Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives
Abstract
1. Introduction
2. Starch Metabolism and Phosphorylase
2.1. Starch Biosynthesis
2.2. Initiation of Starch Granule in Comparison to Glycogen
2.3. Multiple Complex Formation
2.4. Starch Degradation
2.5. Additional Role of PHO1
3. Synthetic and Phosphorolytic Activity of Phosphorylase
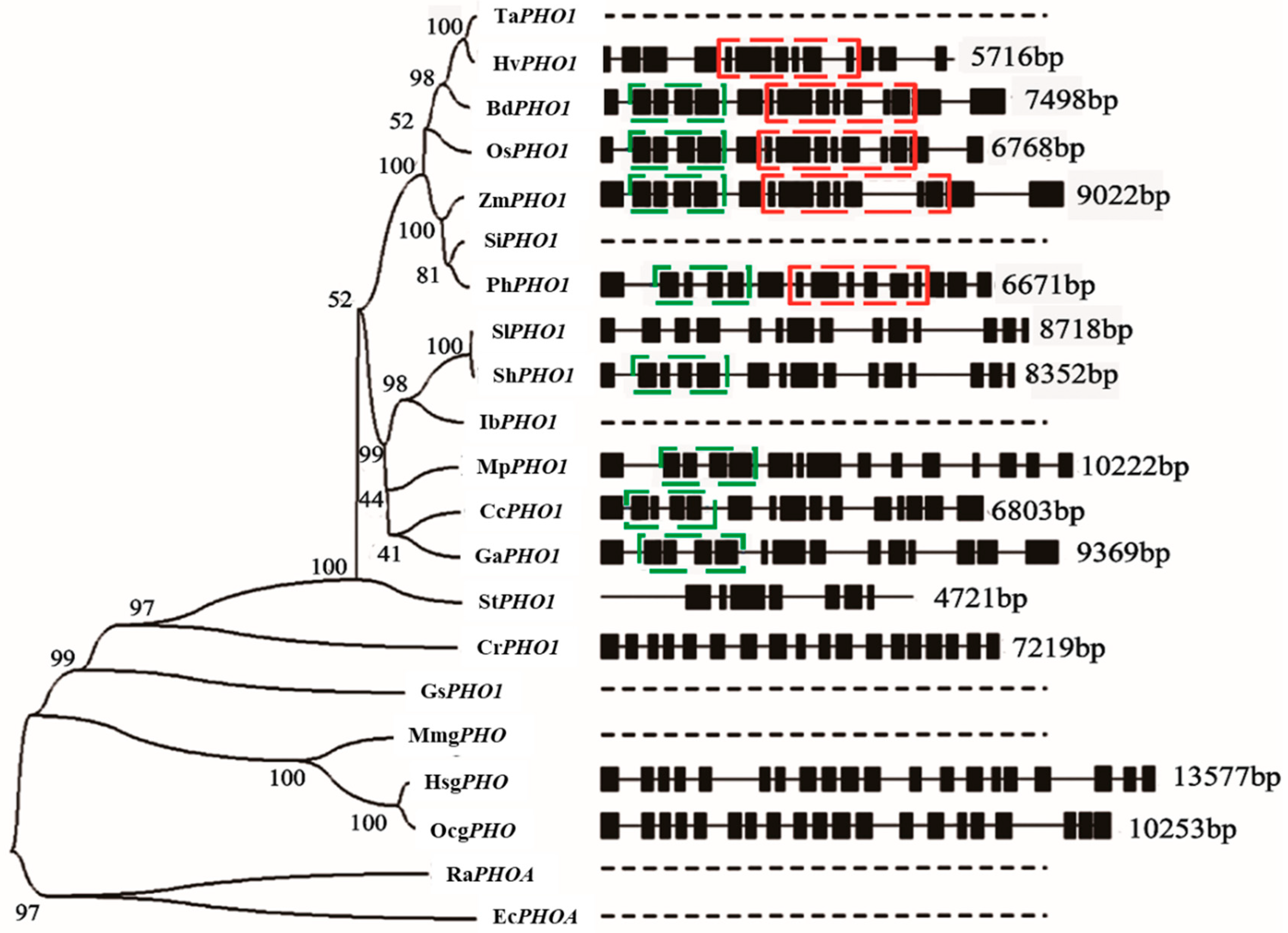
4. Genomics of PHO1
Characteristics of PHO1
| Motif | Sequence | Possible Function and DB ID |
|---|---|---|
| aBrelaterD1 | ACGTG | ABA, drought, and dark-induced regulation (S000414) |
| BIHD1OS | TGTCA | Disease resistance (S000498) |
| MYcatrD22 | CACATG | ABA and drought regulation (S000174) |
| SOrlIP1at | GCCAC | Specific guard cell expression (S000483) |
| taaagStKSt1 | TAAAG | Specific guard cell expression (S000387) |
| tataBOX5 | TTATTT | Light response (S000112) |
| WBOXHVISO1 | TTGAC | Environmental stress response (S000442) |
| WrKY71OS | TGAC | ABA regulation (S000447) |
| acgtaterD1 | ACGT | Drought response (S000415) |
| cUrecOrecr | GTAC | Copper- and oxygen-induced coordinated response (S000493) |
| DOFcOreZM | AAAG | Carbon metabolism (S000265) |
| gataBOX | GATA | Light response and tissue-specific expression (S000039) |
| gt1gMScaM4 | GAAAAA | Pathogen or salt-induced expression (S000453) |
| gtgantg10 | GTGA | Pollen-specific regulation enhancement (S000378) |
| nODcOn2gM | CTCTT | Specific nodule expression (S000462) |
| OSe2rOOtnODUle | CTCTT | Organ-specific expression (S000468) |
| POllen1lelat52 | AGAAA | Response against pollen (S000245) |
| raV1aat | CAACA | Binding to specific bipartite sequence (S000314) |
| rOOtMOtIFtaPOX1 | ATATT | Organ-specific expression (S000098) |
| caatBOX1 | CAAT | Tissue-specific promoter (S000028) |
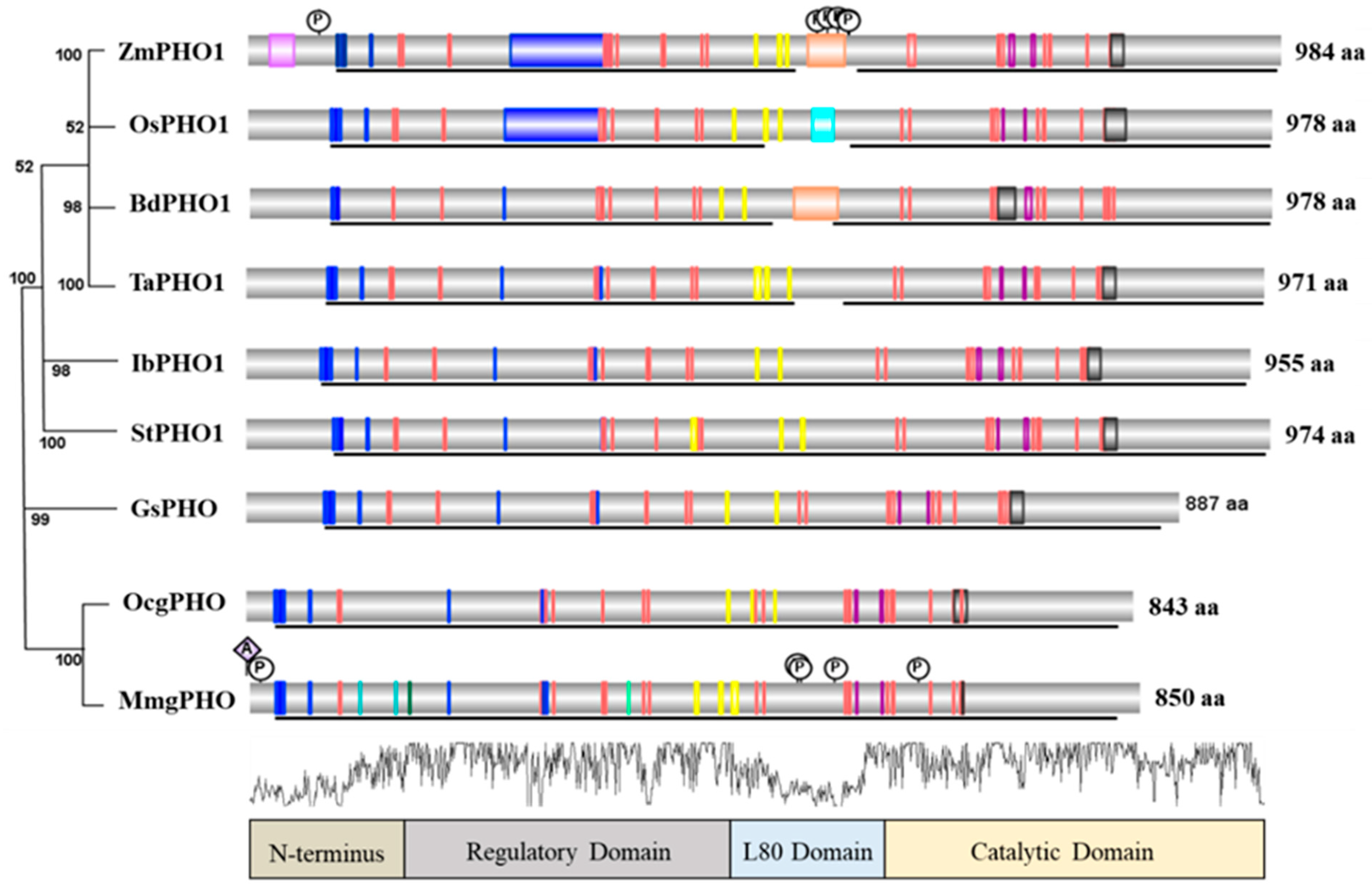
5. Structure and Comparison of PHO1
6. Regulation of Starch Phosphorylase
6.1. Phosphorylation
6.2. Protein–Protein Interaction and PHO1
6.3. Redox Regulation and PHO1
6.4. Transcription Factors (TFs) and PHO1
7. Concluding Comments and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Cori, G.T.; Cori, C.F.; Schmidt, G. The role of glucose-l-phosphate in the formation of blood sugar and synthesis of glycogen in the liver. J. Biol. Chem. 1939, 142, 355–366. [Google Scholar]
- Green, D.E.S.; Stumpf, P.K. Starch Phosphorylase of potato. J. Biol. Chem. 1942, 142, 355–366. [Google Scholar] [CrossRef]
- Hanes, C.S. The breakdown and synthesis of starch by an enzyme system from pea seeds. Proc. R. Soc. Lond. Ser. B Boil. Sci. 1940, 128, 421–450. [Google Scholar] [CrossRef]
- Tsai, C.Y.; Nelson, O.E. Phosphorylases I and II of Maize Endosperm. Plant Physiol. 1968, 43, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Subasinghe, R.M.; Liu, F.; Polack, U.C.; Lee, E.A.; Emes, M.J.; Tetlow, I.J. Multimeric states of starch phosphorylase determine protein–protein interactions with starch biosynthetic enzymes in amyloplasts. Plant Physiol. Biochem. 2014, 83, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Fukui, T. The complete amino acid sequence of potato alpha-glucan phosphorylase. J. Biol. Chem. 1986, 261, 8230–8236. [Google Scholar] [CrossRef]
- Brisson, N.; Giroux, H.; Zollinger, M.; Camirand, A.; Simard, C. Maturation and Subcellular Compartmentation of Potato Starch Phosphorylase. Plant Cell 1989, 1, 559. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Mu, H.H.; Wasserman, B.P.; Carman, G.M. Identification of the Maize Amyloplast Stromal 112-kD Protein as a Plastidic Starch Phosphorylase. Plant Physiol. 2001, 125, 351–359. [Google Scholar] [CrossRef]
- Tickle, P.; Burrell, M.M.; Coates, S.A.; Emes, M.J.; Tetlow, I.J.; Bowsher, C.G. Characterization of plastidial starch phosphorylase in Triticum aestivum L. endosperm. J. Plant Physiol. 2009, 166, 1465–1478. [Google Scholar] [CrossRef]
- Hwang, S.-K.; Singh, S.; Çakir, B.; Satoh, H.; Okita, T.W. The plastidial starch phosphorylase from rice endosperm: Catalytic properties at low temperature. Planta 2016, 243, 999–1009. [Google Scholar] [CrossRef]
- Cuesta-Seijo, J.A.; Nielsen, M.M.; Ruzanski, C.; Krucewicz, K.; Beeren, S.R.; Rydhal, M.G.; Yoshimura, Y.; Striebeck, A.; Motawia, M.S.; Willats, W.G.T.; et al. In Vitro Biochemical Characterization of All Barley Endosperm Starch Synthases. Front. Plant Sci. 2016, 6, 1265. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.; Koper, K.; Okita, T.W. The plastid phosphorylase as a multiple-role player in plant metabolism. Plant Sci. 2020, 290, 110303. [Google Scholar] [CrossRef]
- Albrecht, T.; Greve, B.; Pusch, K.; Kossmann, J.; Buchner, P.; Wobus, U.; Steup, M. Homodimers and heterodimers of Pho1-type phosphorylase isoforms in Solanum tuberosum L. as revealed by sequence-specific antibodies. JBIC J. Biol. Inorg. Chem. 1998, 251, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Chen, H.-M.; Chou, I.-M.; Chen, A.-N.; Chen, C.-P.; Young, G.-H.; Lin, C.-T.; Cheng, C.-H.; Chang, S.-C.; Juang, R.-H. Plastidial Starch Phosphorylase in Sweet Potato Roots Is Proteolytically Modified by Protein-Protein Interaction with the 20S Proteasome. PLoS ONE 2012, 7, e35336. [Google Scholar] [CrossRef]
- Ko, Y.-T.; Chang, J.-Y.; Lee, Y.-T.; Wu, Y.-H. The Identification of Starch Phosphorylase in the Developing Mungbean (Vigna radiata L.). J. Agric. Food Chem. 2005, 53, 5708–5715. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Shibahara, K.; Tokunaga, T.; Nishi, A.; Tasaki, M.; Hwang, S.-K.; Okita, T.; Kaneko, N.; Fujita, N.; Yoshida, M.; et al. Mutation of the Plastidial α-Glucan Phosphorylase Gene in Rice Affects the Synthesis and Structure of Starch in the Endosperm. Plant Cell 2008, 20, 1833–1849. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.-K.; Koper, K.; Satoh, H.; Okita, T.W. Rice Endosperm Starch Phosphorylase (Pho1) Assembles with Disproportionating Enzyme (Dpe1) to Form a Protein Complex That Enhances Synthesis of Malto-oligosaccharides. J. Biol. Chem. 2016, 291, 19994–20007. [Google Scholar] [CrossRef]
- Cuesta-Seijo, J.A.; Ruzanski, C.; Krucewicz, K.; Meier, S.; Hägglund, P.; Svensson, B.; Palcic, M.M. Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development. PLoS ONE 2017, 12, e0175488. [Google Scholar] [CrossRef]
- Liu, F.; Makhmoudova, A.; Lee, E.A.; Wait, R.; Emes, M.J.; Tetlow, I.J. The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 2009, 60, 4423–4440. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Davies, E.J.; Vardy, K.A.; Bowsher, C.G.; Burrell, M.M.; Emes, M.J. Subcellular localization of ADPglucose pyrophosphorylase in developing wheat endosperm and analysis of the properties of a plastidial isoform. J. Exp. Bot. 2003, 54, 715–725. [Google Scholar] [CrossRef]
- Morell, M.K.; Blennow, A.; Kosar-Hashemi, B.; Samuel, M. Differential Expression and Properties of Starch Branching Enzyme Isoforms in Developing Wheat Endosperm. Plant Physiol. 1997, 113, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Maddelein, M.L.; Libessart, N.; Bellanger, F.; Delrue, B.; D’Hulst, C.; Koornhuyse, N.V.D.; Fontaine, T.; Wieruszeski, J.M.; Decq, A.; Ball, S. Toward an understanding of the biogenesis of the starch granule. Determination of granule-bound and soluble starch synthase functions in amylopectin synthesis. J. Biol. Chem. 1994, 269, 25150–25157. [Google Scholar] [CrossRef]
- Putseys, J.A.; Derde, L.J.; Lamberts, L.; Östman, E.; Björck, I.M.; Delcour, J.A. Functionality of Short Chain Amylose-Lipid Complexes in Starch-Water Systems and Their Impact on In Vitro Starch Degradation. J. Agric. Food Chem. 2010, 58, 1939–1945. [Google Scholar] [CrossRef] [PubMed]
- Goren, A.; Ashlock, D.; Tetlow, I.J. Starch formation inside plastids of higher plants. Protoplasma 2018, 255, 1855–1876. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Lv, Y.; Shen, L.; Wang, Y.; Qing, Y.; Wu, N.; Li, Y.; Huang, H.; Zhang, N.; Liu, Y.; et al. The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tag(tm) Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. Int. J. Mol. Sci. 2019, 20, 986. [Google Scholar] [CrossRef]
- Fujita, N.; Kubo, A.; Suh, D.-S.; Wong, K.-S.; Jane, J.-L.; Ozawa, K.; Takaiwa, F.; Inaba, Y.; Nakamura, Y. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol. 2003, 44, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and Characterization of Starch Synthase I Using Mutants in Rice. Plant Physiol. 2006, 140, 1070–1084. [Google Scholar] [CrossRef]
- Hanashiro, I.; Itoh, K.; Kuratomi, Y.; Yamazaki, M.; Igarashi, T.; Matsugasako, J.-I.; Takeda, Y. Granule-Bound Starch Synthase I is Responsible for Biosynthesis of Extra-Long Unit Chains of Amylopectin in Rice. Plant Cell Physiol. 2008, 49, 925–933. [Google Scholar] [CrossRef]
- Ortiz-Marchena, M.I.; Albi, T.; Lucas-Reina, E.; Said, F.E.; Romero-Campero, F.J.; Cano, B.; Ruiz, M.T.; Romero, J.M.; Valverde, F. Photoperiodic Control of Carbon Distribution during the Floral Transition in Arabidopsis. Plant Cell 2014, 26, 565–584. [Google Scholar] [CrossRef] [PubMed]
- Streb, S.; Delatte, T.; Umhang, M.; Eicke, S.; Schorderet, M.; Reinhardt, D.; Zeeman, S.C. Starch Granule Biosynthesis in Arabidopsis Is Abolished by Removal of All Debranching Enzymes but Restored by the Subsequent Removal of an Endoamylase. Plant Cell 2009, 20, 3448–3466. [Google Scholar] [CrossRef]
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016, 73, 2781–2807. [Google Scholar] [CrossRef]
- Delatte, T.; Umhang, M.; Trevisan, M.; Eicke, S.; Thorneycroft, D.; Smith, S.; Zeeman, S. Evidence for Distinct Mechanisms of Starch Granule Breakdown in Plants. J. Biol. Chem. 2006, 281, 12050–12059. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Jiang, Q.-T.; Zhang, X.-W.; Lan, X.-J.; Pu, Z.-E.; Wei, Y.-M.; Liu, C.; Lu, Z.-X.; Zheng, Y.-L. Structure and expression of barley starch phosphorylase genes. Planta 2013, 238, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.E.; Kosar-Hashemi, B.; Li, Z.; Howitt, C.A.; Larroque, O.; Flanagan, B.; Morell, M.K.; Rahman, S. Characterization of starch phosphorylases in barley grains. J. Sci. Food Agric. 2013, 93, 2137–2145. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Thorneycroft, D.; Schupp, N.; Chapple, A.; Weck, M.; Dunstan, H.; Haldimann, P.; Bechtold, N.; Smith, A.M.; Smith, S. Plastidial α-Glucan Phosphorylase Is Not Required for Starch Degradation in Arabidopsis Leaves but Has a Role in the Tolerance of Abiotic Stress. Plant Physiol. 2004, 135, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Bertoft, E. A Review of Starch Biosynthesis in Relation to the Building Block-Backbone Model. Int. J. Mol. Sci. 2020, 21, 7011. [Google Scholar] [CrossRef]
- Roldán, I.; Wattebled, F.; Lucas, M.M.; Delvallé, D.; Planchot, V.; Jiménez, S.; Pérez, R.; Ball, S.; D’Hulst, C.; Mérida, Á. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 2007, 49, 492–504. [Google Scholar] [CrossRef]
- Leterrier, M.; Holappa, L.D.; Broglie, K.E.; Beckles, D.M. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: Functional and evolutionary implications. BMC Plant Biol. 2008, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Roach, P.J.; DePaoli-Roach, A.A.; Hurley, T.; Tagliabracci, V.S. Glycogen and its metabolism: Some new developments and old themes. Biochem. J. 2012, 441, 763–787. [Google Scholar] [CrossRef]
- Burrell, M.M. Starch: The need for improved quality or quantity—An overview. J. Exp. Bot. 2003, 54, 451–456. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 2014, 66, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Crofts, N.; Nakamura, Y.; Fujita, N. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci. 2017, 262, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Illingworth, B.; Brown, D.H.; Cori, C.F. The de novo synthesis of polysaccharide by phosphorylase. Proc. Natl. Acad. Sci. USA 1961, 47, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y.; Nelson, O.E. Two Additional Phosphorylases in Developing Maize Seeds. Plant Physiol. 1969, 44, 159–167. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grimaud, F.; Rogniaux, H.; James, M.G.; Myers, A.; Planchot, V. Proteome and phosphoproteome analysis of starch granule-associated proteins from normal maize and mutants affected in starch biosynthesis. J. Exp. Bot. 2008, 59, 3395–3406. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Beisel, K.G.; Cameron, S.; Makhmoudova, A.; Liu, F.; Bresolin, N.S.; Wait, R.; Morell, M.K.; Emes, M.J. Analysis of Protein Complexes in Wheat Amyloplasts Reveals Functional Interactions among Starch Biosynthetic Enzymes. Plant Physiol. 2008, 146, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.-K.; Nishi, A.; Satoh, H.; Okita, T. Rice endosperm-specific plastidial α-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Arch. Biochem. Biophys. 2010, 495, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Ozbun, J.L.; Hawker, J.S.; Greenberg, E.; Lammel, C.; Preiss, J.; Lee, E.Y.C. Starch Synthetase, Phosphorylase, ADPglucose Pyrophosphorylase, and UDPglucose Pyrophosphorylase in Developing Maize Kernels. Plant Physiol. 1973, 51, 1–5. [Google Scholar] [CrossRef]
- Dauvillée, D.; Chochois, V.; Steup, M.; Haebel, S.; Eckermann, N.; Ritte, G.; Ral, J.-P.; Colleoni, C.; Hicks, G.; Wattebled, F.; et al. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 2006, 48, 274–285. [Google Scholar] [CrossRef]
- Nakamura, Y.; Ono, M.; Sawada, T.; Crofts, N.; Fujita, N.; Steup, M. Characterization of the functional interactions of plastidial starch phosphorylase and starch branching enzymes from rice endosperm during reserve starch biosynthesis. Plant Sci. 2017, 264, 83–95. [Google Scholar] [CrossRef]
- Rathore, R.; Garg, N.; Garg, S.; Kumar, A. Starch phosphorylase: Role in starch metabolism and biotechnological applications. Crit. Rev. Biotechnol. 2009, 29, 214–224. [Google Scholar] [CrossRef]
- Nakamura, Y.; Ono, M.; Utsumi, C.; Steup, M. Functional Interaction Between Plastidial Starch Phosphorylase and Starch Branching Enzymes from Rice During the Synthesis of Branched Maltodextrins. Plant Cell Physiol. 2012, 53, 869–878. [Google Scholar] [CrossRef]
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J.; Emes, M.J.; Nakamura, Y.; Fujita, N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482. [Google Scholar] [CrossRef] [PubMed]
- Colleoni, C.; Dauvillée, D.; Mouille, G.; Morell, M.; Samuel, M.; Slomiany, M.-C.; Liénard, L.; Wattebled, F.; D’Hulst, C.; Ball, S. Biochemical Characterization of the Chlamydomonas reinhardtii α-1,4 Glucanotransferase Supports a Direct Function in Amylopectin Biosynthesis. Plant Physiol. 1999, 120, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Delatte, T.; Messerli, G.; Umhang, M.; Stettler, M.; Mettler, T.; Streb, S.; Reinhold, H.; Kötting, O. Starch breakdown: Recent discoveries suggest distinct pathways and novel mechanisms. Funct. Plant Biol. 2007, 34, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Orzechowski, S. Starch metabolism in leaves. Acta Biochim. Pol. 2008, 55, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Fettke, J.; Eckermann, N.; Kötting, O.; Ritte, G.; Steup, M. Novel starch-related enzymes and carbohydrates. Cell. Mol. Boil. 2007, 52, 883–904. [Google Scholar]
- Mu, H.H.; Yu, Y.; Wasserman, B.P.; Carman, G.M. Purification and Characterization of the Maize Amyloplast Stromal 112-kDa Starch Phosphorylase. Arch. Biochem. Biophys. 2001, 388, 155–164. [Google Scholar] [CrossRef]
- Mizuno, S.; Kamiyoshihara, Y.; Shiba, H.; Shinmachi, F.; Watanabe, K.; Tateishi, A. Plastidial starch phosphorylase is highly associated with starch accumulation process in developing squash (Cucurbita sp.) fruit. Physiol. Plant. 2018, 167, 264–275. [Google Scholar] [CrossRef]
- Tiessen, A.; Nerlich, A.; Faix, B.; Hümmer, C.; Fox, S.; Trafford, K.; Weber, H.; Weschke, W.; Geigenberger, P. Subcellular analysis of starch metabolism in developing barley seeds using a non-aqueous fractionation method. J. Exp. Bot. 2012, 63, 2071–2087. [Google Scholar] [CrossRef] [PubMed]
- Fettke, J.; Albrecht, T.; Hejazi, M.; Mahlow, S.; Nakamura, Y.; Steup, M. Glucose 1-phosphate is efficiently taken up by potato (Solanum tuberosum) tuber parenchyma cells and converted to reserve starch granules. N. Phytol. 2010, 185, 663–675. [Google Scholar] [CrossRef]
- Malinova, I.; Mahlow, S.; Alseekh, S.; Orawetz, T.; Fernie, A.R.; Baumann, O.; Steup, M.; Fettke, J. Double Knockout Mutants of Arabidopsis Grown under Normal Conditions Reveal that the Plastidial Phosphorylase Isozyme Participates in Transitory Starch Metabolism. Plant Physiol. 2014, 164, 907–921. [Google Scholar] [CrossRef]
- Jiang, J.; Yao, C.; Cao, X.; Liu, Y.; Xue, S. Characterization of starch phosphorylase from the marine green microalga (Chlorophyta) Tetraselmis subcordiformis reveals its potential role in starch biosynthesis. J. Plant Physiol. 2017, 218, 84–93. [Google Scholar] [CrossRef]
- Fettke, J.; Leifels, L.; Brust, H.; Herbst, K.; Steup, M. Two carbon fluxes to reserve starch in potato (Solanum tuberosum L.) tuber cells are closely interconnected but differently modulated by temperature. J. Exp. Bot. 2012, 63, 3011–3029. [Google Scholar] [CrossRef] [PubMed]
- Bustos, R.; Fahy, B.; Hylton, C.M.; Seale, R.; Nebane, N.M.; Edwards, A.; Martin, C.; Smith, A.M. Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers. Proc. Natl. Acad. Sci. USA 2004, 101, 2215–2220. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Wait, R.; Lu, Z.; Akkasaeng, R.; Bowsher, C.G.; Esposito, S.; Kosar-Hashemi, B.; Morell, M.K.; Emes, M.J. Protein Phosphorylation in Amyloplasts Regulates Starch Branching Enzyme Activity and Protein–Protein Interactions. Plant Cell 2004, 16, 694–708. [Google Scholar] [CrossRef]
- Subasinghe, R.M. Role and Regulation of Starch Phosphorylase and Starch Synthase IV in Starch Biosynthesis in Maize Endosperm Amyloplast. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2014. [Google Scholar]
- Young, G.-H.; Chen, H.-M.; Lin, C.-T.; Tseng, K.-C.; Wu, J.-S.; Juang, R.-H. Site-specific phosphorylation of L-form starch phosphorylase by the protein kinase activity from sweet potato roots. Planta 2005, 223, 468–478. [Google Scholar] [CrossRef]
- Mori, H.; Tanizawa, K.; Fukui, T. A chimeric alpha-glucan phosphorylase of plant type L and H isozymes. Functional role of 78-residue insertion in type L isozyme. J. Biol. Chem. 1993, 268, 5574–5581. [Google Scholar] [CrossRef]
- Liddle, A.M.; Manners, D.J.; Wright, A. Studies on carbohydrate-metabolizing enzymes. 6. The action of potato phosphorylase (P-enzyme) on starch-type polysaccharides. Biochem. J. 1961, 80, 304–309. [Google Scholar] [CrossRef]
- Shimomura, S.; Nagai, M.; Fukui, T. Comparative Glucan Specificities of Two Types of Spinach Leaf Phosphorylase. J. Biochem. 1982, 91, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.; Benedito, V.A.; Mayer, K.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nat. Cell Biol. 2011, 480, 520–524. [Google Scholar] [CrossRef] [PubMed]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.L.; Brendel, V. The Maize Genome Sequencing Project. Plant Physiol. 2002, 130, 1594–1597. [Google Scholar] [CrossRef] [PubMed]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Nakano, K.; Mori, H.; Fukui, T. Molecular Cloning of cDNA Encoding Potato Amyloplast a-Glucan Phosphorylase and the Structure of Its Transit Peptide. J. Biochem. 1989, 106, 691–695. [Google Scholar] [CrossRef]
- Camirand, A.; St-Pierre, B.; Marineau, C.; Brisson, N. Occurrence of a copia-like transposable element in one of the introns of the potato starch phosphorylase gene. Mol. Genet. Genom. 1990, 224, 33–39. [Google Scholar] [CrossRef]
- Lin, C.-T.; Yeh, K.-W.; Lee, P.-D.; Su, J.-C. Primary Structure of Sweet Potato Starch Phosphorylase Deduced from its cDNA Sequence. Plant Physiol. 1991, 95, 1250–1253. [Google Scholar] [CrossRef]
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.-K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 5443. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Slugina, M.A.; Shchennikova, A.V.; Kochieva, E. The expression pattern of the Pho1a genes encoding plastidic starch phosphorylase correlates with the degradation of starch during fruit ripening in green-fruited and red-fruited tomato species. Funct. Plant Biol. 2019, 46, 1146. [Google Scholar] [CrossRef]
- Long, T.; Xu, B.; Hu, Y.; Wang, Y.; Mao, C.; Wang, Y.; Zhang, J.; Liu, H.; Huang, H.; Liu, Y.; et al. Genome-wide identification of ZmSnRK2 genes and functional analysis of ZmSnRK2.10 in ABA signaling pathway in maize (Zea mays L.). BMC Plant Biol. 2021, 21, 1–17. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kenichi, H.; Yoshihiro, U.; Masao, I.; Tomoko, K. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Walley, J.W.; Shen, Z.; Sartor, R.; Wu, K.J.; Osborn, J.; Smith, L.G.; Briggs, S.P. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc. Natl. Acad. Sci. USA 2013, 110, E4808–E4817. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Carabaza, A.; Ciudad, C.; Baqué, S.; Guinovart, J.J. Glucose has to be phosphorylated to activate glycogen synthase, but not to inactivate glycogen phosphorylase in hepatocytes. FEBS Lett. 1992, 296, 211–214. [Google Scholar] [CrossRef]
- Johnson, L.N. Glycogen phosphorylase: Control by phosphorylation and allosteric effectors. FASEB J. 1992, 6, 2274–2282. [Google Scholar] [CrossRef]
- Sprang, S.R.; Acharya, K.R.; Goldsmith, E.J.; Stuart, D.I.; Varvill, K.; Fletterick, R.J.; Madsen, N.B.; Johnson, L.N. Structural changes in glycogen phosphorylase induced by phosphorylation. Nat. Cell Biol. 1988, 336, 215–221. [Google Scholar] [CrossRef]
- Ahmed, Z.; Tetlow, I.J.; Ahmed, R.; Morell, M.K.; Emes, M.J. Protein–protein interactions among enzymes of starch biosynthesis in high-amylose barley genotypes reveal differential roles of heteromeric enzyme complexes in the synthesis of A and B granules. Plant Sci. 2015, 233, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Hennen-Bierwagen, T.A.; Lin, Q.; Grimaud, F.; Planchot, V.; Keeling, P.L.; James, M.G.; Myers, A.M. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: A model for regulation of carbon allocation in maize amyloplasts. Plant Physiol. 2009, 149, 1541–1559. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Chang, S.-C.; Juang, R.-H. Plastidial α-glucan phosphorylase 1 complexes with disproportionating enzyme 1 in Ipomoea batatas storage roots for elevating malto-oligosaccharide metabolism. PLoS ONE 2017, 12, e0177115. [Google Scholar] [CrossRef] [PubMed]
- Critchley, J.H.; Zeeman, S.; Takaha, T.; Smith, A.M.; Smith, S. A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 2001, 26, 89–100. [Google Scholar] [CrossRef]
- Lütken, H.; Lloyd, J.; Glaring, M.A.; Baunsgaard, L.; Laursen, K.H.; Haldrup, A.; Kossmann, J.; Blennow, A. Repression of both isoforms of disproportionating enzyme leads to higher malto-oligosaccharide content and reduced growth in potato. Planta 2010, 232, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Kakefuda, G.; Duke, S.H. Characterization of Pea Chloroplast D-Enzyme (4-α-d-Glucanotransferase). Plant Physiol. 1989, 91, 136–143. [Google Scholar] [CrossRef]
- Wattebled, F.; Planchot, V.; Dong, Y.; Szydlowski, N.; Pontoire, B.; Devin, A.; Ball, S.; D’Hulst, C. Further Evidence for the Mandatory Nature of Polysaccharide Debranching for the Aggregation of Semicrystalline Starch and for Overlapping Functions of Debranching Enzymes in Arabidopsis Leaves. Plant Physiol. 2008, 148, 1309–1323. [Google Scholar] [CrossRef]
- Pfister, B.; Sánchez-Ferrer, A.; Diaz, A.; Lu, K.; Otto, C.; Holler, M.; Shaik, F.R.; Meier, F.; Mezzenga, R.; Zeeman, S.C. Recreating the synthesis of starch granules in yeast. eLife 2016, 5, e15552. [Google Scholar] [CrossRef]
- Abe, N.; Asai, H.; Yago, H.; Oitome, N.F.; Itoh, R.; Crofts, N.; Nakamura, Y.; Fujita, N. Relationships between starch synthase I and branching enzyme isozymes determined using double mutant rice lines. BMC Plant Biol. 2014, 14, 80. [Google Scholar] [CrossRef] [PubMed]
- Sparla, F.; Falini, G.; Botticella, E.; Pirone, C.; Talamè, V.; Bovina, R.; Salvi, S.; Tuberosa, R.; Sestili, F.; Trost, P. New Starch Phenotypes Produced by TILLING in Barley. PLoS ONE 2014, 9, e107779. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, A.; Çakir, B.; Hwang, S.-K.; Okita, T.W. The role of the large subunit in redox regulation of the rice endosperm ADP-glucose pyrophosphorylase. FEBS J. 2014, 281, 4951–4963. [Google Scholar] [CrossRef]
- Geigenberger, P. Regulation of Starch Biosynthesis in Response to a Fluctuating Environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef]
- Hendriks, J.H.; Kolbe, A.; Gibon, Y.; Stitt, M.; Geigenberger, P. ADP-Glucose Pyrophosphorylase Is Activated by Posttranslational Redox-Modification in Response to Light and to Sugars in Leaves of Arabidopsis and Other Plant Species. Plant Physiol. 2003, 133, 838–849. [Google Scholar] [CrossRef]
- Fu, Y.; Ballicora, M.; Leykam, J.F.; Preiss, J. Mechanism of Reductive Activation of Potato Tuber ADP-glucose Pyrophosphorylase. J. Biol. Chem. 1998, 273, 25045–25052. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.B.; Yu, H.T.; Yan, L.F.; Wang, T. Integrated Proteomic and Cytological Study of Rice Endosperms at the Storage Phase. J. Proteome Res. 2010, 9, 4906–4918. [Google Scholar] [CrossRef] [PubMed]
- Balmer, Y.; Vensel, W.; Cai, N.; Manieri, W.; Schurmann, P.; Hurkman, W.J.; Buchanan, B.B. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc. Natl. Acad. Sci. USA 2006, 103, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- López-González, C.; Juárez-Colunga, S.; Elias, N.C.M.; Tiessen, A. Exploring regulatory networks in plants: Transcription factors of starch metabolism. PeerJ 2019, 7, e6841. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.-F.; Xue, H.-W. Coexpression Analysis Identifies Rice Starch Regulator1, a Rice AP2/EREBP Family Transcription Factor, as a Novel Rice Starch Biosynthesis Regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Wang, J.-C.; Xu, H.; Zhu, Y.; Liu, Q.-Q.; Cai, X.-L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef]
- Stamova, B.S.; Laudencia-Chingcuanco, D.; Beckles, D.M. Transcriptomic Analysis of Starch Biosynthesis in the Developing Grain of Hexaploid Wheat. Int. J. Plant Genom. 2009, 2009, 1–23. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, M.; Zhou, Y.; Wang, Y.; Shen, J.; Chen, H.; Zhang, L.; Lü, B.; Liang, G.; Liang, J. The Rice G Protein gamma Subunit DEP1/qPE9-1 Positively Regulates Grain-Filling Process by Increasing Auxin and Cytokinin Content in Rice Grains. Rice 2019, 12, 91. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Z.; Zhang, Q.; Meng, S.; Wei, C. The NAC Transcription Factors OsNAC20 and OsNAC26 Regulate Starch and Storage Protein Synthesis. Plant Physiol. 2020, 184, 1775–1791. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Chen, M.; Zhou, W.; Dong, Q.; Jiang, H.; Cheng, B. The DOF-Domain Transcription Factor ZmDOF36 Positively Regulates Starch Synthesis in Transgenic Maize. Front. Plant Sci. 2019, 10, 465. [Google Scholar] [CrossRef]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zhang, S.; Yuan, X.; Chen, C.; Wang, X.-F.; Hao, Y.-J. Genome-wide analysis and identification of stress-responsive genes of the NAM–ATAF1,2–CUC2 transcription factor family in apple. Plant Physiol. Biochem. 2013, 71, 11–21. [Google Scholar] [CrossRef]
- Hu, Y.; Li, Y.; Weng, J.; Liu, H.; Yu, G.; Liu, Y.; Xiao, Q.; Huang, H.; Wang, Y.; Wei, B.; et al. Coordinated regulation of starch synthesis in maize endosperm by microRNAs and DNA methylation. Plant J. 2021, 105, 108–123. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Xu, S.; Tian, X.; Li, T.; Wang, L.; Zhong, Y.; Xue, J.; Guo, D. Comparative transcriptomics reveals the difference in early endosperm development between maize with different amylose contents. PeerJ 2019, 7, e7528. [Google Scholar] [CrossRef]
- Cai, C.; Coleman, S.K.; Niemi, K.; Keinänen, K. Selective binding of synapse-associated protein 97 to GluR-A alpha-amino-5-hydroxy-3-methyl-4-isoxazole propionate receptor subunit is determined by a novel sequence motif. J. Biol. Chem. 2002, 277, 31484–31490. [Google Scholar] [CrossRef]
- Song, Y.; Luo, G.; Shen, L.; Yu, K.; Yang, W.; Li, X.; Sun, J.; Zhan, K.; Cui, D.; Liu, D. TubZIP28, a novel bZIP family transcription factor from Triticum urartu, and TabZIP28, its homologue from Triticum aestivum, enhance starch synthesis in wheat. N. Phytol. 2020, 226, 1384–1398. [Google Scholar] [CrossRef]
- Dong, J.; Zheng, Y.; Fu, Y.; Wang, J.; Yuan, S.; Wang, Y.; Zhu, Q.; Ou, X.; Li, G.; Kang, G. PDIL1-2 can indirectly and negatively regulate expression of the AGPL1 gene in bread wheat. Biol. Res. 2019, 52, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Q.; Wang, Y.; Cheng, B.; Zhao, Y.; Zhu, S. A maize NAC transcription factor, ZmNAC34, negatively regulates starch synthesis in rice. Plant Cell Rep. 2019, 38, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-M.; Chang, S.-C.; Wu, C.-C.; Cuo, T.-S.; Wu, J.-S.; Juang, R.-H. Regulation of the catalytic behaviour of L-form starch phosphorylase from sweet potato roots by proteolysis. Physiol. Plant. 2002, 114, 506–515. [Google Scholar] [CrossRef] [PubMed]


| Crop | Approximated Complex Size (kDa) | Enzymes Involved | References |
|---|---|---|---|
| Maize | 300 | PHO1 P, SBEll, SBEllb P, SSlla P | [19] |
| Rice | 230 | PHO1 P, SBEl, SBEllb P | [50] |
| Barley | Not determined | PHO1 P, SBEl, SBEla, SBEllb P | [90] |
| Wheat | 260 | PHO1 P, SSlla, SBElla P, SBEllb P | [46] |
| Maize ae− | 300 | PHO1 P, SBEl, SBElla P, SSl, SSlla P | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shoaib, N.; Liu, L.; Ali, A.; Mughal, N.; Yu, G.; Huang, Y. Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 10450. https://doi.org/10.3390/ijms221910450
Shoaib N, Liu L, Ali A, Mughal N, Yu G, Huang Y. Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives. International Journal of Molecular Sciences. 2021; 22(19):10450. https://doi.org/10.3390/ijms221910450
Chicago/Turabian StyleShoaib, Noman, Lun Liu, Asif Ali, Nishbah Mughal, Guowu Yu, and Yubi Huang. 2021. "Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives" International Journal of Molecular Sciences 22, no. 19: 10450. https://doi.org/10.3390/ijms221910450
APA StyleShoaib, N., Liu, L., Ali, A., Mughal, N., Yu, G., & Huang, Y. (2021). Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives. International Journal of Molecular Sciences, 22(19), 10450. https://doi.org/10.3390/ijms221910450







