Butyrate Protects Pancreatic Beta Cells from Cytokine-Induced Dysfunction
Abstract
1. Introduction
2. Results
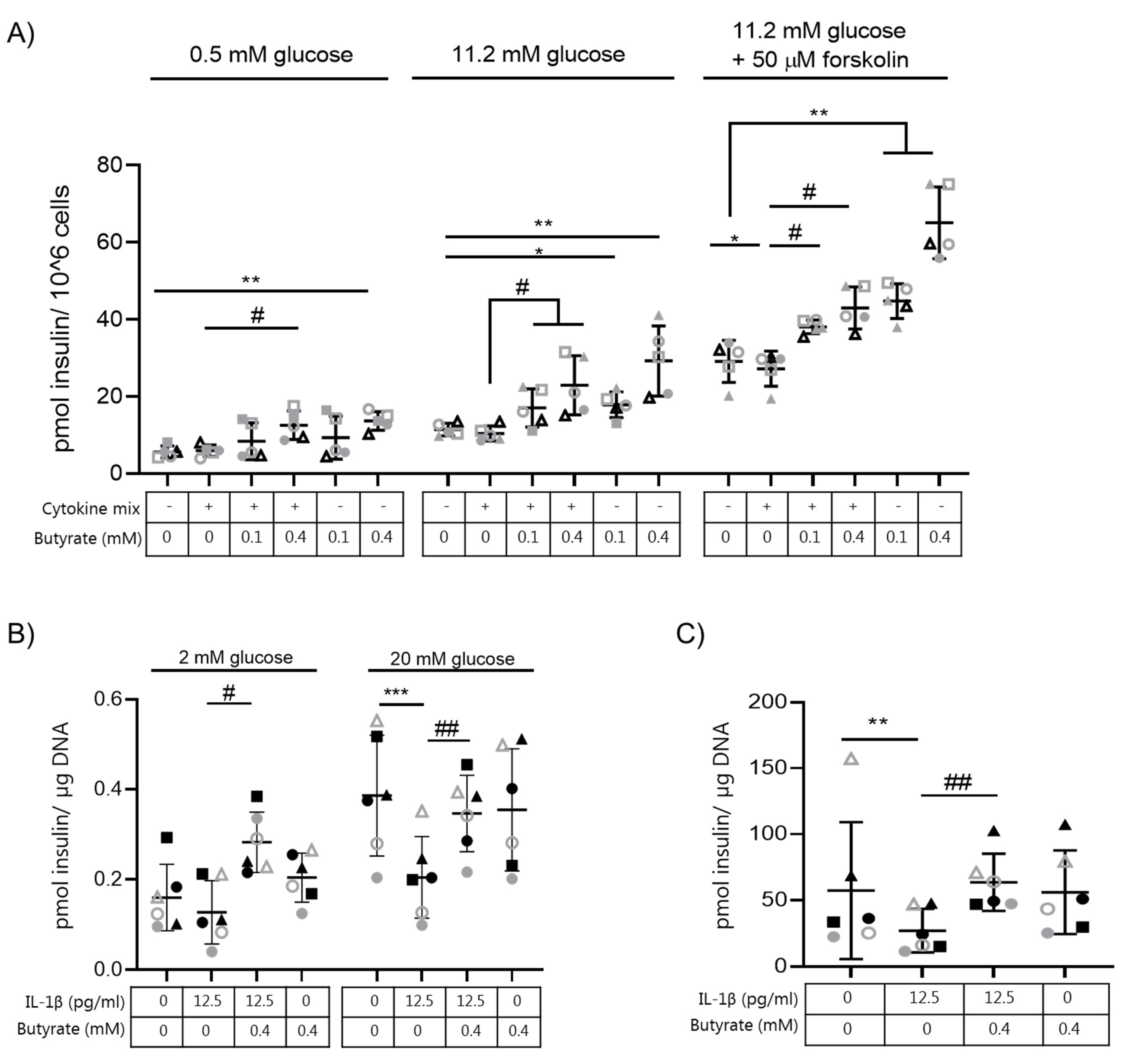
2.1. Butyrate Prevents IL-1β-Induced Impairment of Glucose-Stimulated Insulin Secretion and Insulin Content in Mouse Islets
2.2. Butyrate Prevents Cytokine-Induced Impairment of Glucose-Stimulated Insulin Secretion in Human EndoC-βH1 and Rat INS-1E Beta Cells
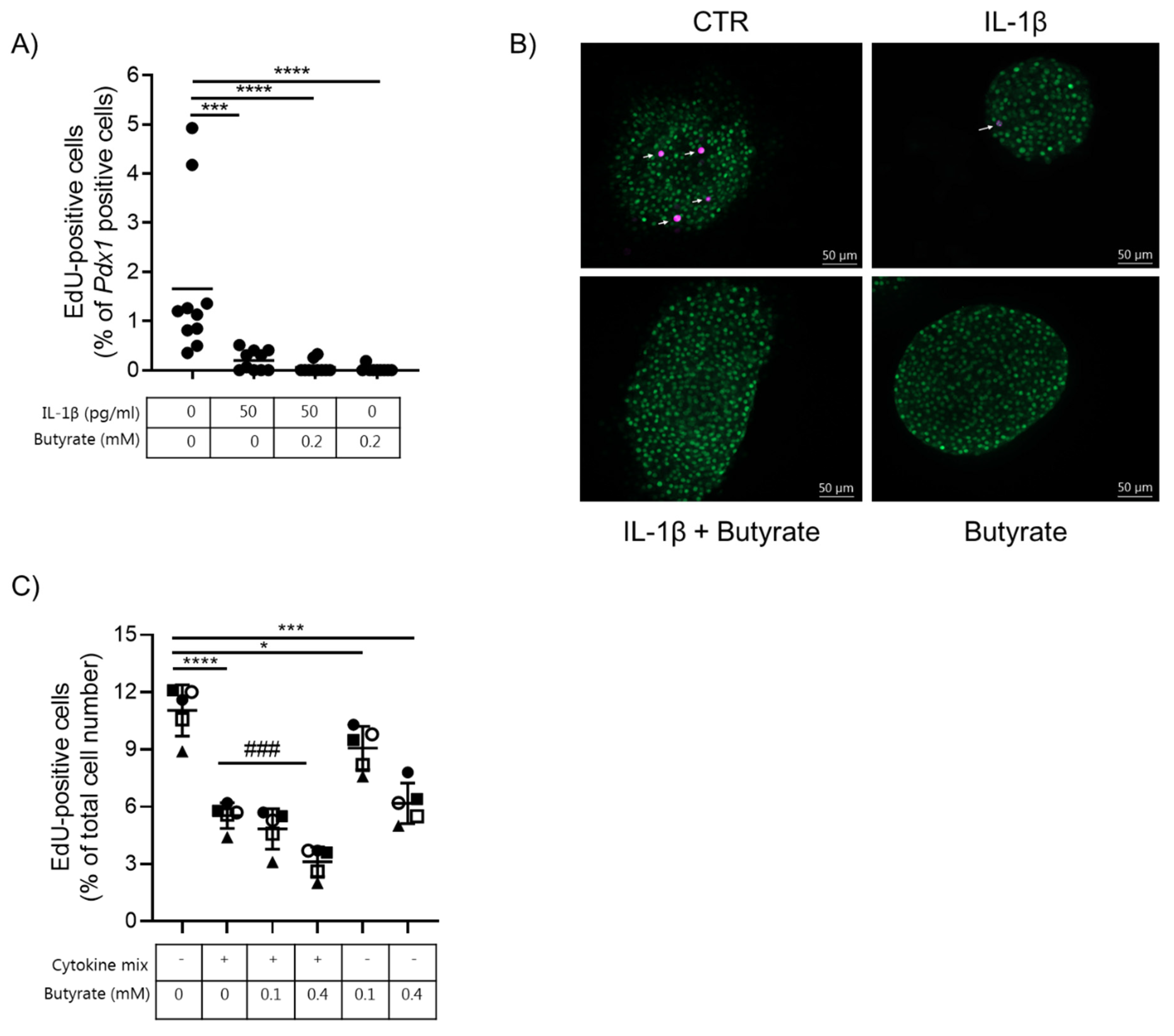
2.3. Butyrate Blocks Beta Cell Proliferation in Mouse Islets and EndoC-βH1 Cells
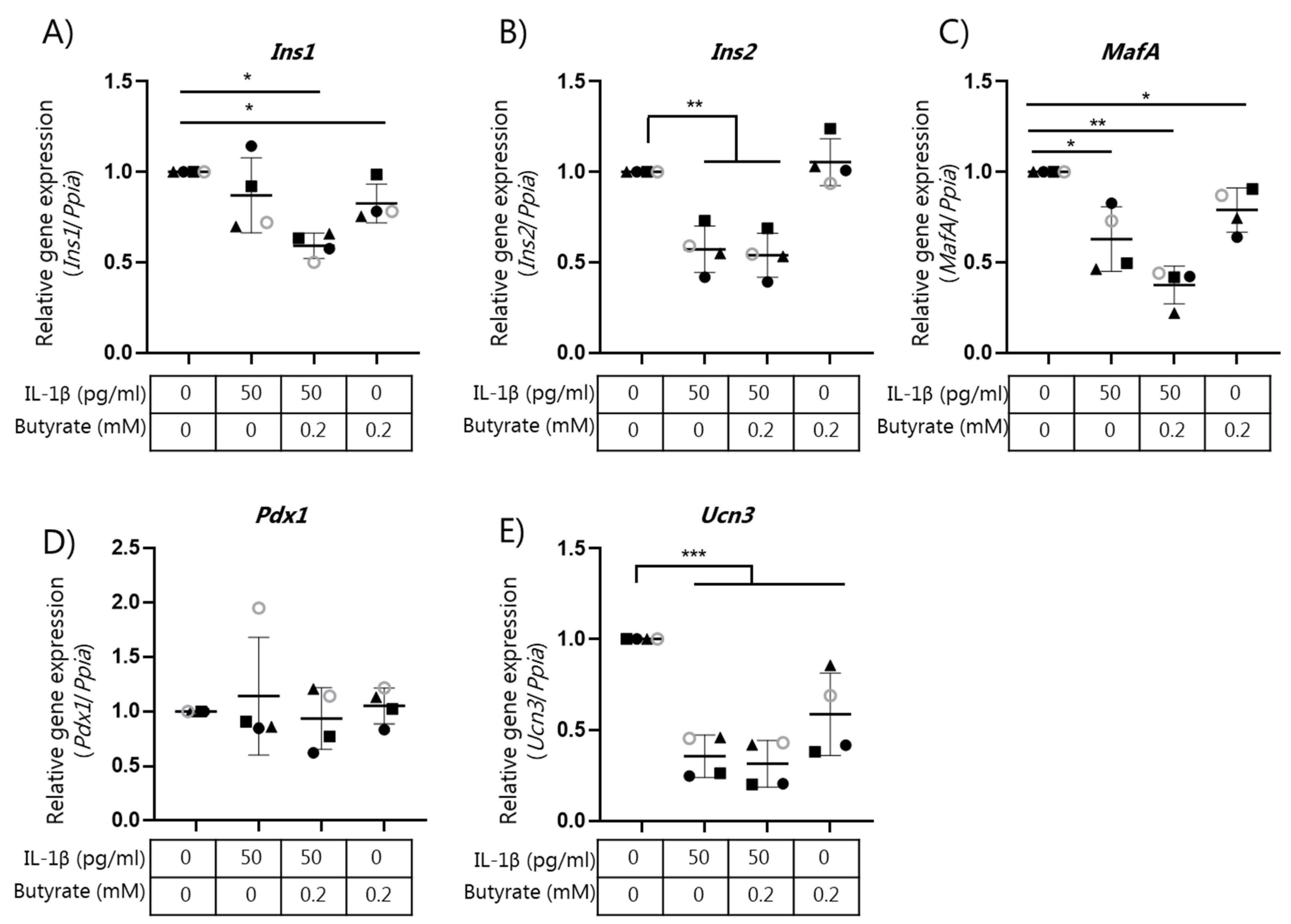
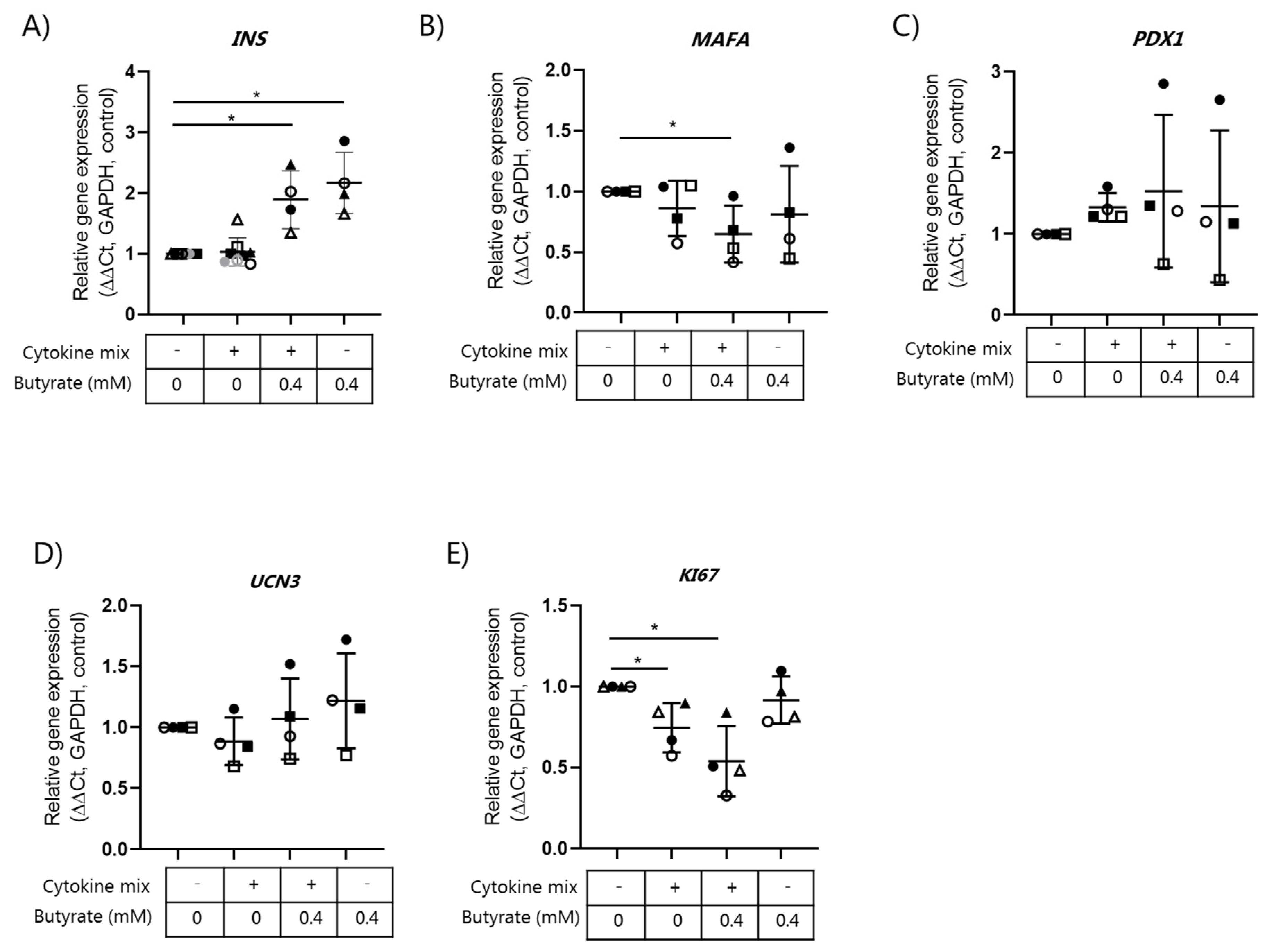
2.4. Butyrate Does Not Affect Beta Cell Mature Gene Expression
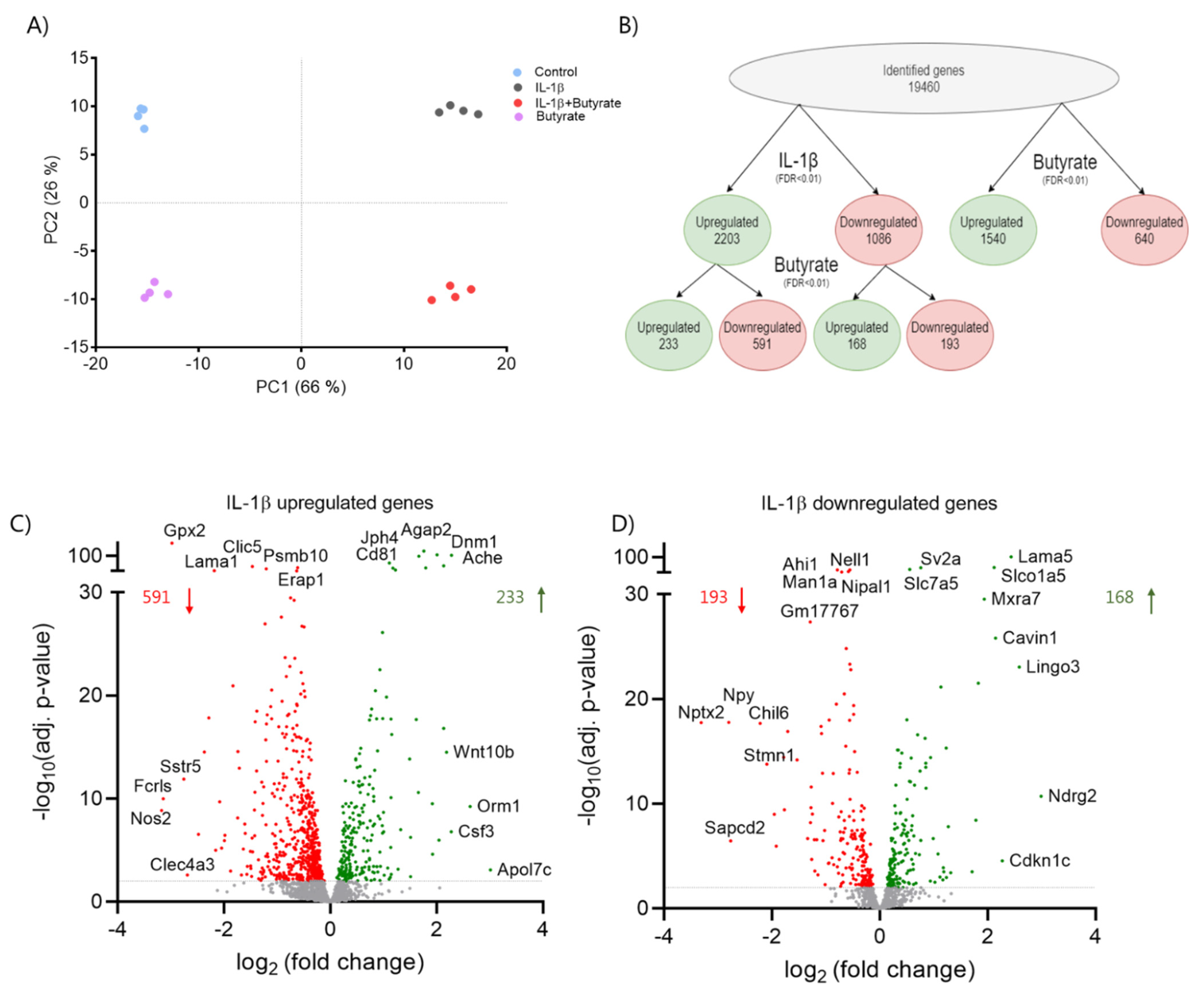
2.5. Butyrate Affects IL-1β Regulated Gene Expression in Mouse Islets
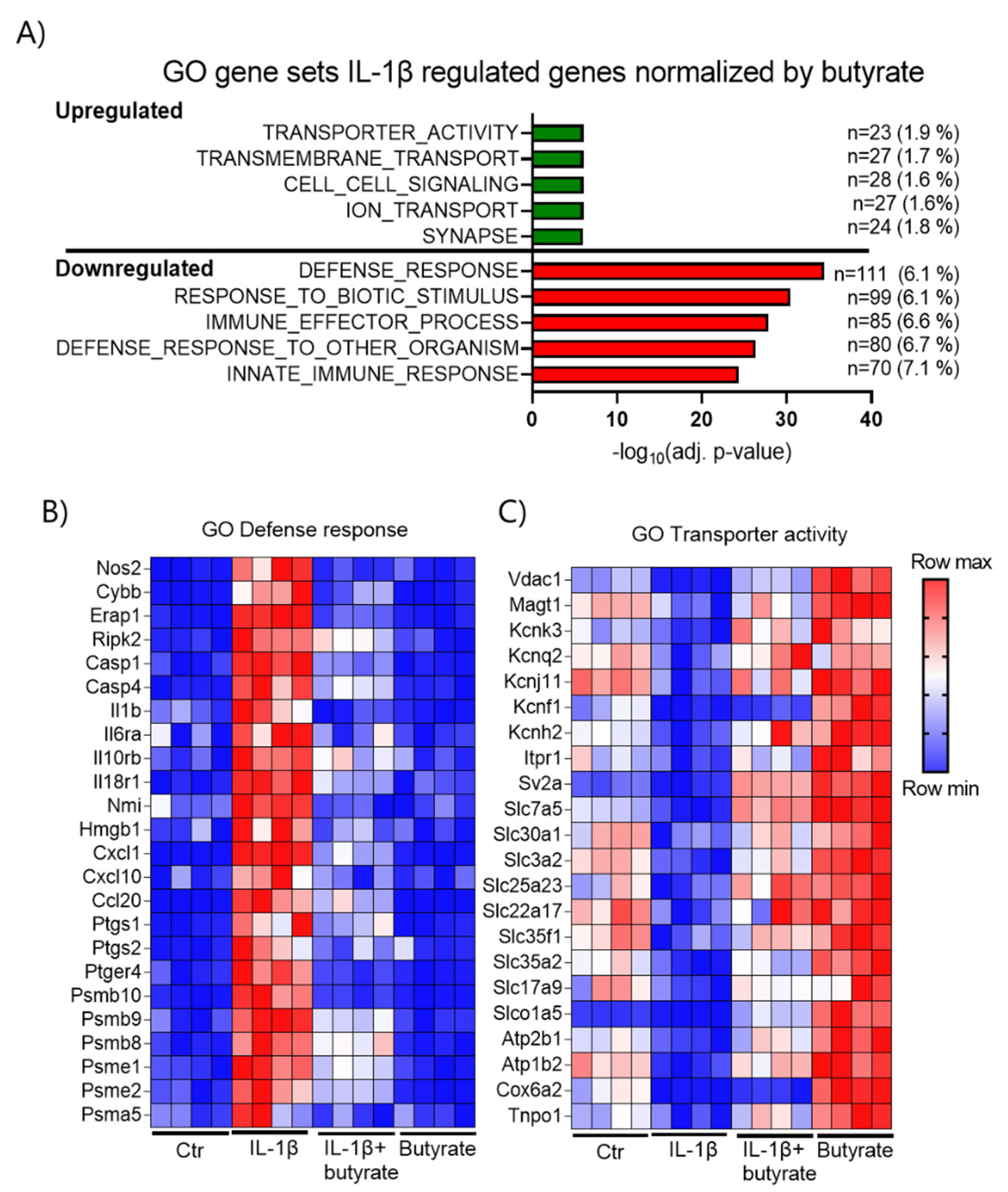
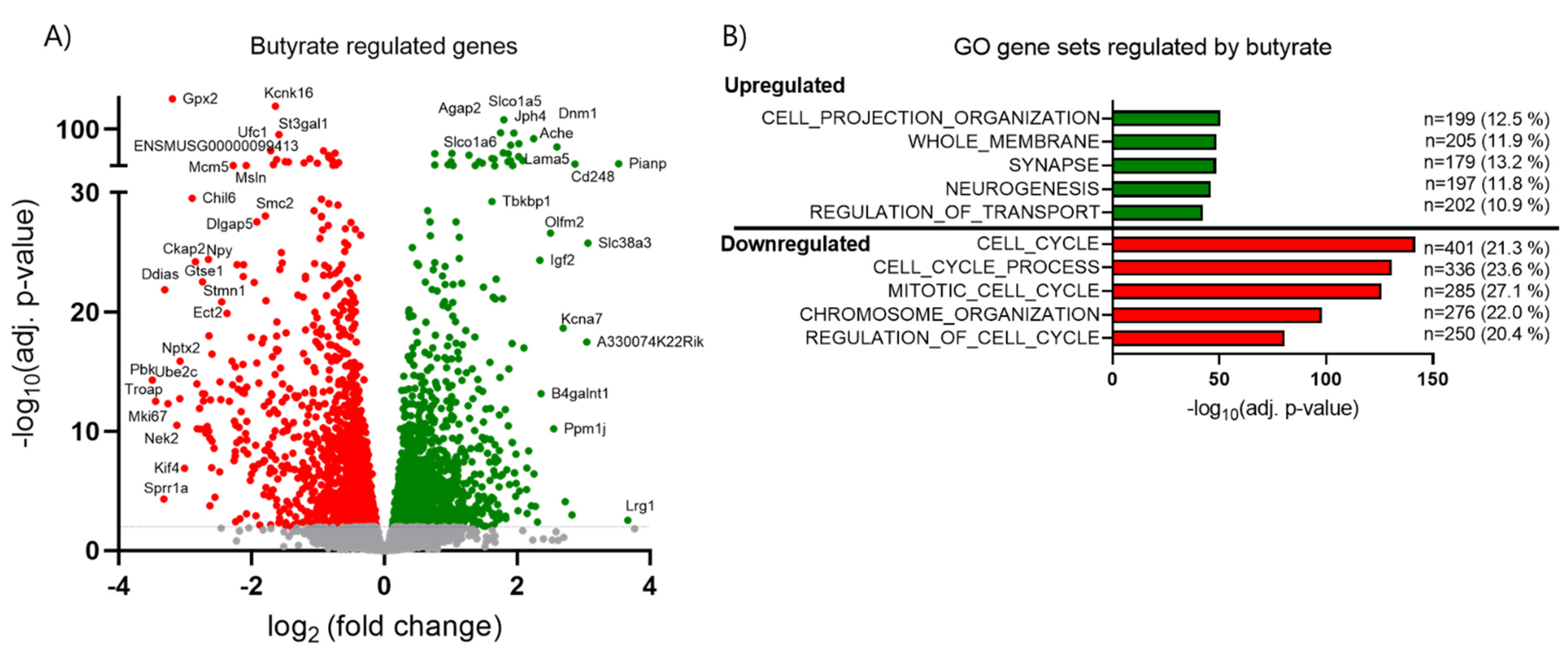
2.6. Butyrate Inhibits Cell Cycle Related Genes and Upregulates Transport-Associated Genes
3. Discussion
4. Materials and Methods
4.1. Mouse Islet Isolation and Beta Cell Line Culture
4.2. Glucose-Stimulated Insulin Release and Insulin Content
4.3. Cell Apoptosis Assay
4.4. Caspase 3/7 Activity
4.5. Proliferation
4.6. Gene Expression Analysis
4.7. RNA Sequencing
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| T2D | Type 2 Diabetes |
| SCFAs | short chain fatty acids |
| HDAC | histone deacetylase |
| GSIS | glucose-stimulated insulin secretion |
| GPCR | G-protein coupled receptors |
| PCA | principal component analysis |
| GO | gene ontology |
References
- Ebrahimi, A.G.; Hollister-Lock, J.; Sullivan, B.A.; Tsuchida, R.; Bonner-Weir, S.; Weir, G.C. Beta cell identity changes with mild hyperglycemia: Implications for function, growth, and vulnerability. Mol. Metab. 2020, 35, 100959. [Google Scholar] [CrossRef] [PubMed]
- Donath, M.Y.; Dalmas, E.; Sauter, N.S.; Böni-Schnetzler, M. Inflammation in Obesity and Diabetes: Islet Dysfunction and Therapeutic Opportunity. Cell Metab. 2013, 17, 860–872. [Google Scholar] [CrossRef]
- Weir, G.C.; Aguayo-Mazzucato, C.; Bonner-Weir, S. β-cell dedifferentiation in diabetes is important, but what is it? Islets 2013, 5, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Cohrs, C.M.; Panzer, J.K.; Drotar, D.M.; Enos, S.J.; Kipke, N.; Chen, C.; Bozsak, R.; Schöniger, E.; Ehehalt, F.; Distler, M.; et al. Dysfunction of Persisting β Cells Is a Key Feature of Early Type 2 Diabetes Pathogenesis. Cell Rep. 2020, 31, 107469. [Google Scholar] [CrossRef]
- Talchai, C.; Xuan, S.; Lin, H.; Sussel, L.; Accili, D. Pancreatic β Cell Dedifferentiation as a Mechanism of Diabetic β Cell Failure. Cell 2012, 150, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, T.M.; Dror, E.; Schulze, F.; Traub, S.; Berishvili, E.; Barbieux, C.; Böni-Schnetzler, M.; Donath, M.Y. The Role of Inflammation in β-cell Dedifferentiation. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Wang, Z.; York, N.W.; Nichols, C.G.; Remedi, M.S. Pancreatic β Cell Dedifferentiation in Diabetes and Redifferentiation following Insulin Therapy. Cell Metab. 2014, 19, 872–882. [Google Scholar] [CrossRef]
- Cinti, F.; Bouchi, R.; Kim-Muller, J.Y.; Ohmura, Y.; Sandoval, P.R.; Masini, M.; Marselli, L.; Suleiman, M.; Ratner, L.E.; Marchetti, P.; et al. Evidence of β-Cell Dedifferentiation in Human Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 1044–1054. [Google Scholar] [CrossRef]
- Dinarello, C.A.; Donath, M.Y.; Mandrup-Poulsen, T. Role of IL-1β in type 2 diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 314–321. [Google Scholar] [CrossRef]
- Eizirik, D.L.; Mandrup-Poulsen, T. A choice of death—The signal-transduction of immune-mediated beta-cell apoptosis. Diabetologia 2001, 44, 2115–2133. [Google Scholar] [CrossRef]
- Berchtold, L.; Prause, M.; Størling, J.; Mandrup-Poulsen, T. Cytokines and Pancreatic β-Cell Apoptosis. Adv. Clin. Chem. 2016, 75, 99–158. [Google Scholar] [CrossRef]
- Urizar, A.I.; Prause, M.; Wortham, M.; Sui, Y.; Thams, P.; Sander, M.; Christensen, G.L.; Billestrup, N. Beta-cell dysfunction induced by non-cytotoxic concentrations of Interleukin-1β is associated with changes in expression of beta-cell maturity genes and associated histone modifications. Mol. Cell. Endocrinol. 2019, 496, 110524. [Google Scholar] [CrossRef] [PubMed]
- McCarville, J.L.; Chen, G.Y.; Cuevas, V.D.; Troha, K.; Ayres, J.S. Microbiota Metabolites in Health and Disease. Annu. Rev. Immunol. 2020, 38, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate Improves Insulin Sensitivity and Increases Energy Expenditure in Mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Khan, S.; Jena, G. Sodium butyrate reduces insulin-resistance, fat accumulation and dyslipidemia in type-2 diabetic rat: A comparative study with metformin. Chem. Interact. 2016, 254, 124–134. [Google Scholar] [CrossRef]
- Li, H.-P.; Chen, X.; Li, M.-Q. Butyrate alleviates metabolic impairments and protects pancreatic β cell function in pregnant mice with obesity. Int. J. Clin. Exp. Pathol. 2013, 6, 1574–1584. [Google Scholar] [PubMed]
- Xu, Y.-H.; Gao, C.-L.; Guo, H.-L.; Zhang, W.-Q.; Huang, W.; Tang, S.-S.; Gan, W.-J.; Xu, Y.; Zhou, H.; Zhu, Q. Sodium butyrate supplementation ameliorates diabetic inflammation in db/db mice. J. Endocrinol. 2018, 238, 231–244. [Google Scholar] [CrossRef]
- Hu, S.; Kuwabara, R.; De Haan, B.J.; Smink, A.M.; De Vos, P. Acetate and Butyrate Improve β-cell Metabolism and Mitochondrial Respiration under Oxidative Stress. Int. J. Mol. Sci. 2020, 21, 1542. [Google Scholar] [CrossRef] [PubMed]
- Lorza-Gil, E.; Kaiser, G.; Ulven, E.R.; König, G.M.; Gerst, F.; Oquendo, M.B.; Birkenfeld, A.L.; Häring, H.-U.; Kostenis, E.; Ulven, T.; et al. FFA2-, but not FFA3-agonists inhibit GSIS of human pseudoislets: A comparative study with mouse islets and rat INS-1E cells. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- McNelis, J.C.; Lee, Y.S.; Mayoral, R.; van der Kant, R.; Johnson, A.M.F.; Wollam, J.; Olefsky, J.M. GPR43 Potentiates β-Cell Function in Obesity. Diabetes 2015, 64, 3203–3217. [Google Scholar] [CrossRef]
- Montague, W.; Taylor, K.W. Regulation of Insulin Secretion by Short Chain Fatty Acids. Nat. Cell Biol. 1968, 217, 853. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, A.; Gonzalez-Abuin, N.; Ruz-Maldonado, I.; Huang, G.C.; Frost, G.; Persaud, S.J. Short chain fatty acids stimulate insulin secretion and reduce apoptosis in mouse and human islets in vitro: Role of free fatty acid receptor 2. Diabetes Obes. Metab. 2019, 21, 330–339. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Sanna, S.; Van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vila, A.V.; Võsa, U.; Mujagic, Z.; Masclee, A.A.M.; Jonkers, D.M.A.E.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, J.; Yuan, Y.; Chen, J.; Cheng, S.; Wang, H.; Xu, Y. Sodium butyrate mitigates type 2 diabetes by inhibiting PERK-CHOP pathway of endoplasmic reticulum stress. Environ. Toxicol. Pharmacol. 2018, 64, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Jena, G. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: Study in juvenile diabetic rat. Chem. Interact. 2014, 213, 1–12. [Google Scholar] [CrossRef]
- Mariño, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.-A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552–562. [Google Scholar] [CrossRef]
- Matheus, V.; Monteiro, L.; Oliveira, R.; Maschio, D.; Collares-Buzato, C. Butyrate reduces high-fat diet-induced metabolic alterations, hepatic steatosis and pancreatic beta cell and intestinal barrier dysfunctions in prediabetic mice. Exp. Biol. Med. 2017, 242, 1214–1226. [Google Scholar] [CrossRef]
- Bruun, C.; Christensen, G.L.; Jacobsen, M.L.B.; Kanstrup, M.B.; Jensen, P.R.; Fjordvang, H.; Mandrup-Poulsen, T.; Billestrup, N. Inhibition of beta cell growth and function by bone morphogenetic proteins. Diabetologia 2014, 57, 2546–2554. [Google Scholar] [CrossRef]
- Ravassard, P.; Hazhouz, Y.; Pechberty, S.; Bricout-Neveu, E.; Armanet, M.; Czernichow, P.; Scharfmann, R. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Investig. 2011, 121, 3589–3597. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 September 2021).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef]
- Caillon, L.; Hoffmann, A.R.F.; Botz, A.; Khemtemourian, L. Molecular Structure, Membrane Interactions, and Toxicity of the Islet Amyloid Polypeptide in Type 2 Diabetes Mellitus. J. Diabetes Res. 2016, 2016, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Blum, B.; Roose, A.N.; Barrandon, O.; Maehr, R.; Arvanites, A.C.; Davidow, L.S.; Davis, J.C.; Peterson, Q.; Rubin, L.; Melton, D.A. Reversal of β cell de-differentiation by a small molecule inhibitor of the TGFβ pathway. eLife 2014, 3, e02809. [Google Scholar] [CrossRef]
- Davie, J. Inhibition of Histone Deacetylase Activity by Butyrate. J. Nutr. 2003, 133, 2485S–2493S. [Google Scholar] [CrossRef]
- Remsberg, J.R.; Ediger, B.N.; Ho, W.Y.; Damle, M.; Li, Z.; Teng, C.; Lanzillotta, C.; Stoffers, D.A.; Lazar, M.A. Deletion of histone deacetylase 3 in adult beta cells improves glucose tolerance via increased insulin secretion. Mol. Metab. 2017, 6, 30–37. [Google Scholar] [CrossRef]
- Lundh, M.; Galbo, T.; Poulsen, S.S.; Mandrup-Poulsen, T. Histone deacetylase 3 inhibition improves glycaemia and insulin secretion in obese diabetic rats. Diabetes Obes. Metab. 2015, 17, 703–707. [Google Scholar] [CrossRef]
- Larsen, L.; Tonnesen, M.; Ronn, S.G.; Størling, J.; Jørgensen, S.; Mascagni, P.; Dinarello, C.A.; Billestrup, N.; Mandrup-Poulsen, T. Inhibition of histone deacetylases prevents cytokine-induced toxicity in beta cells. Diabetologia 2007, 50, 779–789. [Google Scholar] [CrossRef]
- Susick, L.; Senanayake, T.; Veluthakal, R.; Woster, P.; Kowluru, A. A novel histone deacetylase inhibitor prevents IL-1β induced metabolic dysfunction in pancreatic β-cells. J. Cell. Mol. Med. 2009, 13, 1877–1885. [Google Scholar] [CrossRef] [PubMed]
- Lũhrs, H.; Gerke, T.; Boxberger, F.; Backhaus, K.; Melcher, R.; Scheppach, W.; Menzel, T. Butyrate inhibits interleukin-1-mediated nuclear factor-kappa B activation in human epithelial cells. Dig. Dis. Sci. 2001, 46, 1968–1973. [Google Scholar] [CrossRef] [PubMed]
- Segain, J.P.; Raingeard de la Blétière, D.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottière, H.M.; Galmiche, J.P. Butyrate inhibits inflammatory responses through NFkappaB inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. Available online: http://www.ncbi.nlm.nih.gov/pubmed/10940278%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1728045 (accessed on 24 September 2021). [CrossRef] [PubMed]
- Park, J.-S.; Lee, E.-J.; Lee, J.-C.; Kim, W.-K.; Kim, H.-S. Anti-inflammatory effects of short chain fatty acids in IFN-γ-stimulated RAW 264.7 murine macrophage cells: Involvement of NF-κB and ERK signaling pathways. Int. Immunopharmacol. 2007, 7, 70–77. [Google Scholar] [CrossRef]
- Knudsen, K.E.B.; Lærke, H.N.; Hedemann, M.S.; Nielsen, T.S.; Ingerslev, A.K.; Nielsen, D.S.G.; Theil, P.K.; Purup, S.; Hald, S.; Schioldan, A.G.; et al. Impact of Diet-Modulated Butyrate Production on Intestinal Barrier Function and Inflammation. Nutrients 2018, 10, 1499. [Google Scholar] [CrossRef]
- Collier, J.J.; Sparer, T.E.; Karlstad, M.D.; Burke, S.J.; Biomedical, P.; Biomedical, P.; Rouge, B. Pancreatic Islet Inflammation: An Emerging Role for Chemokines. J. Mol. Endocrinol. 2018, 59, 1–23. [Google Scholar] [CrossRef]
- Kutlu, B.; Cardozo, A.K.; Darville, M.I.; Kruhøffer, M.; Magnusson, N.; Ørntoft, T.; Eizirik, D.L. Discovery of gene networks regulating cytokine-induced dysfunction and apoptosis in insulin-producing INS-1 cells. Diabetes 2003, 52, 2701–2719. [Google Scholar] [CrossRef]
- Muhammed, S.J.; Lundquist, I.; Salehi, A. Pancreatic β-cell dysfunction, expression of iNOS and the effect of phosphodiesterase inhibitors in human pancreatic islets of type 2 diabetes. Diabetes Obes. Metab. 2012, 14, 1010–1019. [Google Scholar] [CrossRef]
- Tran, P.O.T.; Gleason, C.E.; Poitout, V.; Robertson, R.P. Prostaglandin E2 Mediates Inhibition of Insulin Secretion by Interleukin-1β. J. Biol. Chem. 1999, 274, 31245–31248. [Google Scholar] [CrossRef][Green Version]
- Li, N.; Li, B.; Brun, T.; Deffert-Delbouille, C.; Mahiout, Z.; Daali, Y.; Ma, X.-J.; Krause, K.-H.; Maechler, P. NADPH Oxidase NOX2 Defines a New Antagonistic Role for Reactive Oxygen Species and cAMP/PKA in the Regulation of Insulin Secretion. Diabetes 2012, 61, 2842–2850. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Chae, C.W.; Lee, H.J.; Jung, Y.H.; Choi, G.E.; Kim, J.S.; Lim, J.R.; Lee, J.E.; Cho, J.H.; Park, H.; et al. Sodium butyrate inhibits high cholesterol-induced neuronal amyloidogenesis by modulating NRF2 stabilization-mediated ROS levels: Involvement of NOX2 and SOD1. Cell Death Dis. 2020, 11, 1–19. [Google Scholar] [CrossRef]
- Mathew, O.P.; Ranganna, K.; Milton, S.G. Involvement of the Antioxidant Effect and Anti-inflammatory Response in Butyrate-Inhibited Vascular Smooth Muscle Cell Proliferation. Pharmaceuticals 2014, 7, 1008–1027. [Google Scholar] [CrossRef] [PubMed]
- Abderrahmani, A. Rest/Nrsf Target Genes in Neuronal and Beta Cells: Pathophysiological and Therapeutic Perspectives for Diabetes and Neurodegenerative Disorders. Eur. Med. J. Diabetes 2015, 3, 87–95. [Google Scholar]
- Abderrahmani, A.; Niederhauser, G.; Plaisance, V.; Haefliger, J.-A.; Regazzi, R.; Waeber, G. Neuronal traits are required for glucose-induced insulin secretion. FEBS Lett. 2004, 565, 133–138. [Google Scholar] [CrossRef]
- Gauthier, B.R.; Wollheim, C.B. Synaptotagmins bind calcium to release insulin. Am. J. Physiol. Metab. 2008, 295, E1279–E1286. [Google Scholar] [CrossRef]
- Speidel, D.; Salehi, A.; Obermueller, S.; Lundquist, I.; Brose, N.; Renström, E.; Rorsman, P. CAPS1 and CAPS2 Regulate Stability and Recruitment of Insulin Granules in Mouse Pancreatic β Cells. Cell Metab. 2008, 7, 57–67. [Google Scholar] [CrossRef]
- Hellsten, S.V.; Hägglund, M.G.; Eriksson, M.M.; Fredriksson, R. The neuronal and astrocytic protein SLC38A10 transports glutamine, glutamate, and aspartate, suggesting a role in neurotransmission. FEBS Open Bio 2017, 7, 730–746. [Google Scholar] [CrossRef]
- Gammelsaeter, R.; Jenstad, M.; Bredahl, M.; Gundersen, V.; Chaudhry, F. Complementary expression of SN1 and SAT2 in the islets of Langerhans suggests concerted action of glutamine transport in the regulation of insulin secretion. Biochem. Biophys. Res. Commun. 2009, 381, 378–382. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Wang, Y.; Liu, X.; Zhou, L.; Zhang, C.; Shao, L. Histone deacetylase inhibition by MS-275 potentiates glucose-stimulated insulin secretion without affecting glucose oxidation. Life Sci. 2020, 257, 118073. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, K.-N.; Vitetta, L. Effects of Intestinal Microbial–Elaborated Butyrate on Oncogenic Signaling Pathways. Nutrients 2019, 11, 1026. [Google Scholar] [CrossRef]
- Philippe, J.; Drucker, D.J.; Chick, W.L.; Habener, J.F. Transcriptional regulation of genes encoding insulin, glucagon, and angiotensinogen by sodium butyrate in a rat islet cell line. Mol. Cell. Biol. 1987, 7, 560–563. [Google Scholar] [CrossRef]
- Dirice, E.; Ng, R.; Martinez, R.; Hu, J.; Wagner, F.F.; Holson, E.B.; Wagner, B.K.; Kulkarni, R.N. Isoform-selective inhibitor of histone deacetylase 3 (HDAC3) limits pancreatic islet infiltration and protects female nonobese diabetic mice from diabetes. J. Biol. Chem. 2017, 292, 17598–17608. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Puri, S.; Roy, N.; Russ, H.A.; Leonhardt, L.; French, E.K.; Roy, R.; Bengtsson, H.; Scott, D.K.; Stewart, A.F.; Hebrok, M. Replication confers β cell immaturity. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Ahmed, K.; Gille, A.; Lu, S.; Gröne, H.-J.; Tunaru, S.; Offermanns, S. Loss of FFA2 and FFA3 increases insulin secretion and improves glucose tolerance in type 2 diabetes. Nat. Med. 2015, 21, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-L.; Segovia, I.; Yuan, X.-L.; Gao, Z.-H. Controversial Roles of Gut Microbiota-Derived Short-Chain Fatty Acids (SCFAs) on Pancreatic β-Cell Growth and Insulin Secretion. Int. J. Mol. Sci. 2020, 21, 910. [Google Scholar] [CrossRef] [PubMed]
- De Ruijter, A.J.; Van Gennip, A.H.; Caron, H.N.; Kemp, S.; Van Kuilenburg, A.B. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prause, M.; Pedersen, S.S.; Tsonkova, V.; Qiao, M.; Billestrup, N. Butyrate Protects Pancreatic Beta Cells from Cytokine-Induced Dysfunction. Int. J. Mol. Sci. 2021, 22, 10427. https://doi.org/10.3390/ijms221910427
Prause M, Pedersen SS, Tsonkova V, Qiao M, Billestrup N. Butyrate Protects Pancreatic Beta Cells from Cytokine-Induced Dysfunction. International Journal of Molecular Sciences. 2021; 22(19):10427. https://doi.org/10.3390/ijms221910427
Chicago/Turabian StylePrause, Michala, Signe Schultz Pedersen, Violeta Tsonkova, Min Qiao, and Nils Billestrup. 2021. "Butyrate Protects Pancreatic Beta Cells from Cytokine-Induced Dysfunction" International Journal of Molecular Sciences 22, no. 19: 10427. https://doi.org/10.3390/ijms221910427
APA StylePrause, M., Pedersen, S. S., Tsonkova, V., Qiao, M., & Billestrup, N. (2021). Butyrate Protects Pancreatic Beta Cells from Cytokine-Induced Dysfunction. International Journal of Molecular Sciences, 22(19), 10427. https://doi.org/10.3390/ijms221910427






