Discovery of Novel Polycyclic Polyprenylated Acylphloroglucinols from the Fruits of Garcinia xanthochymus as Antitumor Agents by Suppressing the STAT3 Signaling
Abstract
:1. Introduction
2. Results
2.1. Structural Elucidation of Isolated Compounds
2.2. Anti-Proliferative Activity of the Isolated PPAPs on Human Tumor Cell Lines
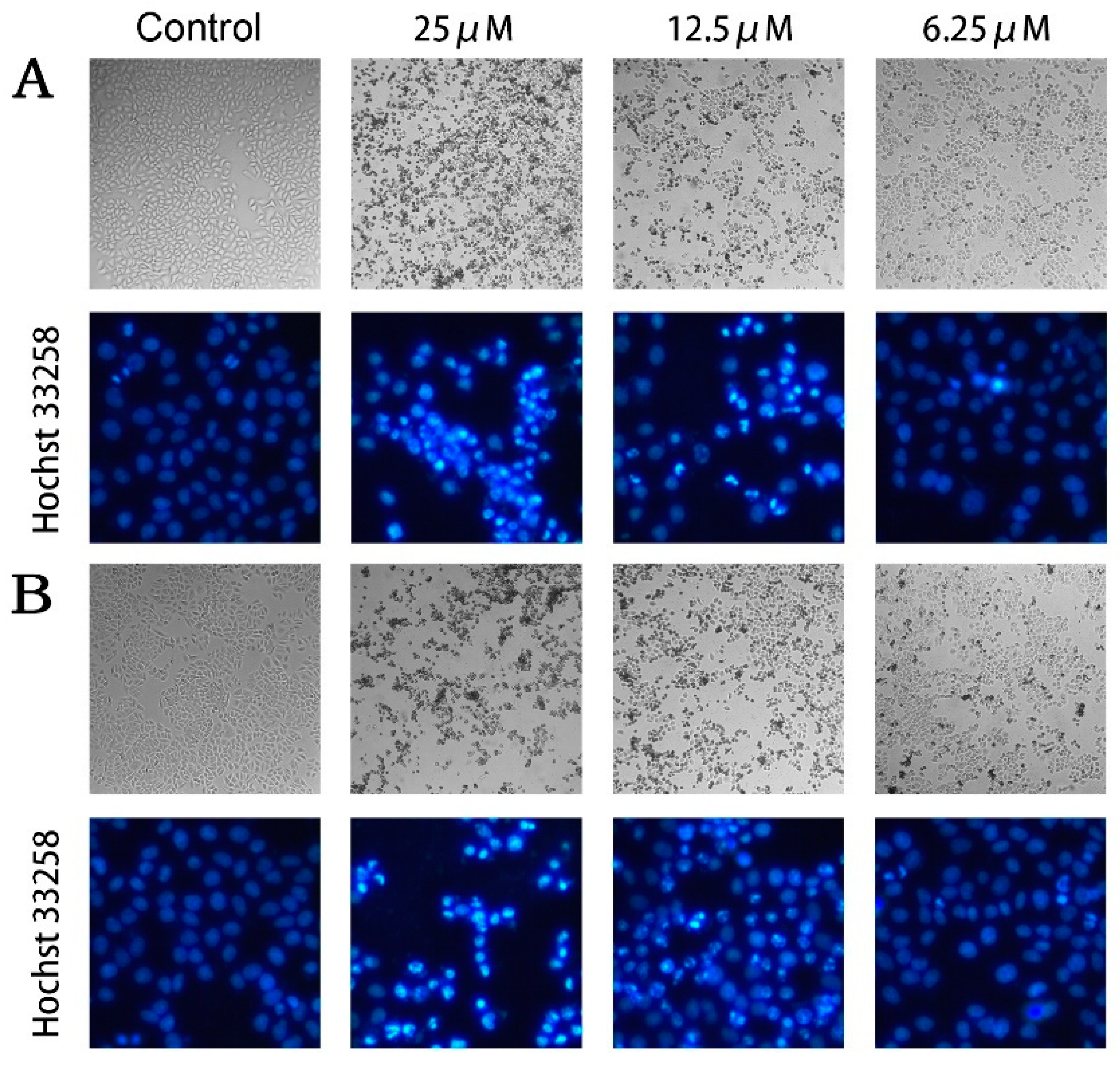
2.3. Compounds 2 and 5 Induced Cellular Apoptosis of MCF-7
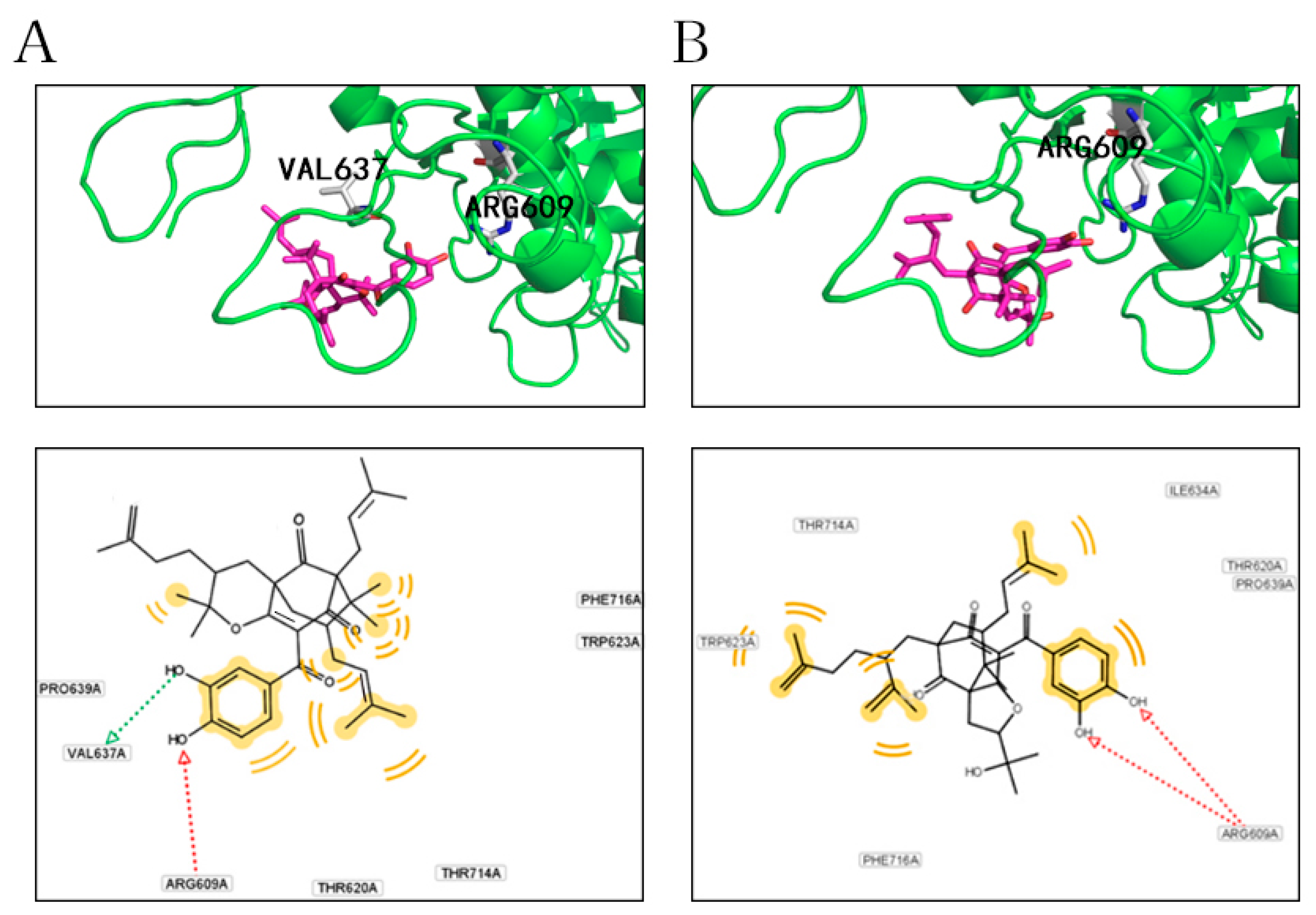
2.4. Docking Analysis of Compounds 2 and 5 with STAT3
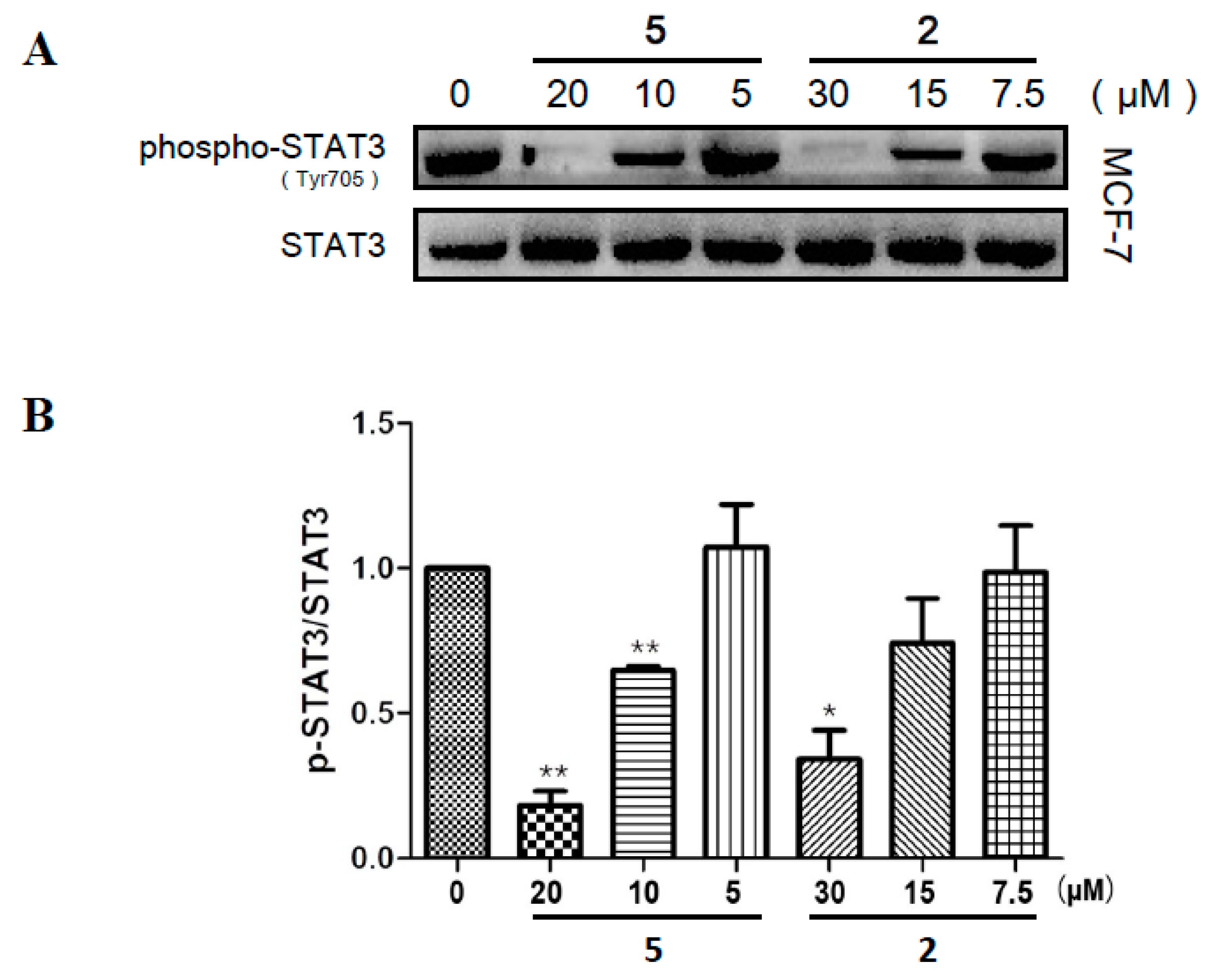
2.5. Compounds 2 and 5 Suppressed the Phosphorylation of STAT3 in MCF-7
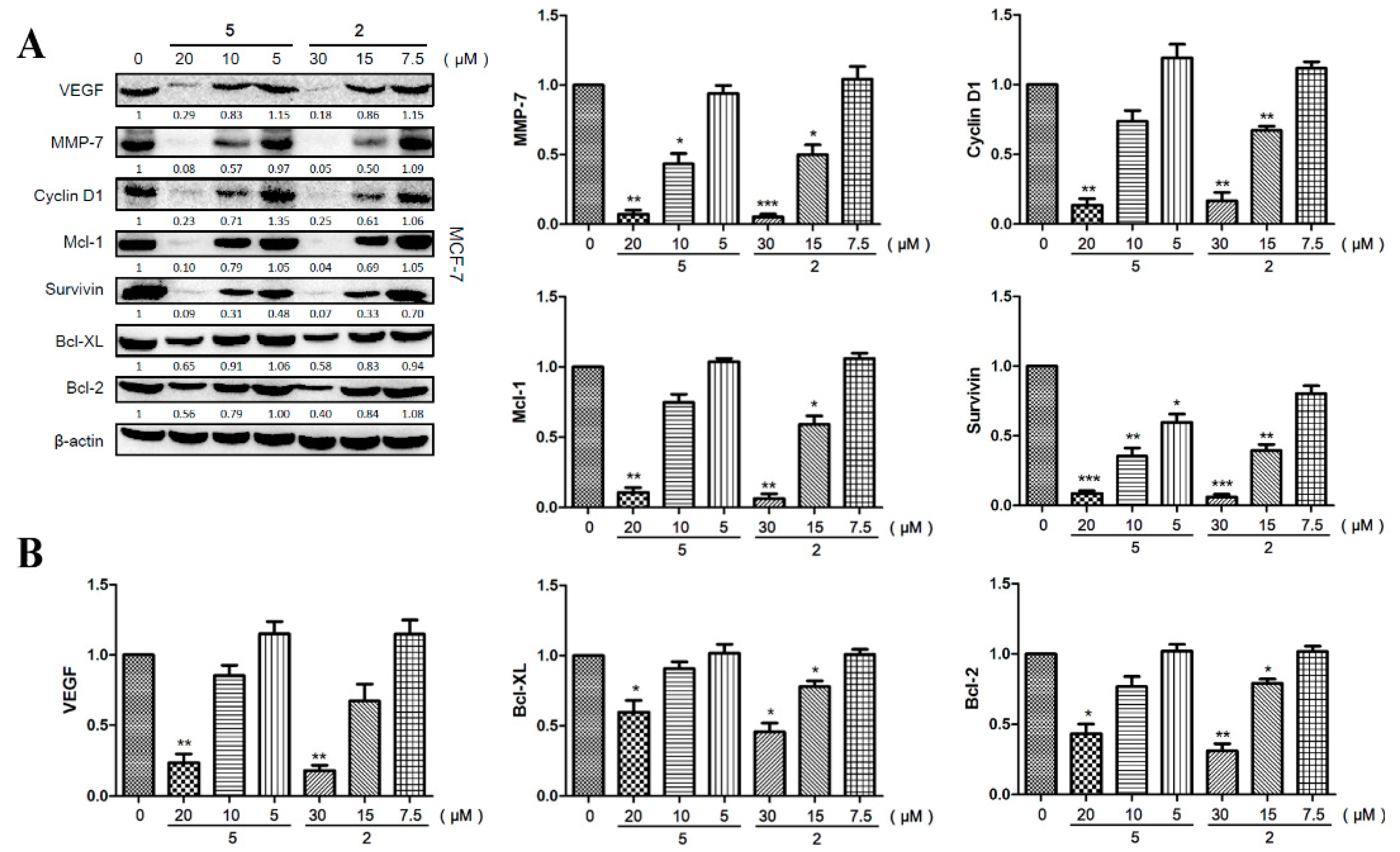
2.6. Compounds 2 and 5 Downregulated the Expression of Effector Genes Downstream of STAT3 Signaling
3. Discussion
4. Materials and Methods
4.1. General
4.2. Plant Material
4.3. Regents
4.4. Extraction and Isolation
4.5. Cell Culture and Treatment
4.6. MTT Assay
4.7. Docking Analysis
4.8. Western Blotting Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Jemal, A.; Grey, N.; Ferlay, J.; Forman, D. Global cancer transitions according to the Human Development Index (2008–2030): A population-based study. Lancet Oncol. 2012, 13, 790–801. [Google Scholar] [CrossRef]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharm. 2018, 834, 188–196. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.; Garcia, R.; Turkson, J.; Jove, R. STATs in oncogenesis. Oncogene 2000, 19, 2474–2488. [Google Scholar] [CrossRef] [Green Version]
- Sriuranpong, V.; Park, J.I.; Amornphimoltham, P.; Patel, V.; Nelkin, B.D.; Gutkind, J.S. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res 2003, 63, 2948–2956. [Google Scholar] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yue, P.; Fletcher, S.; Zhao, W.; Gunning, P.T.; Turkson, J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem. Pharm. 2010, 79, 1398–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, Z.; Xu, D.; Ji, J.; Guo, W.; Zhu, W.; He, J. JAK/STAT signal pathway activation promotes progression and survival of human oesophageal squamous cell carcinoma. Clin. Transl. Oncol. 2012, 14, 143–149. [Google Scholar] [CrossRef]
- Koskela, H.L.; Eldfors, S.; Ellonen, P.; van Adrichem, A.J.; Kuusanmäki, H.; Andersson, E.I.; Lagström, S.; Clemente, M.J.; Olson, T.; Jalkanen, S.E.; et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 2012, 366, 1905–1913. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, F.C.; Cheng, G.; Lin, J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem. Biophys. Res. Commun. 2005, 335, 292–299. [Google Scholar] [CrossRef]
- Wang, Z.; Hui, C.; Xie, Y. Natural STAT3 inhibitors: A mini perspective. Bioorg. Chem. 2021, 115, 105169. [Google Scholar] [CrossRef] [PubMed]
- Flora Reipublicae Popularis Sinicae. Available online: http://www.iplant.cn/info/Garcinia%20xanthochymus?t=z (accessed on 22 August 2021).
- Liu, B.; Zhang, X.; Bussmann, R.W.; Hart, R.H.; Li, P.; Bai, Y.; Long, C. Garcinia in Southern China: Ethnobotany, Management, and Niche Modeling. Econ. Bot. 2017, 70, 416–430. [Google Scholar] [CrossRef]
- Yapwattanaphun, C.; Subhadrabandhu, S.; Sugiura, A.; Yonemori, K.; Utsunomiya, N. Utilization of some garcinia species in thailand. Acta Hortic. 2002, 575, 563–570. [Google Scholar] [CrossRef]
- Wu, S.B.; Long, C.; Kennelly, E.J. Structural diversity and bioactivities of natural benzophenones. Nat. Prod. Rep. 2014, 31, 1158–1174. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, S.; Chattopadhyay, S.K. The potential health benefit of polyisoprenylated benzophenones from Garcinia and related genera: Ethnobotanical and therapeutic importance. Fitoterapia 2013, 89, 86–125. [Google Scholar] [CrossRef]
- Che Hassan, N.K.N.; Taher, M.; Susanti, D. Phytochemical constituents and pharmacological properties of Garcinia xanthochymus—A review. Biomed. Pharm. 2018, 106, 1378–1389. [Google Scholar] [CrossRef]
- Ciochina, R.; Grossman, R.B. Polycyclic polyprenylated acylphloroglucinols. Chem. Rev. 2006, 106, 3963–3986. [Google Scholar] [CrossRef]
- Yang, H.; Figueroa, M.; To, S.; Baggett, S.; Jiang, B.; Basile, M.J.; Weinstein, I.B.; Kennelly, E.J. Benzophenones and biflavonoids from Garcinia livingstonei fruits. J. Agric. Food Chem. 2010, 58, 4749–4755. [Google Scholar] [CrossRef]
- Xu, J.; Jin, S.; Gan, F.; Xiong, H.; Mei, Z.; Chen, Y.; Yang, G. Polycyclic polyprenylated acylphloroglucinols from Garcinia xanthochymus fruits exhibit antitumor effects through inhibition of the STAT3 signaling pathway. Food Funct. 2020, 11, 10568–10579. [Google Scholar] [CrossRef]
- Jin, S.; Shi, K.; Liu, L.; Chen, Y.; Yang, G. Xanthones from the Bark of Garcinia xanthochymus and the Mechanism of Induced Apoptosis in Human Hepatocellular Carcinoma HepG2 Cells via the Mitochondrial Pathway. Int. J. Mol. Sci. 2019, 20, 4803. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Gan, F.; Jin, S.; Liu, H.; Wu, S.; Yang, W.; Yang, G. Adamantyl derivatives and rearranged benzophenones from Garcinia xanthochymus fruits. RSC Adv. 2017, 7, 17289–17296. [Google Scholar] [CrossRef] [Green Version]
- Roux, D.; Hadi, H.A.; Thoret, S.; Guénard, D.; Thoison, O.; Païs, M.; Sévenet, T. Structure-activity relationship of polyisoprenyl benzophenones from Garcinia pyrifera on the tubulin/microtubule system. J. Nat. Prod. 2000, 63, 1070–1076. [Google Scholar] [CrossRef]
- Phang, Y.; Wang, X.; Lu, Y.; Fu, W.; Zheng, C.; Xu, H. Bicyclic polyprenylated acylphloroglucinols and their derivatives: Structural modification, structure-activity relationship, biological activity and mechanism of action. Eur. J. Med. Chem. 2020, 205, 112646. [Google Scholar] [CrossRef] [PubMed]
- Socolsky, C.; Plietker, B. Total synthesis and absolute configuration assignment of MRSA active garcinol and isogarcinol. Chemistry 2015, 21, 3053–3061. [Google Scholar] [CrossRef]
- Xia, Z.X.; Zhang, D.D.; Liang, S.; Lao, Y.Z.; Zhang, H.; Tan, H.S.; Chen, S.L.; Wang, X.H.; Xu, H.X. Bioassay-guided isolation of prenylated xanthones and polycyclic acylphloroglucinols from the leaves of Garcinia nujiangensis. J. Nat. Prod. 2012, 75, 1459–1464. [Google Scholar] [CrossRef]
- Marti, G.; Eparvier, V.; Moretti, C.; Susplugas, S.; Prado, S.; Grellier, P.; Retailleau, P.; Gueritte, F.; Litaudon, M. Antiplasmodial benzophenones from the trunk latex of Moronobea coccinea (Clusiaceae). Phytochemistry 2009, 70, 75–85. [Google Scholar] [CrossRef]
- Tian, W.-J.; Qiu, Y.-Q.; Jin, X.-J.; Chen, H.-F.; Yao, X.-J.; Dai, Y.; Yao, X.-S. Hypersampsones S–W, new polycyclic polyprenylated acylphloroglucinols from Hypericum sampsonii. RSC Adv. 2016, 6, 50887–50894. [Google Scholar] [CrossRef]
- Masullo, M.; Bassarello, C.; Bifulco, G.; Piacente, S. Polyisoprenylated benzophenone derivatives from the fruits of Garcinia cambogia and their absolute configuration by quantum chemical circular dichroism calculations. Tetrahedron 2010, 66, 139–145. [Google Scholar] [CrossRef]
- Yang, X.W.; Grossman, R.B.; Xu, G. Research Progress of Polycyclic Polyprenylated Acylphloroglucinols. Chem. Rev. 2018, 118, 3508–3558. [Google Scholar] [CrossRef] [PubMed]
- Baggett, S.; Protiva, P.; Mazzola, E.P.; Yang, H.; Ressler, E.T.; Basile, M.J.; Weinstein, I.B.; Kennelly, E.J. Bioactive benzophenones from Garcinia xanthochymus fruits. J. Nat. Prod. 2005, 68, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, A.E.; Nakamura, H.; Inoue, D.; Hirasawa, Y.; Wong, C.P.; Kaneda, T.; Hadi, A.H.A.; Morita, H. Polyisoprenylated Acylphloroglucinols from Garcinia nervosa. Nat. Prod. Commun. 2018, 13, 1934578X1801300324. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yu, T.; Gao, X.M.; Zhou, Y.; Qiao, C.F.; Peng, Y.; Chen, S.L.; Luo, K.Q.; Xu, H.X. Apoptotic effects of polyprenylated benzoylphloroglucinol derivatives from the twigs of Garcinia multiflora. J. Nat. Prod. 2010, 73, 1355–1359. [Google Scholar] [CrossRef]
- Fu, W.; Wu, M.; Zhu, L.; Lao, Y.; Wang, L.; Tan, H.; Yuan, Q.; Xu, H. Prenylated benzoylphloroglucinols and biphenyl derivatives from the leaves of Garcinia multiflora Champ. RSC Adv. 2015, 5, 78259–78267. [Google Scholar] [CrossRef]
- Marti, G.; Eparvier, V.; Moretti, C.; Prado, S.; Grellier, P.; Hue, N.; Thoison, O.; Delpech, B.; Gueritte, F.; Litaudon, M. Antiplasmodial benzophenone derivatives from the root barks of Symphonia globulifera (Clusiaceae). Phytochemistry 2010, 71, 964–974. [Google Scholar] [CrossRef]
- Yang, X.W.; Li, M.M.; Liu, X.; Ferreira, D.; Ding, Y.; Zhang, J.J.; Liao, Y.; Qin, H.B.; Xu, G. Polycyclic Polyprenylated Acylphloroglucinol Congeners Possessing Diverse Structures from Hypericum henryi. J. Nat. Prod. 2015, 78, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Behera, A.K.; Swamy, M.M.; Natesh, N.; Kundu, T.K. Garcinol and Its Role in Chronic Diseases. Adv. Exp. Med. Biol. 2016, 928, 435–452. [Google Scholar] [PubMed]
- Parasramka, M.A.; Gupta, S.V. Synergistic effect of garcinol and curcumin on antiproliferative and apoptotic activity in pancreatic cancer cells. J. Oncol. 2012, 2012, 709739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, F.H.; Pardo-Andreu, G.L.; Nunez-Figueredo, Y.; Cuesta-Rubio, O.; Marin-Prida, J.; Uyemura, S.A.; Curti, C.; Alberici, L.C. Clusianone, a naturally occurring nemorosone regioisomer, uncouples rat liver mitochondria and induces HepG2 cell death. Chem. Biol. Interact 2014, 212, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Ho, P.C.; Wong, F.C.; Sethi, G.; Wang, L.Z.; Goh, B.C. Garcinol: Current status of its anti-oxidative, anti-inflammatory and anti-cancer effects. Cancer Lett. 2015, 362, 8–14. [Google Scholar] [CrossRef]
- Avalle, L.; Camporeale, A.; Camperi, A.; Poli, V. STAT3 in cancer: A double edged sword. Cytokine 2017, 98, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Chen, Z.; Zhang, L.; Huang, Z.; Chen, Y.; Xu, J.; Zhang, J.; Shu, X. Screening and biological evaluation of a novel STAT3 signaling pathway inhibitor against cancer. Bioorg. Med. Chem. Lett. 2016, 26, 5172–5176. [Google Scholar] [CrossRef] [PubMed]
- Siveen, K.S.; Sikka, S.; Surana, R.; Dai, X.; Zhang, J.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim. Biophys. Acta 2014, 1845, 136–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sethi, G.; Chatterjee, S.; Rajendran, P.; Li, F.; Shanmugam, M.K.; Wong, K.F.; Kumar, A.P.; Senapati, P.; Behera, A.K.; Hui, K.M.; et al. Inhibition of STAT3 dimerization and acetylation by garcinol suppresses the growth of human hepatocellular carcinoma in vitro and in vivo. Mol. Cancer 2014, 13, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]

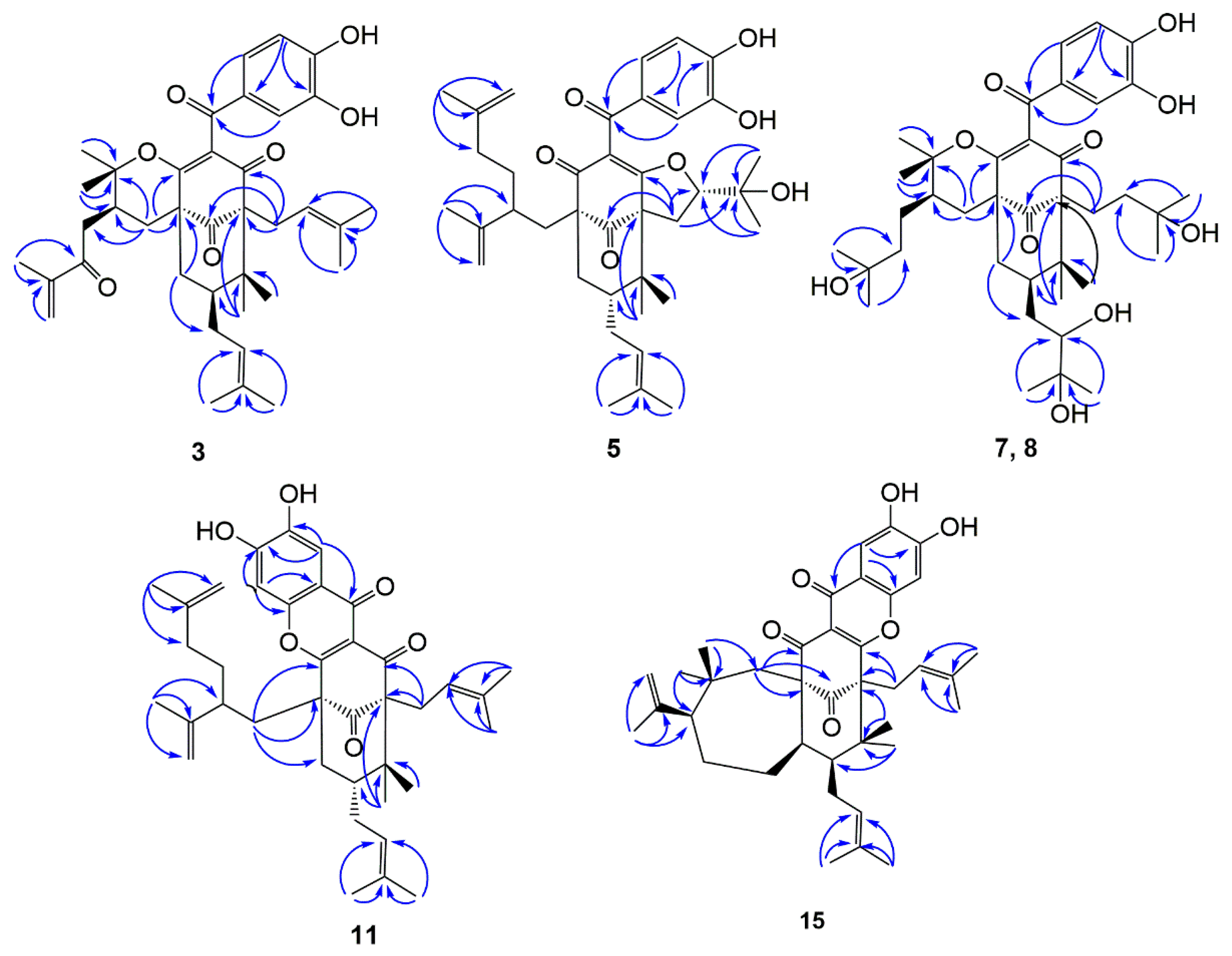
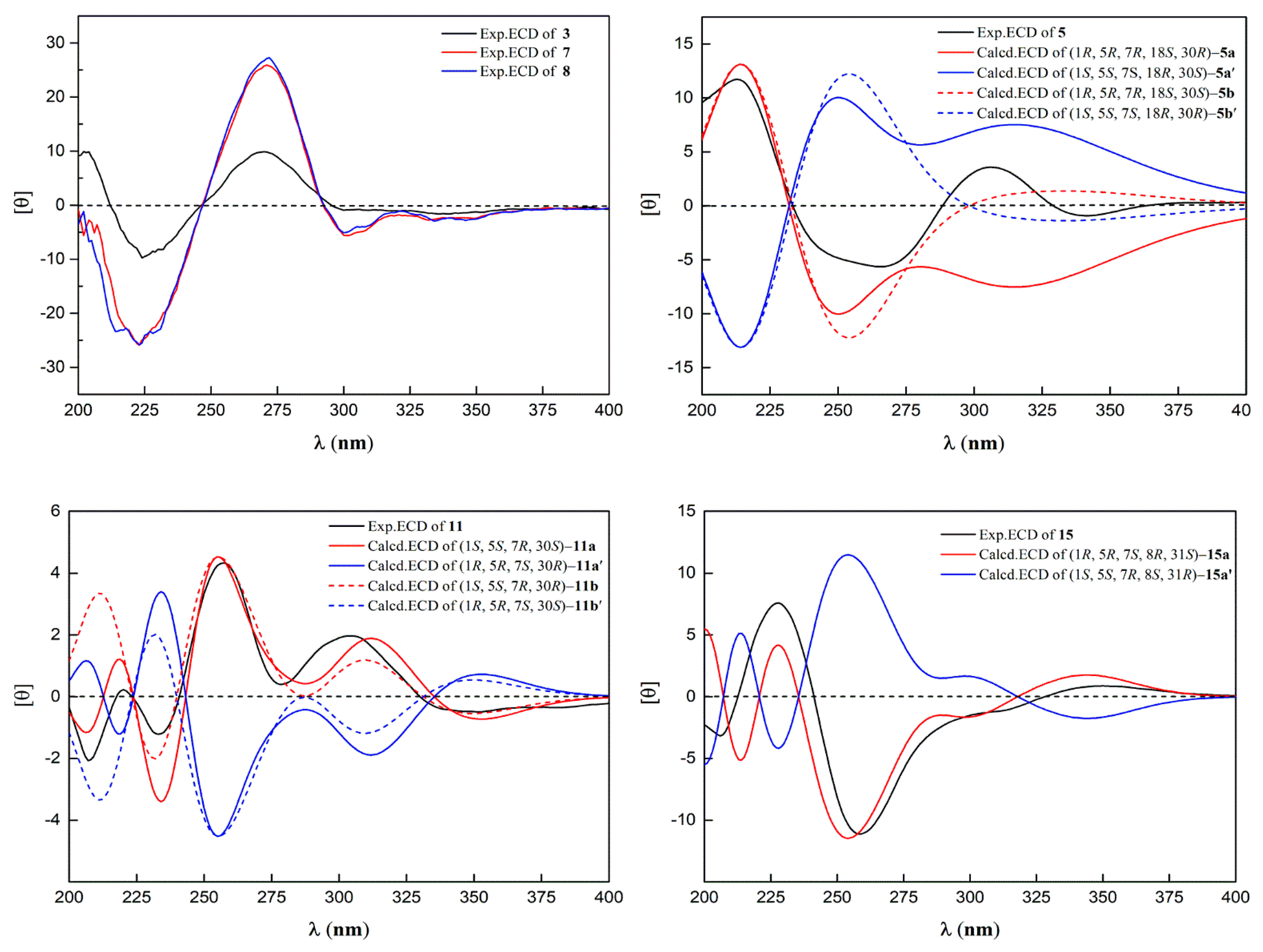
| Position | 3 a | 5 b | 7 c | 8 c | 11 b | 15 b |
|---|---|---|---|---|---|---|
| 7 | 1.52 m | 2.40 m | 1.82 m | 1.83 m | 1.91 m | 1.56 m |
| 8 | 2.33 d (14.5) 2.02 m | 2.33 m 1.63 m | 2.28 d (14.5) 2.20 m | 2.44 d (14.0) 2.35 dd (14.5, 6.5) | 2.42 dd (13.5, 4.0) 1.74 m | 2.11 m |
| 12 | 7.39 d (2.0) | 8.18 d (2.0) | 7.22 d (2.0) | 7.23 d (2.0) | 8.11 s | 8.10 s |
| 15 | 6.84 d (8.0) | 7.29 d (8.0) | 6.76 d (8.0) | 6.77 d (8.0) | 7.26 s | 7.45 s |
| 16 | 7.14 dd (8.0, 2.0) | 7.88 dd (8.0, 2.0) | 7.10 dd (8.0, 2.0) | 7.11 dd (8.0, 2.0) | ||
| 17 | 2.63 dd (13.6, 8.0) 2.43 m | 3.21 dd (14.0, 8.0) 2.48 dd (14.0, 7.5) | 1.89 m | 1.88 m | 2.97 dd (13.5, 6.5) 2.86 dd (13.5, 6.5) | 3.02 m |
| 18 | 5.01 m | 4.82 t (8.0) | 1.43 m 1.08 m | 1.44 m 1.10 m | 5.20 br s | 5.04 br s |
| 20 | 1.57 s | 1.34 s | 1.16 s | 1.17 s | 1.85 s | 1.37 s |
| 21 | 1.56 s | 1.35 s | 1.17 s | 1.18 s | 1.55 s | 1.78 s |
| 22 | 0.97 s | 0.95 s | 1.02 s | 1.02 s | 0.85 s | 1.15 s |
| 23 | 1.14 s | 1.18 s | 1.14 s | 1.13 s | 1.21 s | 1.27 s |
| 24 | 2.75 m 1.29 m | 2.21 m 1.75 m | 2.16 m 1.44 m | 2.05 dd (14.5, 5.5) 1.49 m | 2.08 m 1.79 m | 2.09 m 1.97 m |
| 25 | 5.01 m | 5.18 t (6.5) | 3.18 d (10.5) | 3.12 m | 4.92 br s | 5.01 br s |
| 26 | ||||||
| 27 | 1.68 s | 1.58 s | 1.21 s | 1.23 s | 1.51 s | 1.64 s |
| 28 | 1.69 s | 1.62 s | 1.22 s | 1.20 s | 1.51 s | 1.51 s |
| 29 | 3.01 dd (14.0, 4.0) 1.19 dd (14.0, 9.5) | 2.32 m 2.03 m | 3.13 dd (14.0, 3.5) 1.09 m | 3.12 m 1.09 m | 2.36 dd (14.0, 9.5) 2.15 dd (14.0, 2.5) | 2.39 m 2.13 m |
| 30 | 2.10 m | 2.96 m | 1.27 m | 1.26 m | 2.65 m | |
| 31 | 2.48 m | |||||
| 32 | 1.30 s | 4.92 s 4.88 s | 1.29 s | 1.30 s | 4.44 s 4.38 s | 2.23 m 1.91 m |
| 33 | 0.91 s | 1.67 s | 0.95 s | 0.95 s | 1.63 s | 2.29 m 2.21 m |
| 34 | 2.80 m 2.73 m | 1.59 m | 1.52 m 1.09 m | 1.52 m 1.12 m | 1.73 m 1.60 m | |
| 35 | 1.94 m | 1.80 m 1.42 m | 1.80 m 1.42 m | 1.91 m | 4.80 br s 4.90 br s | |
| 36 | 1.65 s | |||||
| 37 | 6.09 s 5.88 s | 4.82 s 4.76 s | 1.22 s | 1.20 s | 4.77 s | 1.01 s |
| 38 | 1.89 s | 1.70 s | 1.22 s | 1.22 s | 1.71 s | 1.20 s |
| Position | 3 a | 5 b | 7 c | 8 c | 11 b | 15 b |
|---|---|---|---|---|---|---|
| 1 | 51.4 | 62.5 | 53.0 | 53.3 | 57.9 | 66.3 |
| 2 | 170.9 | 195.2 | 174.0 | 174.4 | 176.8 | 193.1 |
| 3 | 126.3 | 119.9 | 127.1 | 127.3 | 119.8 | 119.2 |
| 4 | 194.3 | 174.5 | 196.8 | 197.1 | 191.9 | 177.0 |
| 5 | 69.0 | 70.2 | 69.6 | 69.8 | 74.4 | 63.6 |
| 6 | 46.7 | 47.4 | 47.7 | 48.8 | 49.2 | 48.8 |
| 7 | 47.0 | 42.0 | 43.5 | 44.7 | 42.9 | 47.3 |
| 8 | 39.9 | 44.2 | 40.3 | 46.1 | 44.3 | 38.2 |
| 9 | 206.9 | 207.6 | 208.2 | 208.3 | 207.9 | 207.7 |
| 10 | 192.3 | 191.7 | 194.8 | 194.8 | 172.4 | 172.8 |
| 11 | 131.3 | 131.0 | 131.0 | 131.2 | 118.7 | 118.0 |
| 12 | 115.9 | 117.1 | 116.7 | 116.7 | 110.0 | 109.6 |
| 13 | 146.0 | 147.9 | 146.8 | 146.8 | 149.8 | 146.9 |
| 14 | 151.3 | 153.8 | 153.1 | 152.9 | 154.8 | 155.9 |
| 15 | 115.6 | 116.2 | 116.0 | 116.0 | 104.5 | 104.5 |
| 16 | 123.6 | 125.2 | 124.4 | 124.3 | 147.6 | 150.6 |
| 17 | 26.4 | 26.6 | 22.8 | 22.5 | 26.2 | 27.4 |
| 18 | 121.6 | 93.7 | 39.8 | 39.7 | 122.0 | 119.9 |
| 19 | 134.2 | 71.5 | 71.9 | 72.0 | 133.6 | 135.3 |
| 20 | 26.4 | 25.9 | 29.2 | 29.2 | 26.4 | 26.2 |
| 21 | 18.3 | 26.1 | 29.2 | 29.3 | 18.8 | 18.8 |
| 22 | 27.0 | 16.4 | 27.2 | 27.1 | 16.4 | 26.9 |
| 23 | 22.8 | 23.4 | 23.1 | 22.8 | 23.7 | 22.4 |
| 24 | 29.8 | 29.4 | 33.5 | 37.4 | 28.7 | 30.1 |
| 25 | 126.4 | 123.5 | 78.6 | 83.2 | 123.5 | 124.6 |
| 26 | 133.3 | 133.8 | 74.4 | 74.5 | 134.0 | 135.3 |
| 27 | 26.2 | 26.3 | 24.6 | 24.6 | 26.1 | 26.3 |
| 28 | 18.6 | 18.2 | 25.9 | 25.8 | 18.0 | 18.2 |
| 29 | 29.3 | 37.1 | 29.2 | 28.9 | 36.5 | 40.0 |
| 30 | 38.9 | 44.0 | 45.0 | 45.1 | 44.4 | 41.5 |
| 31 | 86.0 | 148.1 | 89.2 | 89.4 | 147.8 | 49.4 |
| 32 | 22.2 | 114.6 | 21.7 | 21.7 | 113.0 | 29.4 |
| 33 | 29.0 | 18.3 | 29.1 | 29.1 | 18.2 | 31.8 |
| 34 | 39.7 | 32.4 | 26.6 | 26.6 | 32.6 | 146.9 |
| 35 | 199.8 | 36.4 | 42.8 | 42.8 | 35.9 | 109.6 |
| 36 | 145.5 | 146.6 | 71.4 | 71.4 | 146.1 | 24.3 |
| 37 | 125.5 | 110.4 | 29.3 | 29.3 | 111.0 | 24.7 |
| 38 | 17.9 | 23.1 | 29.7 | 29.7 | 22.9111.0 | 25.7 |
| Compound | IC50 in μmol·L−1 | |||
|---|---|---|---|---|
| SGC7901 | A549 | Hep G2 | MCF-7 | |
| Doxorubicin a | 7.54 ± 1.11 | 14.03 ± 0.21 | 6.52 ± 0.13 | 4.40 ± 1.17 |
| 1 | 13.96 ± 2.88 | 6.06 ± 0.04 | 9.60 ± 0.39 | 27.33 ± 0.22 |
| 2 | 11.41 ± 1.22 | 7.06 ± 0.20 | 9.55 ± 0.63 | 29.41 ± 0.85 |
| 3 | 11.00 ± 1.96 | 10.66 ± 1.85 | 14.45 ± 1.06 | 25.85 ± 4.07 |
| 4 | 23.57 ± 1.06 | 11.95 ± 0.01 | 17.84 ± 0.06 | 25.27 ± 1.53 |
| 5 | 14.22 ± 0.47 | 8.29 ± 0.31 | 9.22 ± 0.81 | 21.77 ± 0.46 |
| 6 | 15.26 ± 2.26 | 4.55 ± 0.34 | 8.62 ± 0.32 | 16.93 ± 0.41 |
| 7 | >50 | >50 | >50 | >50 |
| 8 | >50 | >50 | >50 | >50 |
| 9 | 19.08 ± 3.71 | 21.44 ± 2.12 | 19.09 ± 1.97 | 22.29 ± 0.12 |
| 10 | 17.58 ± 0.92 | 19.28 ± 3.58 | 18.23 ± 4.18 | 25.93 ± 0.76 |
| 11 | >50 | >50 | >50 | >50 |
| 12 | 2.83 ± 0.66 | 0.89 ± 0.26 | 4.91 ± 1.61 | 3.28 ± 2.17 |
| 13 | 18.53 ± 6.46 | 9.17 ± 2.10 | >50 | >50 |
| 14 | 36.98 ± 9.72 | 32.76 ± 0.41 | >50 | >50 |
| 15 | >50 | >50 | >50 | >50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, S.; Wang, W.; Gan, F.; Xie, W.; Xu, J.; Chen, Y.; Mei, Z.; Yang, G. Discovery of Novel Polycyclic Polyprenylated Acylphloroglucinols from the Fruits of Garcinia xanthochymus as Antitumor Agents by Suppressing the STAT3 Signaling. Int. J. Mol. Sci. 2021, 22, 10365. https://doi.org/10.3390/ijms221910365
Jin S, Wang W, Gan F, Xie W, Xu J, Chen Y, Mei Z, Yang G. Discovery of Novel Polycyclic Polyprenylated Acylphloroglucinols from the Fruits of Garcinia xanthochymus as Antitumor Agents by Suppressing the STAT3 Signaling. International Journal of Molecular Sciences. 2021; 22(19):10365. https://doi.org/10.3390/ijms221910365
Chicago/Turabian StyleJin, Shan, Wen Wang, Fei Gan, Wenli Xie, Jing Xu, Yu Chen, Zhinan Mei, and Guangzhong Yang. 2021. "Discovery of Novel Polycyclic Polyprenylated Acylphloroglucinols from the Fruits of Garcinia xanthochymus as Antitumor Agents by Suppressing the STAT3 Signaling" International Journal of Molecular Sciences 22, no. 19: 10365. https://doi.org/10.3390/ijms221910365
APA StyleJin, S., Wang, W., Gan, F., Xie, W., Xu, J., Chen, Y., Mei, Z., & Yang, G. (2021). Discovery of Novel Polycyclic Polyprenylated Acylphloroglucinols from the Fruits of Garcinia xanthochymus as Antitumor Agents by Suppressing the STAT3 Signaling. International Journal of Molecular Sciences, 22(19), 10365. https://doi.org/10.3390/ijms221910365







