temperature and Blue-Light Sensing in Acinetobacter baumannii Through the BlsA Sensor. Photochem. Photobiol. 2017, 93, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Tuttobene, M.R.; Pérez, J.F.; Pavesi, E.S.; Mora, B.P.; Biancotti, D.; Cribb, P.; Altilio, M.; Müller, G.L.; Gramajo, H.; Tamagno, G.; et al. Light modulates important pathogenic determinants and virulence in ESKAPE pathogens Acinetobacter baumannii, Pseudomonas aeruginosa, and Staphylococcus aureus. J. Bacteriol. 2020, 203, e00566-20. [Google Scholar] [CrossRef]
- Tasler, R.; Moises, T.; Frankenberg-Dinkel, N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 2005, 272, 1927–1936. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Jemielita, M.; Stergioula, V.; Tikhonov, M.; Bassler, B.L. Photosensing and quorum sensing are integrated to control Pseudomonas aeruginosa collective behaviors. PLoS Biol. 2019, 17, e3000579. [Google Scholar] [CrossRef]
- Fila, G.; Kawiak, A.; Grinholc, M.S. Blue light treatment of Pseudomonas aeruginosa: Strong bactericidal activity, synergism with antibiotics and inactivation of virulence factors. Virulence 2017, 8, 938–958. [Google Scholar] [CrossRef]
- Kahl, L.J.; Price-Whelan, A.; Dietrich, L.E.P. Light-mediated decreases in cyclic di-gmp levels inhibit structure formation in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2020, 202, e00117-20. [Google Scholar] [CrossRef]
- Okegbe, C.; Fields, B.L.; Cole, S.J.; Beierschmitt, C.; Morgan, C.J.; Price-Whelan, A.; Stewart, R.C.; Lee, V.T.; Dietrich, L.E.P. Electron-shuttling antibiotics structure bacterial communities by modulating cellular levels of c-di-GMP. Proc. Natl. Acad. Sci. USA 2017, 114, E5236–E5245. [Google Scholar] [CrossRef]
- Wright, S.; Walia, B.; Parkinson, J.S.; Khan, S. Differential activation of Escherichia coli chemoreceptors by blue-light stimuli. J. Bacteriol. 2006, 188, 3962–3971. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perlova, T.; Gruebele, M.; Chemla, Y.R. Blue light is a universal signal for Escherichia coli chemoreceptors. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.L. Aer on the inside looking out: Paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 2007, 65, 1415–1424. [Google Scholar] [CrossRef]
- Santamaría-Hernando, S.; Cerna-Vargas, J.P.; Martínez-García, P.M.; de Francisco-de Polanco, S.; Nebreda, S.; Rodríguez-Palenzuela, P.; Rodríguez-Herva, J.J.; López-Solanilla, E. Blue-light perception by epiphytic Pseudomonas syringae drives chemoreceptor expression, enabling efficient plant infection. Mol. Plant Pathol. 2020, 21, 1606–1619. [Google Scholar] [CrossRef] [PubMed]
- Rapacka-Zdonczyk, A.; Wozniak, A.; Pieranski, M.; Woziwodzka, A.; Bielawski, K.P.; Grinholc, M. Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci. Rep. 2019, 9, 9423. [Google Scholar] [CrossRef]
- Pieranski, M.; Sitkiewicz, I.; Grinholc, M. Increased photoinactivation stress tolerance of Streptococcus agalactiae upon consecutive sublethal phototreatments. Free Radic. Biol. Med. 2020, 160, 657–669. [Google Scholar] [CrossRef]
- Jiang, Q.; Karata, K.; Woodgate, R.; Cox, M.M.; Goodman, M.F. The active form of DNA polymerase v is UmuD′ 2 C-RecA-ATP. Nature 2009, 460, 359–363. [Google Scholar] [CrossRef]
- Gruber, A.J.; Erdem, A.L.; Sabat, G.; Karata, K.; Jaszczur, M.M.; Vo, D.D.; Olsen, T.M.; Woodgate, R.; Goodman, M.F.; Cox, M.M. A RecA Protein Surface Required for Activation of DNA Polymerase V. PLoS Genet. 2015, 11, e1005066. [Google Scholar] [CrossRef]
- Hui, J.G.K.; Mai-Prochnow, A.; Kjelleberg, S.; McDougald, D.; Rice, S.A. Environmental cues and genes involved in establishment of the superinfective Pf4 phage of Pseudomonas aeruginosa. Front. Microbiol. 2014, 5, 654. [Google Scholar] [CrossRef]
- Hocquet, D.; Llanes, C.; Thouverez, M.; Kulasekara, H.D.; Bertrand, X.; Plésiat, P.; Mazel, D.; Miller, S.I. Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog. 2012, 8, 1002778. [Google Scholar] [CrossRef]
- Irazoki, O.; Mayola, A.; Campoy, S.; Barbé, J. SOS system induction inhibits the assembly of chemoreceptor signaling clusters in Salmonella enterica. PLoS ONE 2016, 11, e0146685. [Google Scholar] [CrossRef] [PubMed]
- Mayola, A.; Irazoki, O.; Martínez, I.A.; Petrov, D.; Menolascina, F.; Stocker, R.; Reyes-Darias, J.A.; Krell, T.; Barbé, J.; Campoy, S. RecA protein plays a role in the chemotactic response and chemoreceptor clustering of Salmonella enterica. PLoS ONE 2014, 9, e105578. [Google Scholar] [CrossRef]
- Cirz, R.T.; Jones, M.B.; Gingles, N.A.; Minogue, T.D.; Jarrahi, B.; Peterson, S.N.; Romesberg, F.E. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 2007, 189, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Aranda, J.; Poza, M.; Shingu-Vázquez, M.; Cortés, P.; Boyce, J.D.; Adler, B.; Barbé, J.; Bou, G. Identification of a DNA-Damage-Inducible regulon in Acinetobacter baumannii. J. Bacteriol. 2013, 195, 5577–5582. [Google Scholar] [CrossRef] [PubMed]
- Mérida-Floriano, A.; Rowe, W.P.M.; Casadesús, J. Genome-Wide Identification and Expression Analysis of SOS Response Genes in Salmonella enterica serovar Typhimurium. Cells 2021, 10, 943. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Alberola, N.; Campoy, S.; Barbé, J.; Erill, I. Analysis of the SOS response of Vibrio and other bacteria with multiple chromosomes. BMC Genom. 2012, 13, 58. [Google Scholar] [CrossRef]
- Krin, E.; Pierlé, S.A.; Sismeiro, O.; Jagla, B.; Dillies, M.A.; Varet, H.; Irazoki, O.; Campoy, S.; Rouy, Z.; Cruveiller, S.; et al. Expansion of the SOS regulon of Vibrio cholerae through extensive transcriptome analysis and experimental validation. BMC Genom. 2018, 19, 373. [Google Scholar] [CrossRef]
- Cirz, R.T.; O’Neill, B.M.; Hammond, J.A.; Head, S.R.; Romesberg, F.E. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J. Bacteriol. 2006, 188, 7101–7110. [Google Scholar] [CrossRef]
- Norton, M.D.; Spilkia, A.J.; Godoy, V.G. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J. Bacteriol. 2013, 195, 1335–1345. [Google Scholar] [CrossRef]
- Hare, J.M.; Adhikari, S.; Lambert, K.V.; Hare, A.E.; Grice, A.N. The Acinetobacter regulatory UmuDAb protein cleaves in response to DNA damage with chimeric LexA/UmuD characteristics. FEMS Microbiol. Lett. 2012, 334, 57–65. [Google Scholar] [CrossRef]
- Painter, K.L.; Strange, E.; Parkhill, J.; Bamford, K.B.; Armstrong-James, D.; Edwards, A.M. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect. Immun. 2015, 83, 1830–1844. [Google Scholar] [CrossRef] [PubMed]
- Baharoglu, Z.; Mazel, D. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: A route towards multiresistance. Antimicrob. Agents Chemother. 2011, 55, 2438–2441. [Google Scholar] [CrossRef] [PubMed]
- Dosselli, R.; Millioni, R.; Puricelli, L.; Tessari, P.; Arrigoni, G.; Franchin, C.; Segalla, A.; Teardo, E.; Reddi, E. Molecular targets of antimicrobial photodynamic therapy identified by a proteomic approach. J. Proteom. 2012, 77, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Tomb, R.M.; Maclean, M.; Coia, J.E.; MacGregor, S.J.; Anderson, J.G. Assessment of the potential for resistance to antimicrobial violet-blue light in Staphylococcus aureus. Antimicrob. Resist. Infect. Control 2017, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, V.T.; Martegani, E.; Bolognese, F. Catalase A is involved in the response to photooxidative stress in Pseudomonas aeruginosa. Photodiagnosis Photodyn. Ther. 2018, 22, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Martegani, E.; Bolognese, F.; Trivellin, N.; Orlandi, V.T. Effect of blue light at 410 and 455 nm on Pseudomonas aeruginosa biofilm. J. Photochem. Photobiol. B Biol. 2020, 204, 111790. [Google Scholar] [CrossRef] [PubMed]
- Leanse, L.G.; Goh, X.S.; Cheng, J.X.; Hooper, D.C.; Dai, T. Dual-wavelength photo-killing of methicillin-resistant Staphylococcus aureus. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.T.; Mohammad, H.; Hui, J.; Leanse, L.G.; Li, J.; Liang, L.; Dai, T.; Seleem, M.N.; Cheng, J.X. Photolysis of Staphyloxanthin in Methicillin-Resistant Staphylococcus aureus Potentiates Killing by Reactive Oxygen Species. Adv. Sci. 2019, 6, 1900030. [Google Scholar] [CrossRef]
- Djouiai, B.; Thwaite, J.E.; Laws, T.R.; Commichau, F.M.; Setlow, B.; Setlow, P.; Moeller, R. Role of DNA repair and protective components in Bacillus subtilis spore resistance to inactivation by 400-nm-wavelength blue light. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Tardu, M.; Bulut, S.; Kavakli, I.H. MerR and ChrR mediate blue light induced photo-oxidative stress response at the transcriptional level in Vibrio cholerae. Sci. Rep. 2017, 7, 40817. [Google Scholar] [CrossRef] [PubMed]
- Worthington, E.N.; Kavakli, I.H.; Berrocal-Tito, G.; Bondo, B.E.; Sancar, A. Purification and characterization of three members of the photolyase/cryptochrome family blue-light photoreceptors from Vibrio cholerae. J. Biol. Chem. 2003, 278, 39143–39154. [Google Scholar] [CrossRef] [PubMed]
- Losi, A.; Gärtner, W. A light life together: Photosensing in the plant microbiota. Photochem. Photobiol. Sci. 2021, 20, 451–473. [Google Scholar] [CrossRef] [PubMed]
- Metz, S.; Haberzettl, K.; Frühwirth, S.; Teich, K.; Hasewinkel, C.; Klug, G. Interaction of two photoreceptors in the regulation of bacterial photosynthesis genes. Nucleic Acids Res. 2012, 40, 5901–5909. [Google Scholar] [CrossRef]
- Yakimov, A.; Pobegalov, G.; Bakhlanova, I.; Khodorkovskii, M.; Petukhov, M.; Baitin, D. Blocking the RecA activity and SOS-response in bacteria with a short α-helical peptide. Nucleic Acids Res. 2017, 45, 9788–9796. [Google Scholar] [CrossRef]
- Yakimov, A.; Bakhlanova, I.; Baitin, D. Targeting evolution of antibiotic resistance by SOS response inhibition. Comput. Struct. Biotechnol. J. 2021, 19, 777–783. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Species | Blue Light | Photosensitizer a | Light Dosage (Joule/cm2) *, a | Bactericidal Efficacy **, a | Reference |
|---|---|---|---|---|---|
| Streptococcus agalactiae | 450 nm (pulsed) | CP III (0.08 mg/mL) PP IX (0.08 mg/mL) | 7.6 | 8.89 log CFU/mL 9.54 log CFU/mL | [47] |
| Streptococcus mutans | 360–550 nm | curcumin (2 µM) | 54 | 2 log cells *** | [48] |
| Streptococcus pneumoniae (planktonic or biofilm) | 405 nm | chlorin e6 (10 µM) | 12 or 90 | approximately 5 or 6.5 log CFU/mL | [49] |
| Enterococcus faecalis (planktonic or biofilm) | 405–500 nm | eosin Y (5 or 10 µM) rose bengal (1 or 2 µM) curcumin (5 or 1 µM) | 108 | 4.9 or 13.8 log CFU/mL 7.3 or 13.8 log CFU/mL 7.6 or 13.7 log CFU/mL | [50] |
| Enterococcus faecium | 405 nm | curcumin (1 µg/mL) PP IX (0.1 µg/mL) | 25.3 | significant drop in optical density (655 nm) at p < 0.001 | [42] |
| Bacterial Species (Phylum/Class) | NCBI Accession Number of Parent Protein | LOV Domain | Number of Amino Acids |
|---|---|---|---|
| Listeria monocytogenes, Listeria innocua, Bacillus subtilis, Bacillus cereus (Firmicutes) | WP_003738433.1 (LM), WP_003761135.1 (LI), WP_047183059.1 (BS) or AUZ27534.1 (BC) | LOV+STAS | 253 (LM and LI) or 261 (BS and BC) |
| Acinetobacter baumannii 1294596 (ɣ-Proteobacteria) | EXF56192.1 | GAF+PAS+LOV+GGDEAL+EAL | 855 |
| Shewenella putrefaciens (ɣ-Proteobacteria) | WP_025008327.1 | MASE+CHASE+PAS+LOV+GGDEF+EAL | 1216 |
| Arthrospira maxima (Cyanobacteria) | WP_006668677.1 | GAF+PAS+LOV+PAS+GAF+Kinase | 1184 |
| Nakamurella multipartita (Actinobacteria) | WP_015749472.1 | PAS+RR+LOV | 365 |
| Brucella abortus (α-Proteobacteria) | Q2YKK7.2 | LOV+PAS+HK | 489 |
| Virulent Factors | Description | Reference |
|---|---|---|
| ActA | ActA functions as a bacterial defense against autophagy and is controlled by the transcription factor PrfA. Inside the host cell cytoplasm, ActA recruits host cell cytoskeletal proteins to inhibit ubiquitin and p62, which renders L. monocytogenes unrecognizable during the autophagic process, allowing the bacteria to proliferate. | [81,82] |
| InlA and InlB | Invasins that are essential for internalization into the host cell. The efficacy of InlA and InlB varies across L. monocytogenes strains and types of host cell (particularly the expression of relevant receptors on different host cells). It is also known that InlA and InlB are regulated by σB, with all three components needed for an effective infection of L. monocytogenes. | [83,84,85] |
| Listeriolysin O | Cholesterol-dependent cytolysin that primarily plays a critical role in breaking the membrane of phagosomes post-internalization, allowing L. monocytogenes to invade the cytosol. Pore-forming activity of listeriolysin O may also facilitate the internalization of L. monocytogenes at the early stage of infection in a calcium ion- and potassium ion-dependent manner. | [84,86] |
| Bacterial Species | NCBI Accession Number of Parent Protein | BLUF Domain (Predicted Function) | Number of Amino Acids |
|---|---|---|---|
| Escherichia coli | ARH96915.1 | BLUF+EAL (regulation of diguanylate cyclases and phosphodiesterase activity) | 403 |
| Rhodobacter sphaeroides ATCC 17025 | ABP71929.1 | BLUF+B12-binding (enhancement of photosensing capability) | 448 |
| Leptonoma illini DSM 21528 | EHQ08139.1 | BLUF+CHD (regulation of diguanylate cyclases) | 309 |
| Methylobacterium radiotolerans | WP_012321331.1 | BLUF+PRK09039 superfamily (regulation of phosphoribulokinase, uridine kinase and panthothenate kinase activity) | 309 |
| Hymenobacter sp. PAMC 26554 | AMR27912.1 | BLUF+REC (regulation of chemotaxis) | 292 |
| Curtobacterium luteum | WP_058726129.1 | BLUF+AcrR (regulation of antibiotic resistance) | 335 |
| Thiocystis violascens DSM 198 | AFL74487.1 | BLUF+EAL+GGDEF (regulation of c-di-GMP level) | 597 |
| Legionella steelei | WP_058511962.1 | BLUF+PAS (regulation of cellular signaling) | 402 |
| Bacterial Species | SOS Activator | Physiological Manifestation | Reference |
|---|---|---|---|
| Staphylococcus aerus | H2O2 | formation of gentamicin-resistant and H2O2-tolerant SCVs due to enhanced catalase production mediated by pol V, RecA and RexAB proteins | [154] |
| Acinetobacter baumannii | UV, MMS (alkylation), ciprofloxacin and dessication | increased prevalence of rifampin-resistant mutants as mediated by RecA protein | [152] |
| S. enterica subsp. enterica serovar Typhimurium | MMC | decreased swarming motility to avoid lethal MMC, as controlled by RecA protein, but not other proteins involved in SOS response | [144] |
| Pseudomonas aeruginosa | H2O2, nitric oxide and MMC | formation of superinfective phage Pf4, resulting in the appearance of SCV within the bacterial biofilm | [142] |
| Vibrio cholerae | Antibiotics 1 | increased prevalence of rifampin-resistant mutants | [155] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadi, J.; Wu, S.; Soni, A.; Gardner, A.; Brightwell, G. Genetic Factors Affect the Survival and Behaviors of Selected Bacteria during Antimicrobial Blue Light Treatment. Int. J. Mol. Sci. 2021, 22, 10452. https://doi.org/10.3390/ijms221910452
Hadi J, Wu S, Soni A, Gardner A, Brightwell G. Genetic Factors Affect the Survival and Behaviors of Selected Bacteria during Antimicrobial Blue Light Treatment. International Journal of Molecular Sciences. 2021; 22(19):10452. https://doi.org/10.3390/ijms221910452
Chicago/Turabian StyleHadi, Joshua, Shuyan Wu, Aswathi Soni, Amanda Gardner, and Gale Brightwell. 2021. "Genetic Factors Affect the Survival and Behaviors of Selected Bacteria during Antimicrobial Blue Light Treatment" International Journal of Molecular Sciences 22, no. 19: 10452. https://doi.org/10.3390/ijms221910452
APA StyleHadi, J., Wu, S., Soni, A., Gardner, A., & Brightwell, G. (2021). Genetic Factors Affect the Survival and Behaviors of Selected Bacteria during Antimicrobial Blue Light Treatment. International Journal of Molecular Sciences, 22(19), 10452. https://doi.org/10.3390/ijms221910452








