Dynamic Changes of Brain Cilia Transcriptomes across the Human Lifespan
Abstract
1. Introduction
2. Results
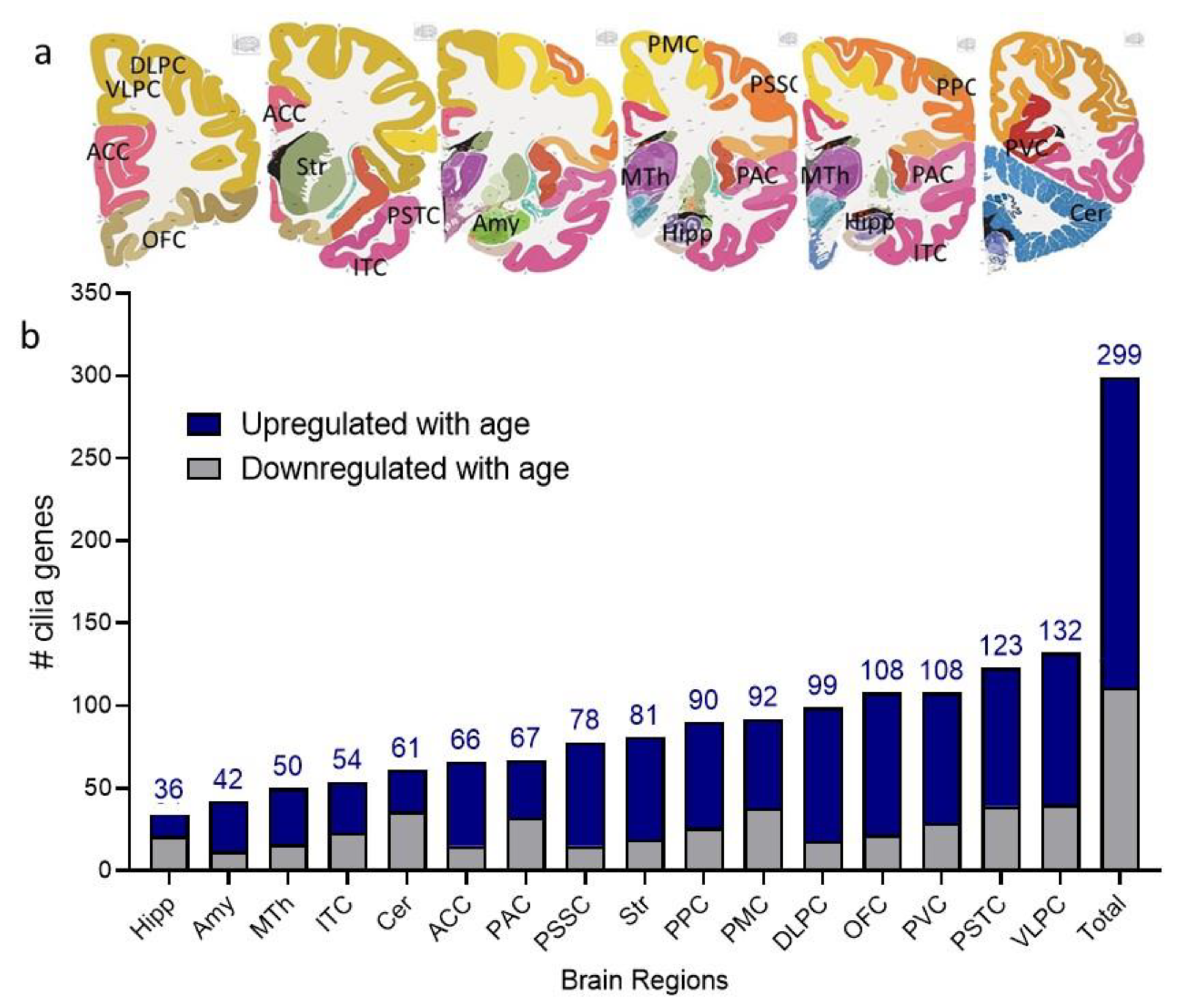
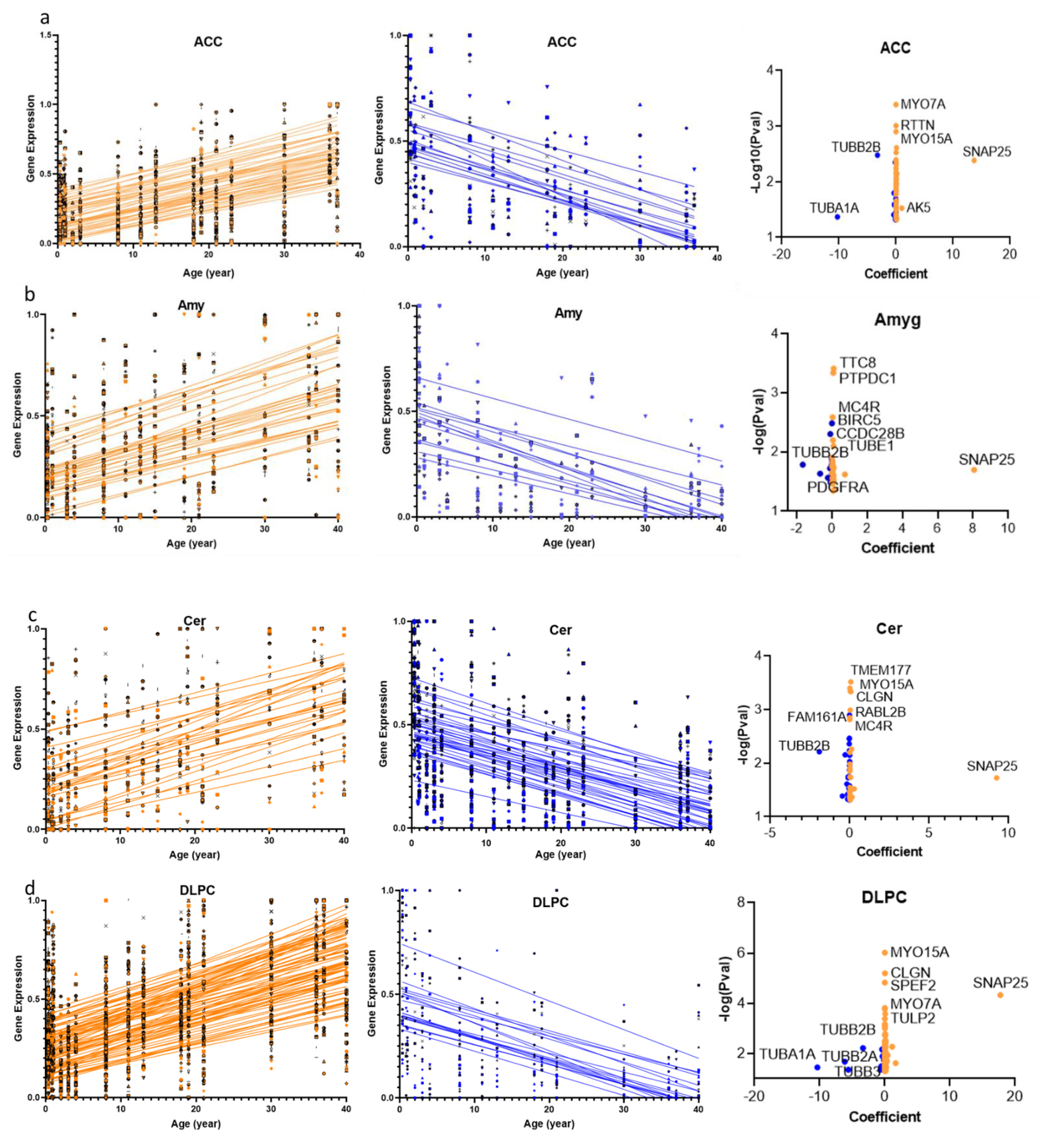
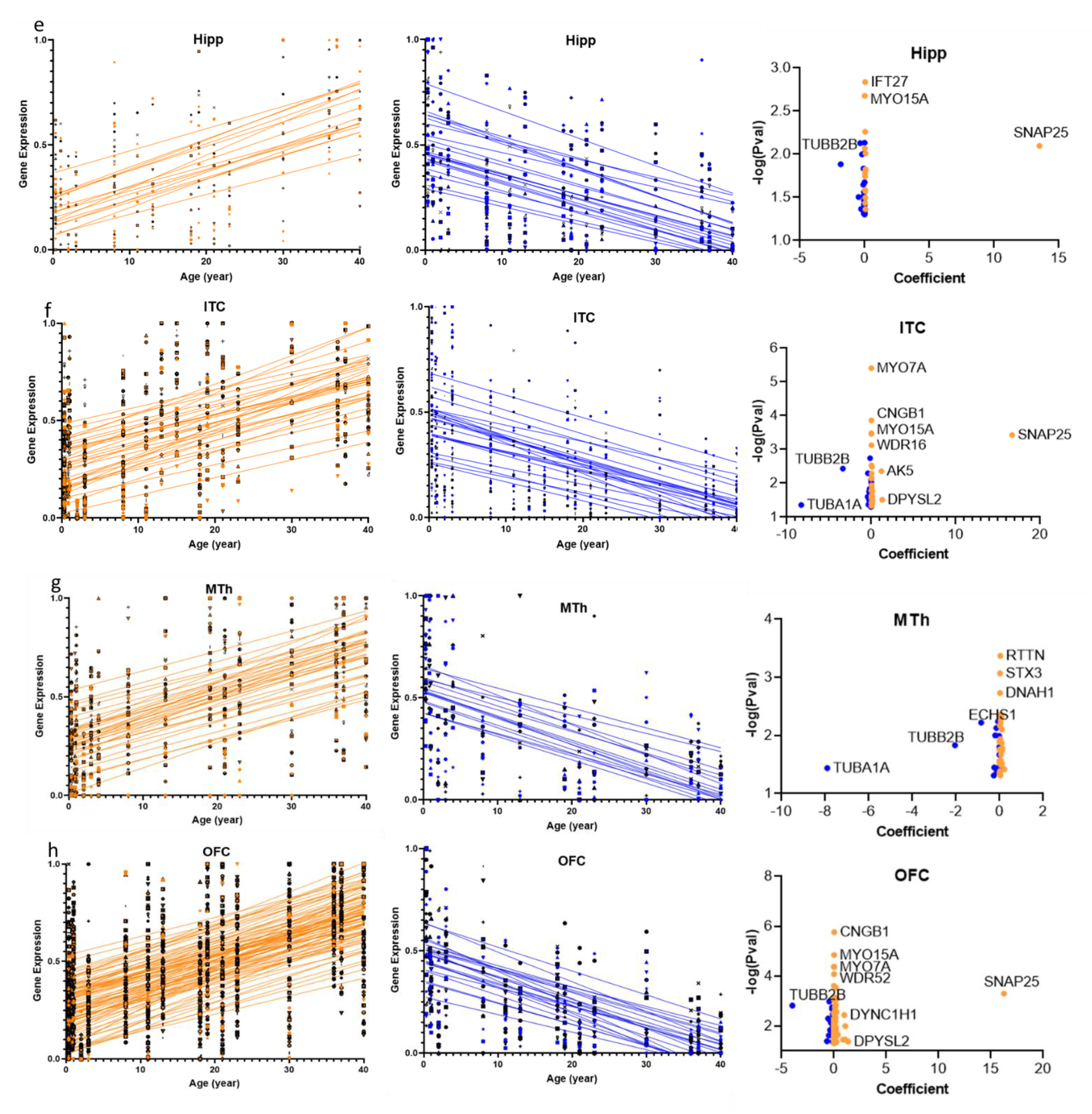
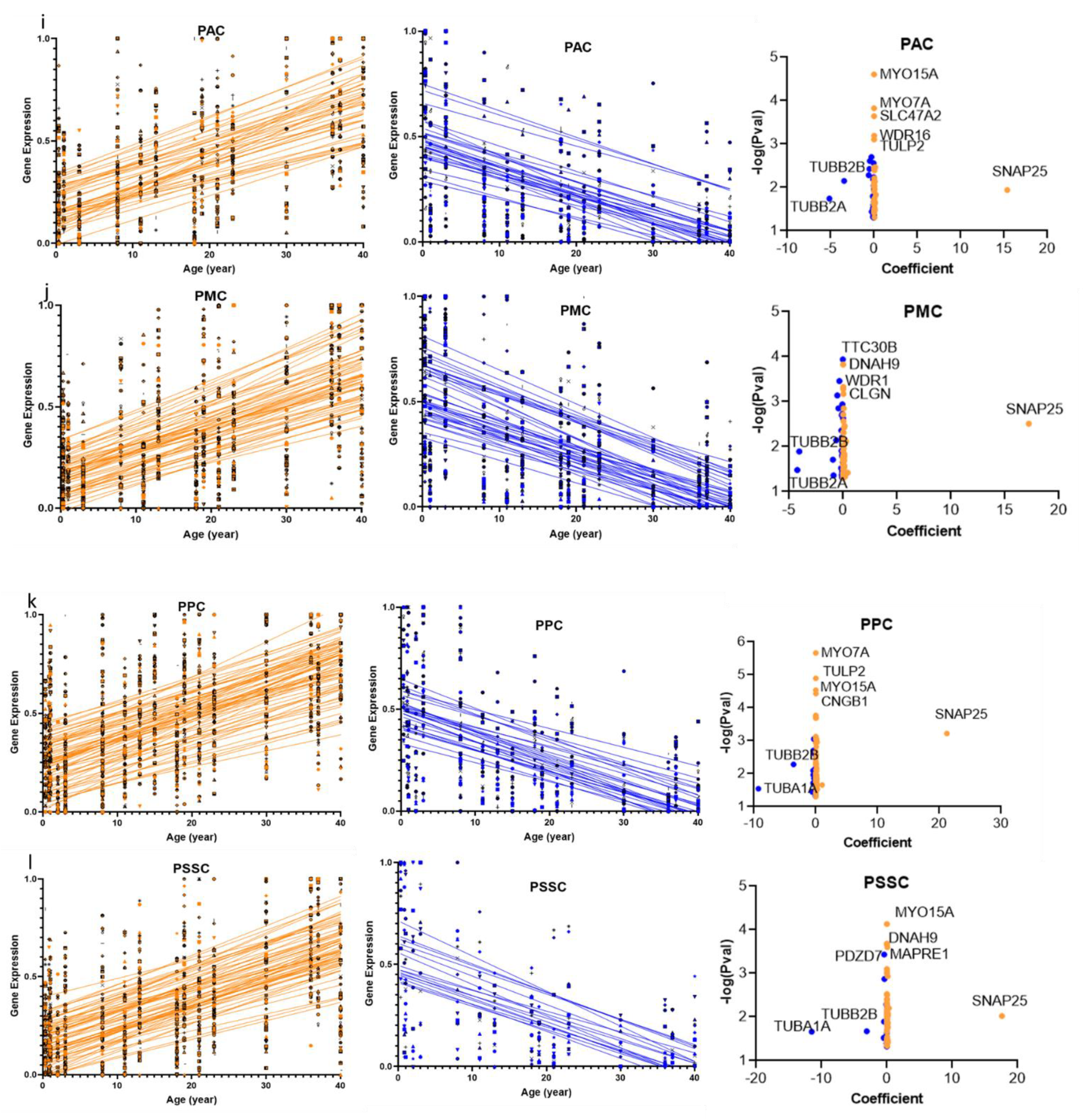
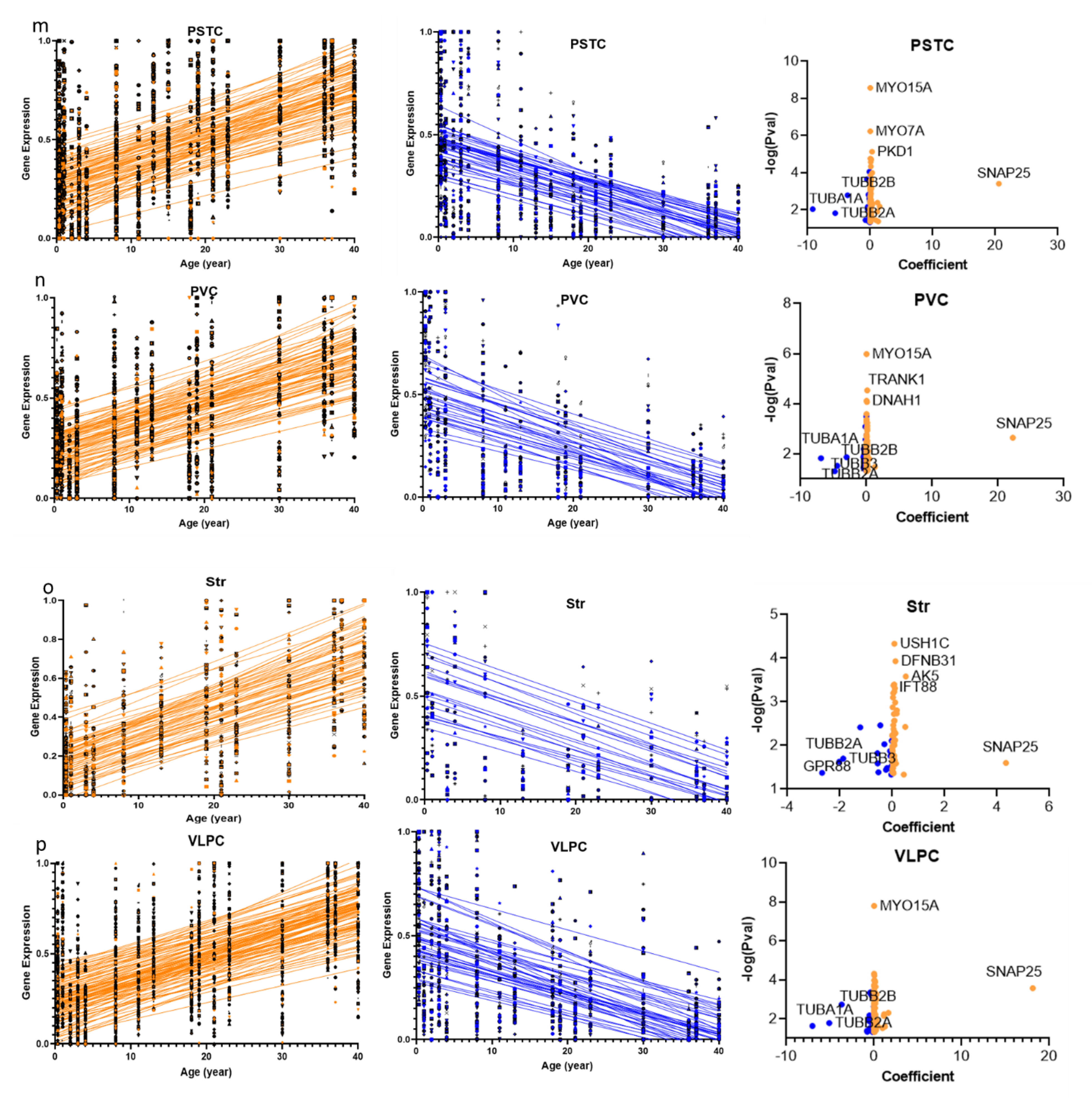
2.1. Cilia Transcripts Exhibit High Age-Dependent Expression
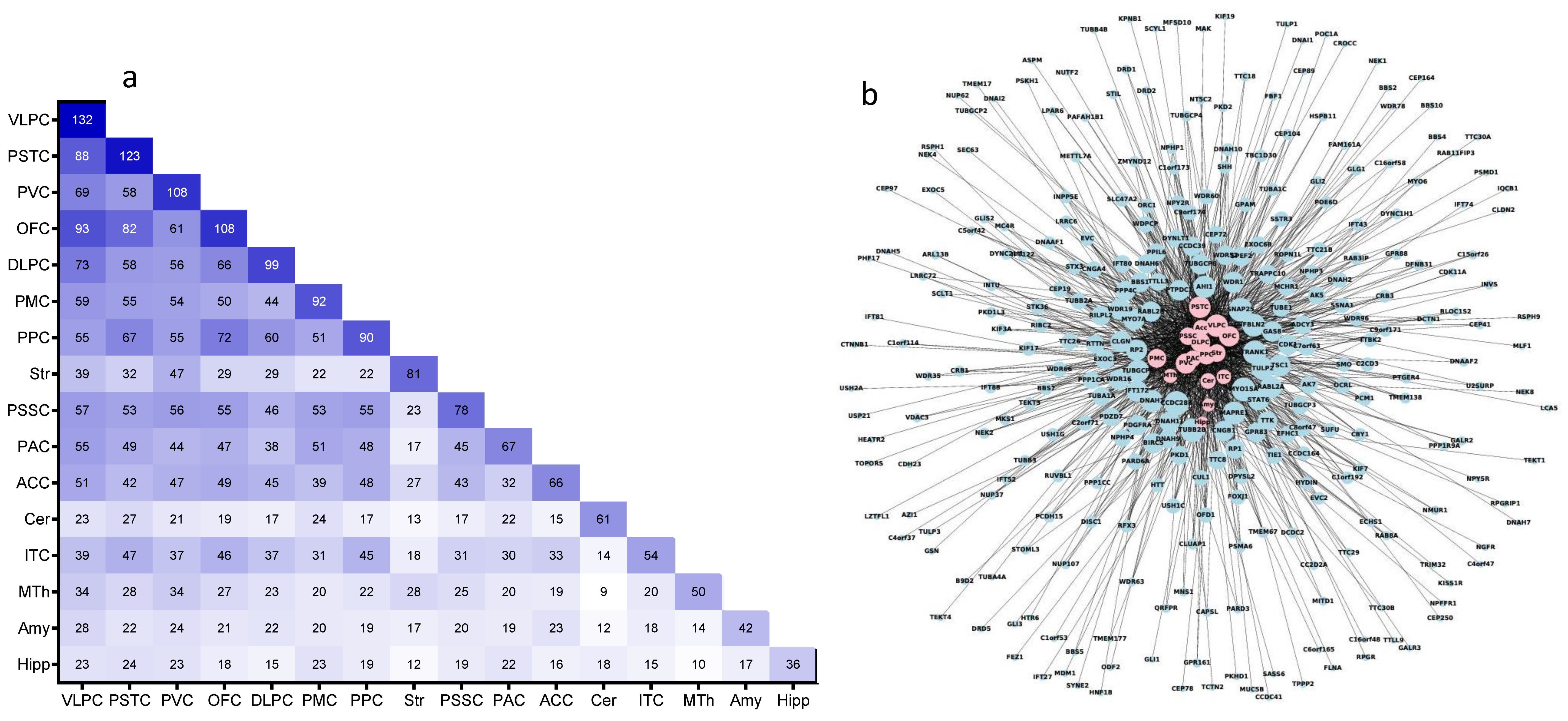
2.2. Overlap of the Expressions of Age-Regulated Cilia Transcripts across Brain Regions
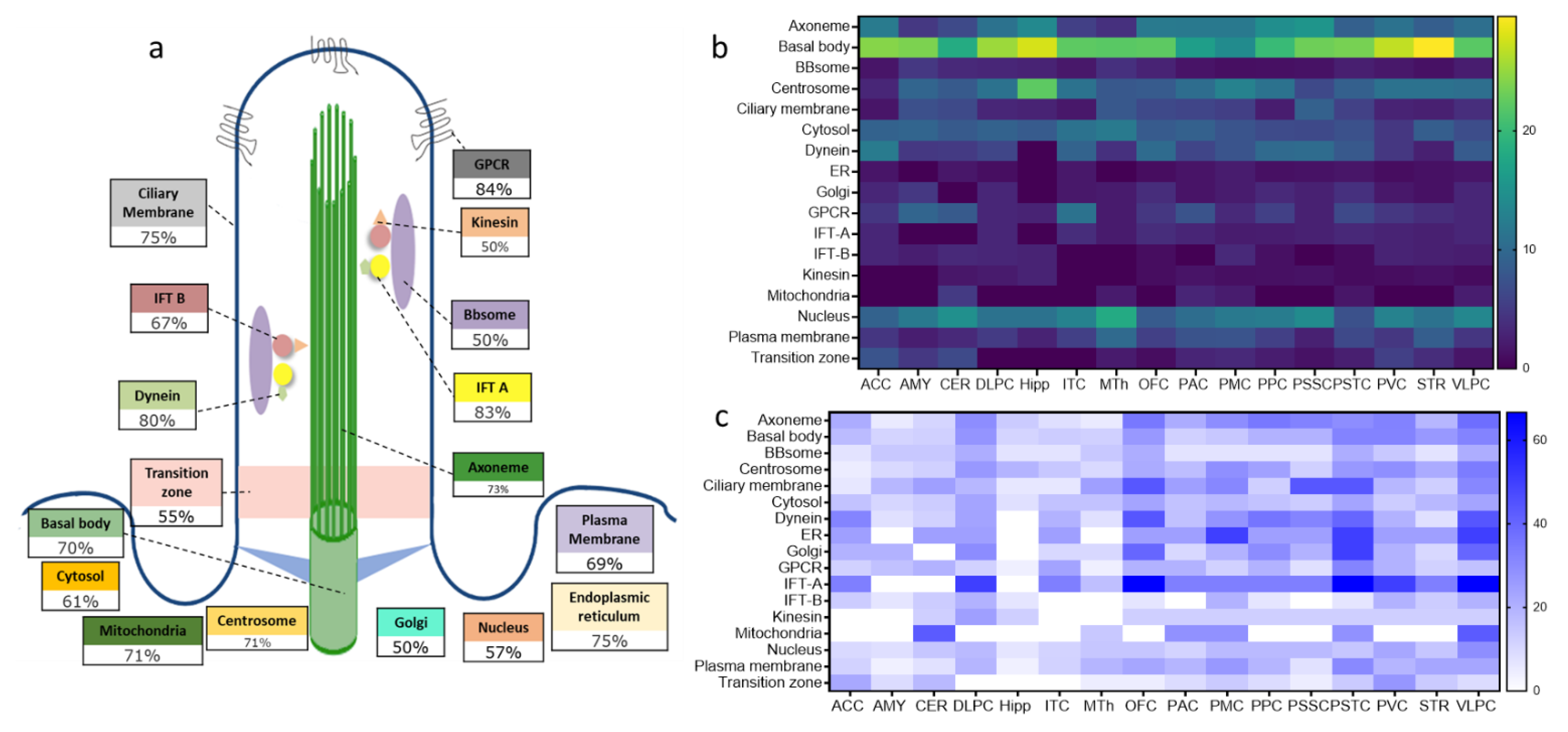
2.3. Structural and Functional Organization of Age-Regulated Cilia Genes
3. Discussion
3.1. Cilia Transcripts Are Dynamic, and Their Age-Regulated Expressions Are Region-Specific
3.2. Distinct Age-Dependent Dynamics of Cilia Structural and Functional Components
4. Methods and Experimental Designs
4.1. Cilia Genes’ List
4.2. Brain Lifespan’ Gene Expression
4.3. Fitting the Data into a Linear Regression Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Z.; Tu, H.; Kang, Y.; Xue, Y.; Ma, D.; Zhao, C.; Li, H.; Wang, L.; Liu, F. Primary cilia regulate hematopoietic stem and progenitor cell specification through Notch signaling in zebrafish. Nat. Commun. 2019, 10, 1839. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Otis, J.M.; Higginbotham, H.; Monckton, C.; Cheng, J.; Asokan, A.; Mykytyn, K.; Caspary, T.; Stuber, G.D.; Anton, E.S. Primary Cilia Signaling Shapes the Development of Interneuronal Connectivity. Dev. Cell 2017, 42, 286–300.e4. [Google Scholar] [CrossRef]
- Nechipurenko, I.V.; Doroquez, D.B.; Sengupta, P. Primary cilia and dendritic spines: Different but similar signaling compart-ments. Mol. Cells 2013, 36, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Breunig, J.J.; Sarkisian, M.R.; Arellano, J.I.; Morozov, Y.M.; Ayoub, A.E.; Sojitra, S.; Wang, B.; Flavell, R.A.; Rakic, P.; Town, T. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 13127–13132. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.G.; Spassky, N.; Romaguera-Ros, M.; Garcia-Verdugo, J.M.; Aguilar, A.; Schneider-Maunoury, S.; Alvarez-Buylla, A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat. Neurosci. 2008, 11, 277–284. [Google Scholar] [CrossRef]
- Oishi, I.; Kawakami, Y.; Raya, A.; Callol-Massot, C.; Izpisúa Belmonte, J.C. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nat. Genet. 2006, 38, 1316–1322. [Google Scholar] [CrossRef] [PubMed]
- Youn, Y.H.; Han, Y.G. Primary Cilia in Brain Development and Diseases. Am. J. Pathol. 2018, 188, 11–22. [Google Scholar] [CrossRef]
- Guemez-Gamboa, A.; Coufal, N.G.; Gleeson, J.G. Primary cilia in the developing and mature brain. Neuron 2014, 82, 511–521. [Google Scholar] [CrossRef]
- Juric-Sekhar, G.; Adkins, J.; Doherty, D.; Hevner, R.F. Joubert syndrome: Brain and spinal cord malformations in genotyped cases and implications for neurodevelopmental functions of primary cilia. Acta Neuropathol. 2012, 123, 695–709. [Google Scholar] [CrossRef]
- Bowie, E.; Goetz, S.C. TTBK2 and primary cilia are essential for the connectivity and survival of cerebellar Purkinje neurons. eLife 2020, 9, e51166. [Google Scholar] [CrossRef]
- Valente, E.M.; Rosti, R.O.; Gibbs, E.; Gleeson, J.G. Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 2014, 10, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Caspary, T.; Larkins, C.E.; Anderson, K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. cell 2007, 12, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Alhassen, W.; Chen, S.; Vawter, M.; Robbins, B.K.; Nguyen, H.; Myint, T.N.; Saito, Y.; Schulmann, A.; Nauli, S.M.; Civelli, O.; et al. Patterns of cilia gene dysregulations in major psychiatric disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 109, 110255. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.K.; Alvarez-Buylla, A. SnapShot: Adult neurogenesis in the V-SVZ. Neuron 2014, 81, 220–220.e221. [Google Scholar] [CrossRef] [PubMed]
- Berbari, N.F.; O’Connor, A.K.; Haycraft, C.J.; Yoder, B.K. The primary cilium as a complex signaling center. Curr. Biol. 2009, 19, R526–R535. [Google Scholar] [CrossRef]
- Wheatley, D.N. Nanobiology of the primary cilium—Paradigm of a multifunctional nanomachine complex. Methods Cell Biol. 2008, 90, 139–156. [Google Scholar] [CrossRef]
- Brailov, I.; Bancila, M.; Brisorgueil, M.J.; Miquel, M.C.; Hamon, M.; Verge, D. Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000, 872, 271–275. [Google Scholar] [CrossRef]
- Berbari, N.F.; Lewis, J.S.; Bishop, G.A.; Askwith, C.C.; Mykytyn, K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA 2008, 105, 4242–4246. [Google Scholar] [CrossRef]
- Domire, J.S.; Green, J.A.; Lee, K.G.; Johnson, A.D.; Askwith, C.C.; Mykytyn, K. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. CMLS 2011, 68, 2951–2960. [Google Scholar] [CrossRef]
- Handel, M.; Schulz, S.; Stanarius, A.; Schreff, M.; Erdtmann-Vourliotis, M.; Schmidt, H.; Wolf, G.; Hollt, V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 1999, 89, 909–926. [Google Scholar] [CrossRef]
- Omori, Y.; Chaya, T.; Yoshida, S.; Irie, S.; Tsujii, T.; Furukawa, T. Identification of G Protein-Coupled Receptors (GPCRs) in Primary Cilia and Their Possible Involvement in Body Weight Control. PLoS ONE 2015, 10, e0128422. [Google Scholar] [CrossRef]
- Varela, L.; Horvath, T.L. Neuronal Cilia: Another Player in the Melanocortin System. Trends Mol. Med. 2018, 24, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Baldi, P.; Alhassen, W.; Chen, S.; Nguyen, H.; Khoudari, M.; Alachkar, A. Large-scale analysis reveals spatiotemporal circadian patterns of cilia transcriptomes in the primate brain. J. Neurosci. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Alhassen, W.; Kobayashi, Y.; Su, J.; Robbins, B.; Ngyuen, H.; Myint, T.; Yu, M.; Nauli, S.M.; Saito, Y.; Alachkar, A. Regulation of Brain Primary Cilia Length by MCH Signaling: Evidence from Pharmacological, Genetic, Optogenetic and Chemogenic Manipulations. bioRxiv 2021. [Google Scholar] [CrossRef]
- Miller, J.A.; Ding, S.L.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Ebbert, A.; Riley, Z.L.; Royall, J.J.; Aiona, K.; et al. Transcriptional landscape of the prenatal human brain. Nature 2014, 508, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Mikula, S.; Trotts, I.; Stone, J.M.; Jones, E.G. Internet-enabled high-resolution brain mapping and virtual microscopy. Neuroimage 2007, 35, 9–15. [Google Scholar] [CrossRef]
- Cardenas-Rodriguez, M.; Irigoin, F.; Osborn, D.P.; Gascue, C.; Katsanis, N.; Beales, P.L.; Badano, J.L. The Bardet-Biedl syndrome-related protein CCDC28B modulates mTORC2 function and interacts with SIN1 to control cilia length independently of the mTOR complex. Hum. Mol. Genet. 2013, 22, 4031–4042. [Google Scholar] [CrossRef]
- Cardenas-Rodriguez, M.; Osborn, D.P.; Irigoin, F.; Grana, M.; Romero, H.; Beales, P.L.; Badano, J.L. Characterization of CCDC28B reveals its role in ciliogenesis and provides insight to understand its modifier effect on Bardet-Biedl syndrome. Hum. Genet. 2013, 132, 91–105. [Google Scholar] [CrossRef]
- Spalding, K.L.; Bergmann, O.; Alkass, K.; Bernard, S.; Salehpour, M.; Huttner, H.B.; Bostrom, E.; Westerlund, I.; Vial, C.; Buchholz, B.A.; et al. Dynamics of hippocampal neurogenesis in adult humans. Cell 2013, 153, 1219–1227. [Google Scholar] [CrossRef]
- Christensen, S.T.; Pedersen, L.B.; Schneider, L.; Satir, P. Sensory cilia and integration of signal transduction in human health and disease. Traffic 2007, 8, 97–109. [Google Scholar] [CrossRef]
- Satir, P.; Christensen, S.T. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 2007, 69, 377–400. [Google Scholar] [CrossRef]
- Alvarado, J.A.; Dhande, O.S.; Prosseda, P.P.; Kowal, T.J.; Ning, K.; Jabbehdari, S.; Hu, Y.; Sun, Y. Developmental distribution of primary cilia in the retinofugal visual pathway. J. Comp. Neurol. 2021, 529, 1442–1455. [Google Scholar] [CrossRef] [PubMed]
- Schroter, J.; Doring, J.H.; Garbade, S.F.; Hoffmann, G.F.; Kolker, S.; Ries, M.; Syrbe, S. Cross-sectional quantitative analysis of the natural history of TUBA1A and TUBB2B tubulinopathies. Genet. Med. Off. J. Am. Coll. Med Genet. 2021, 23, 516–523. [Google Scholar] [CrossRef]
- Aiken, J.; Buscaglia, G.; Aiken, A.S.; Moore, J.K.; Bates, E.A. Tubulin mutations in brain development disorders: Why haploinsufficiency does not explain TUBA1A tubulinopathies. Cytoskeleton 2020, 77, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Cushion, T.D.; Paciorkowski, A.R.; Pilz, D.T.; Mullins, J.G.; Seltzer, L.E.; Marion, R.W.; Tuttle, E.; Ghoneim, D.; Christian, S.L.; Chung, S.K.; et al. De novo mutations in the beta-tubulin gene TUBB2A cause simplified gyral patterning and infantile-onset epilepsy. Am. J. Hum. Genet. 2014, 94, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, M.R.; Morrison, E.; Sirey, T.; Molnar, Z.; Hayward, B.E.; Carr, I.M.; Springell, K.; Woods, C.G.; Ahmed, M.; Hattingh, L.; et al. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am. J. Hum. Genet. 2009, 85, 737–744. [Google Scholar] [CrossRef]
- Keays, D.A.; Tian, G.; Poirier, K.; Huang, G.J.; Siebold, C.; Cleak, J.; Oliver, P.L.; Fray, M.; Harvey, R.J.; Molnar, Z.; et al. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell 2007, 128, 45–57. [Google Scholar] [CrossRef]
- Jaglin, X.H.; Poirier, K.; Saillour, Y.; Buhler, E.; Tian, G.; Bahi-Buisson, N.; Fallet-Bianco, C.; Phan-Dinh-Tuy, F.; Kong, X.P.; Bomont, P.; et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet. 2009, 41, 746–752. [Google Scholar] [CrossRef]
- Poirier, K.; Saillour, Y.; Bahi-Buisson, N.; Jaglin, X.H.; Fallet-Bianco, C.; Nabbout, R.; Castelnau-Ptakhine, L.; Roubertie, A.; Attie-Bitach, T.; Desguerre, I.; et al. Mutations in the neuronal ss-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum. Mol. Genet. 2010, 19, 4462–4473. [Google Scholar] [CrossRef]
- Poirier, K.; Lebrun, N.; Broix, L.; Tian, G.; Saillour, Y.; Boscheron, C.; Parrini, E.; Valence, S.; Pierre, B.S.; Oger, M.; et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013, 45, 639–647. [Google Scholar] [CrossRef]
- Bahi-Buisson, N.; Poirier, K.; Fourniol, F.; Saillour, Y.; Valence, S.; Lebrun, N.; Hully, M.; Bianco, C.F.; Boddaert, N.; Elie, C.; et al. The wide spectrum of tubulinopathies: What are the key features for the diagnosis? Brain J. Neurol. 2014, 137, 1676–1700. [Google Scholar] [CrossRef]
- Labisso, W.L.; Raulin, A.C.; Nwidu, L.L.; Kocon, A.; Wayne, D.; Erdozain, A.M.; Morentin, B.; Schwendener, D.; Allen, G.; Enticott, J.; et al. The Loss of alpha- and beta-Tubulin Proteins Are a Pathological Hallmark of Chronic Alcohol Consumption and Natural Brain Ageing. Brain Sci. 2018, 8, 175. [Google Scholar] [CrossRef]
- Nguyen, M.M.; McCracken, C.J.; Milner, E.S.; Goetschius, D.J.; Weiner, A.T.; Long, M.K.; Michael, N.L.; Munro, S.; Rolls, M.M. Gamma-tubulin controls neuronal microtubule polarity independently of Golgi outposts. Mol. Biol. Cell 2014, 25, 2039–2050. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Sonobe, S.; Baskin, T.I.; Hyodo, S.; Hasezawa, S.; Nagata, T.; Horio, T.; Hasebe, M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 2005, 7, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Moritz, M.; Braunfeld, M.B.; Sedat, J.W.; Alberts, B.; Agard, D.A. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 1995, 378, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.R.; Oakley, C.E.; Yoon, Y.; Jung, M.K. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell 1990, 61, 1289–1301. [Google Scholar] [CrossRef]
- Joshi, H.C.; Palacios, M.J.; McNamara, L.; Cleveland, D.W. Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature 1992, 356, 80–83. [Google Scholar] [CrossRef]
- Kollman, J.M.; Merdes, A.; Mourey, L.; Agard, D.A. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011, 12, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Joshi, H.C. gamma-tubulin is a minus end-specific microtubule binding protein. J. Cell Biol. 1995, 131, 207–214. [Google Scholar] [CrossRef]
- Zhang, J.; Megraw, T.L. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell 2007, 18, 4037–4049. [Google Scholar] [CrossRef]
- Tovey, C.A.; Conduit, P.T. Microtubule nucleation by gamma-tubulin complexes and beyond. Essays Biochem. 2018, 62, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, M.; Shi, J.; Zhou, Y.; Wan, Z.; Wang, Y.; Wan, Y.; Li, J.; Wang, Z.; Fei, J. Brain-Specific SNAP-25 Deletion Leads to Elevated Extracellular Glutamate Level and Schizophrenia-Like Behavior in Mice. Neural Plast. 2017, 2017, 4526417. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.M.; Kelley, M.; Yao, J.; Tsai, G.; van Kammen, D.P. Elevated cerebrospinal fluid SNAP-25 in schizophrenia. Biol. Psychiatry 2003, 53, 1132–1137. [Google Scholar] [CrossRef]
- Thompson, P.M.; Egbufoama, S.; Vawter, M.P. SNAP-25 reduction in the hippocampus of patients with schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2003, 27, 411–417. [Google Scholar] [CrossRef]
- Barakauskas, V.E.; Moradian, A.; Barr, A.M.; Beasley, C.L.; Rosoklija, G.; Mann, J.J.; Ilievski, B.; Stankov, A.; Dwork, A.J.; Falkai, P.; et al. Quantitative mass spectrometry reveals changes in SNAP-25 isoforms in schizophrenia. Schizophr. Res. 2016, 177, 44–51. [Google Scholar] [CrossRef][Green Version]
- Thompson, P.M.; Rosenberger, C.; Qualls, C. CSF SNAP-25 in schizophrenia and bipolar illness. A pilot study. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 1999, 21, 717–722. [Google Scholar] [CrossRef]
- Scarr, E.; Gray, L.; Keriakous, D.; Robinson, P.J.; Dean, B. Increased levels of SNAP-25 and synaptophysin in the dorsolateral prefrontal cortex in bipolar I disorder. Bipolar Disord. 2006, 8, 133–143. [Google Scholar] [CrossRef]
- Houenou, J.; Boisgontier, J.; Henrion, A.; d’Albis, M.A.; Dumaine, A.; Linke, J.; Wessa, M.; Daban, C.; Hamdani, N.; Delavest, M.; et al. A Multilevel Functional Study of a SNAP25 At-Risk Variant for Bipolar Disorder and Schizophrenia. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 10389–10397. [Google Scholar] [CrossRef]
- Ye, C.; Hu, Z.; Wu, E.; Yang, X.; Buford, U.J.; Guo, Z.; Saveanu, R.V. Two SNAP-25 genetic variants in the binding site of multiple microRNAs and susceptibility of ADHD: A meta-analysis. J. Psychiatr. Res. 2016, 81, 56–62. [Google Scholar] [CrossRef]
- Kim, J.W.; Biederman, J.; Arbeitman, L.; Fagerness, J.; Doyle, A.E.; Petty, C.; Perlis, R.H.; Purcell, S.; Smoller, J.W.; Faraone, S.V.; et al. Investigation of variation in SNAP-25 and ADHD and relationship to co-morbid major depressive disorder. Am. J. Med Genetics. B Neuropsychiatr. Genet. 2007, 144B, 781–790. [Google Scholar] [CrossRef]
- Mill, J.; Xu, X.; Ronald, A.; Curran, S.; Price, T.; Knight, J.; Craig, I.; Sham, P.; Plomin, R.; Asherson, P. Quantitative trait locus analysis of candidate gene alleles associated with attention deficit hyperactivity disorder (ADHD) in five genes: DRD4, DAT1, DRD5, SNAP-25, and 5HT1B. Am. J. Med Genetics. B Neuropsychiatr. Genet. 2005, 133B, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Miguel, A.; Gicas, K.; Alamri, J.; Beasley, C.L.; Dwork, A.J.; Mann, J.J.; Rosoklija, G.; Cai, F.; Song, W.; Barr, A.M.; et al. Reduced SNAP25 Protein Fragmentation Contributes to SNARE Complex Dysregulation in Schizophrenia Postmortem Brain. Neuroscience 2019, 420, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Gigg, J.; McEwan, F.; Smausz, R.; Neill, J.; Harte, M.K. Synaptic biomarker reduction and impaired cognition in the sub-chronic PCP mouse model for schizophrenia. J. Psychopharmacol. 2020, 34, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Najera, K.; Fagan, B.M.; Thompson, P.M. SNAP-25 in Major Psychiatric Disorders: A Review. Neuroscience 2019, 420, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Cupertino, R.B.; Kappel, D.B.; Bandeira, C.E.; Schuch, J.B.; da Silva, B.S.; Muller, D.; Bau, C.H.; Mota, N.R. SNARE complex in developmental psychiatry: Neurotransmitter exocytosis and beyond. J. Neural Transm. 2016, 123, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Antonucci, F.; Corradini, I.; Morini, R.; Fossati, G.; Menna, E.; Pozzi, D.; Pacioni, S.; Verderio, C.; Bacci, A.; Matteoli, M. Reduced SNAP-25 alters short-term plasticity at developing glutamatergic synapses. EMBO Rep. 2013, 14, 645–651. [Google Scholar] [CrossRef]
- Corradini, I.; Verderio, C.; Sala, M.; Wilson, M.C.; Matteoli, M. SNAP-25 in neuropsychiatric disorders. Ann. N. Y. Acad. Sci. 2009, 1152, 93–99. [Google Scholar] [CrossRef]
- Scherk, H.; Backens, M.; Zill, P.; Schneider-Axmann, T.; Wobrock, T.; Usher, J.; Reith, W.; Falkai, P.; Moller, H.J.; Bondy, B.; et al. SNAP-25 genotype influences NAA/Cho in left hippocampus. J. Neural Transm. 2008, 115, 1513–1518. [Google Scholar] [CrossRef]
- Lenart, J.; Bratek, E.; Lazarewicz, J.W.; Zieminska, E. Changes in the Expression of SNAP-25 Protein in the Brain of Juvenile Rats in Two Models of Autism. J. Mol. Neurosci. 2020, 70, 1313–1320. [Google Scholar] [CrossRef]
- Braida, D.; Guerini, F.R.; Ponzoni, L.; Corradini, I.; De Astis, S.; Pattini, L.; Bolognesi, E.; Benfante, R.; Fornasari, D.; Chiappedi, M.; et al. Association between SNAP-25 gene polymorphisms and cognition in autism: Functional consequences and potential therapeutic strategies. Transl. Psychiatry 2015, 5, e500. [Google Scholar] [CrossRef]
- Guerini, F.R.; Bolognesi, E.; Chiappedi, M.; Manca, S.; Ghezzo, A.; Agliardi, C.; Sotgiu, S.; Usai, S.; Matteoli, M.; Clerici, M. SNAP-25 single nucleotide polymorphisms are associated with hyperactivity in autism spectrum disorders. Pharmacol. Res. 2011, 64, 283–288. [Google Scholar] [CrossRef]
- Boschert, U.; O’Shaughnessy, C.; Dickinson, R.; Tessari, M.; Bendotti, C.; Catsicas, S.; Pich, E.M. Developmental and plasticity-related differential expression of two SNAP-25 isoforms in the rat brain. J. Comp. Neurol. 1996, 367, 177–193. [Google Scholar] [CrossRef]
- Bark, I.C.; Hahn, K.M.; Ryabinin, A.E.; Wilson, M.C. Differential expression of SNAP-25 protein isoforms during divergent vesicle fusion events of neural development. Proc. Natl. Acad. Sci. USA 1995, 92, 1510–1514. [Google Scholar] [CrossRef]
- Bark, I.C.; Wilson, M.C. Human cDNA clones encoding two different isoforms of the nerve terminal protein SNAP-25. Gene 1994, 139, 291–292. [Google Scholar] [CrossRef]
- Etain, B.; Dumaine, A.; Mathieu, F.; Chevalier, F.; Henry, C.; Kahn, J.P.; Deshommes, J.; Bellivier, F.; Leboyer, M.; Jamain, S. A SNAP25 promoter variant is associated with early-onset bipolar disorder and a high expression level in brain. Mol. Psychiatry 2010, 15, 748–755. [Google Scholar] [CrossRef]
- Furuya, T.K.; Silva, P.N.; Payao, S.L.; Bertolucci, P.H.; Rasmussen, L.T.; De Labio, R.W.; Braga, I.L.; Chen, E.S.; Turecki, G.; Mechawar, N.; et al. Analysis of SNAP25 mRNA expression and promoter DNA methylation in brain areas of Alzheimer’s Disease patients. Neuroscience 2012, 220, 41–46. [Google Scholar] [CrossRef]
- Greber, S.; Lubec, G.; Cairns, N.; Fountoulakis, M. Decreased levels of synaptosomal associated protein 25 in the brain of patients with Down syndrome and Alzheimer’s disease. Electrophoresis 1999, 20, 928–934. [Google Scholar] [CrossRef]
- Novas, R.; Cardenas-Rodriguez, M.; Lepanto, P.; Fabregat, M.; Rodao, M.; Fariello, M.I.; Ramos, M.; Davison, C.; Casanova, G.; Alfaya, L.; et al. Kinesin 1 regulates cilia length through an interaction with the Bardet-Biedl syndrome related protein CCDC28B. Sci. Rep. 2018, 8, 3019. [Google Scholar] [CrossRef]
- Pedersen, L.B.; Rosenbaum, J.L. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr. Top. Dev. Biol. 2008, 85, 23–61. [Google Scholar] [CrossRef]
- Mourao, A.; Christensen, S.T.; Lorentzen, E. The intraflagellar transport machinery in ciliary signaling. Curr. Opin. Struct. Biol. 2016, 41, 98–108. [Google Scholar] [CrossRef]
- Taschner, M.; Lorentzen, E. The Intraflagellar Transport Machinery. Cold Spring Harb. Perspect. Biol. 2016, 8, a028092. [Google Scholar] [CrossRef]
- Nakayama, K.; Katoh, Y. Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 2018, 163, 155–164. [Google Scholar] [CrossRef]
- Boonsawat, P.; Joset, P.; Steindl, K.; Oneda, B.; Gogoll, L.; Azzarello-Burri, S.; Sheth, F.; Datar, C.; Verma, I.C.; Puri, R.D.; et al. Elucidation of the phenotypic spectrum and genetic landscape in primary and secondary microcephaly. Genet. Med. Off. J. Am. Coll. Med Genet. 2019, 21, 2043–2058. [Google Scholar] [CrossRef]
- Brancati, F.; Dallapiccola, B.; Valente, E.M. Joubert Syndrome and related disorders. Orphanet J. Rare Dis. 2010, 5, 20. [Google Scholar] [CrossRef]
- Robinson, B.V.; Faundez, V.; Lerit, D.A. Understanding microcephaly through the study of centrosome regulation in Drosophila neural stem cells. Biochem. Soc. Trans. 2020, 48, 2101–2115. [Google Scholar] [CrossRef]
- Mykytyn, K.; Askwith, C. G-Protein-Coupled Receptor Signaling in Cilia. Cold Spring Harb. Perspect. Biol. 2017, 9, a028183. [Google Scholar] [CrossRef]
- Hirano, T.; Katoh, Y.; Nakayama, K. Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein-coupled receptors. Mol. Biol. Cell 2017, 28, 429–439. [Google Scholar] [CrossRef]
- Schou, K.B.; Pedersen, L.B.; Christensen, S.T. Ins and outs of GPCR signaling in primary cilia. EMBO Rep. 2015, 16, 1099–1113. [Google Scholar] [CrossRef]
- Green, J.A.; Gu, C.; Mykytyn, K. Heteromerization of ciliary G protein-coupled receptors in the mouse brain. PLoS ONE 2012, 7, e46304. [Google Scholar] [CrossRef]
- van Dam, T.J.P.; Townsend, M.J.; Turk, M.; Schlessinger, A.; Sali, A.; Field, M.C.; Huynen, M.A. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc. Natl. Acad. Sci. USA 2013, 110, 6943–6948. [Google Scholar] [CrossRef]
- Arnaiz, O.; Cohen, J.; Tassin, A.M.; Koll, F. Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 2014, 3, 9. [Google Scholar] [CrossRef]
- Arnaiz, O.; Malinowska, A.; Klotz, C.; Sperling, L.; Dadlez, M.; Koll, F.; Cohen, J. Cildb: A knowledgebase for centrosomes and cilia. Database J. Biol. Databases Curation 2009, 2009, bap022. [Google Scholar] [CrossRef]
- van Dam, T.J.P.; Kennedy, J.; van der Lee, R.; de Vrieze, E.; Wunderlich, K.A.; Rix, S.; Dougherty, G.W.; Lambacher, N.J.; Li, C.; Jensen, V.L.; et al. CiliaCarta: An integrated and validated compendium of ciliary genes. PLoS ONE 2019, 14, e0216705. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Alhassen, W.; Vakil Monfared, R.; Vachirakorntong, B.; Nauli, S.M.; Baldi, P.; Alachkar, A. Dynamic Changes of Brain Cilia Transcriptomes across the Human Lifespan. Int. J. Mol. Sci. 2021, 22, 10387. https://doi.org/10.3390/ijms221910387
Chen S, Alhassen W, Vakil Monfared R, Vachirakorntong B, Nauli SM, Baldi P, Alachkar A. Dynamic Changes of Brain Cilia Transcriptomes across the Human Lifespan. International Journal of Molecular Sciences. 2021; 22(19):10387. https://doi.org/10.3390/ijms221910387
Chicago/Turabian StyleChen, Siwei, Wedad Alhassen, Roudabeh Vakil Monfared, Benjamin Vachirakorntong, Surya M. Nauli, Pierre Baldi, and Amal Alachkar. 2021. "Dynamic Changes of Brain Cilia Transcriptomes across the Human Lifespan" International Journal of Molecular Sciences 22, no. 19: 10387. https://doi.org/10.3390/ijms221910387
APA StyleChen, S., Alhassen, W., Vakil Monfared, R., Vachirakorntong, B., Nauli, S. M., Baldi, P., & Alachkar, A. (2021). Dynamic Changes of Brain Cilia Transcriptomes across the Human Lifespan. International Journal of Molecular Sciences, 22(19), 10387. https://doi.org/10.3390/ijms221910387





