GmFULc Is Induced by Short Days in Soybean and May Accelerate Flowering in Transgenic Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
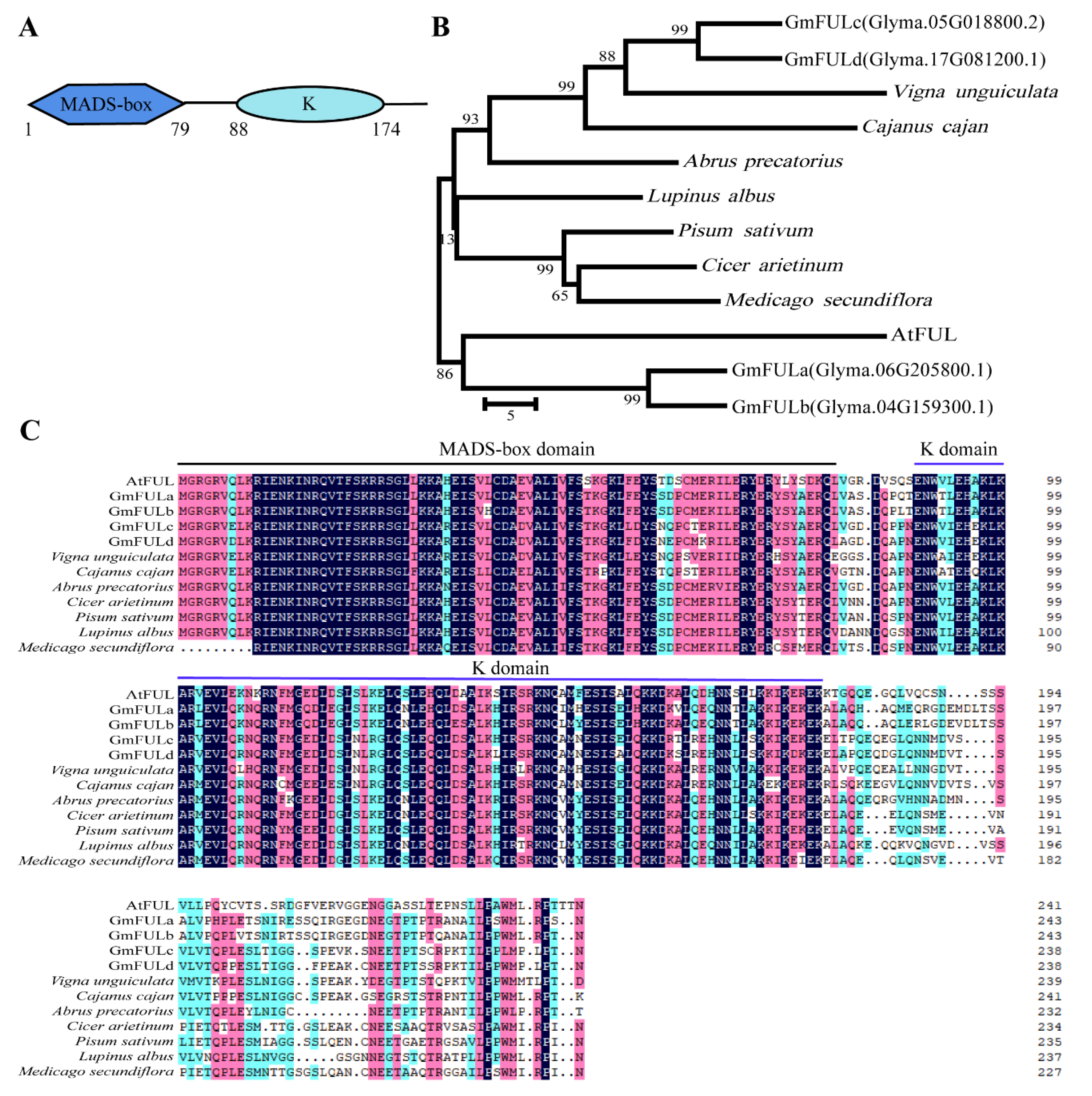
2.1. Sequence Analysis of the GmFULc
2.2. Photoperiod and the Circadian Clock Regulate the Expression of GmFULc
2.3. The tissue-specific Expression Patterns of GmFULc in Soybean
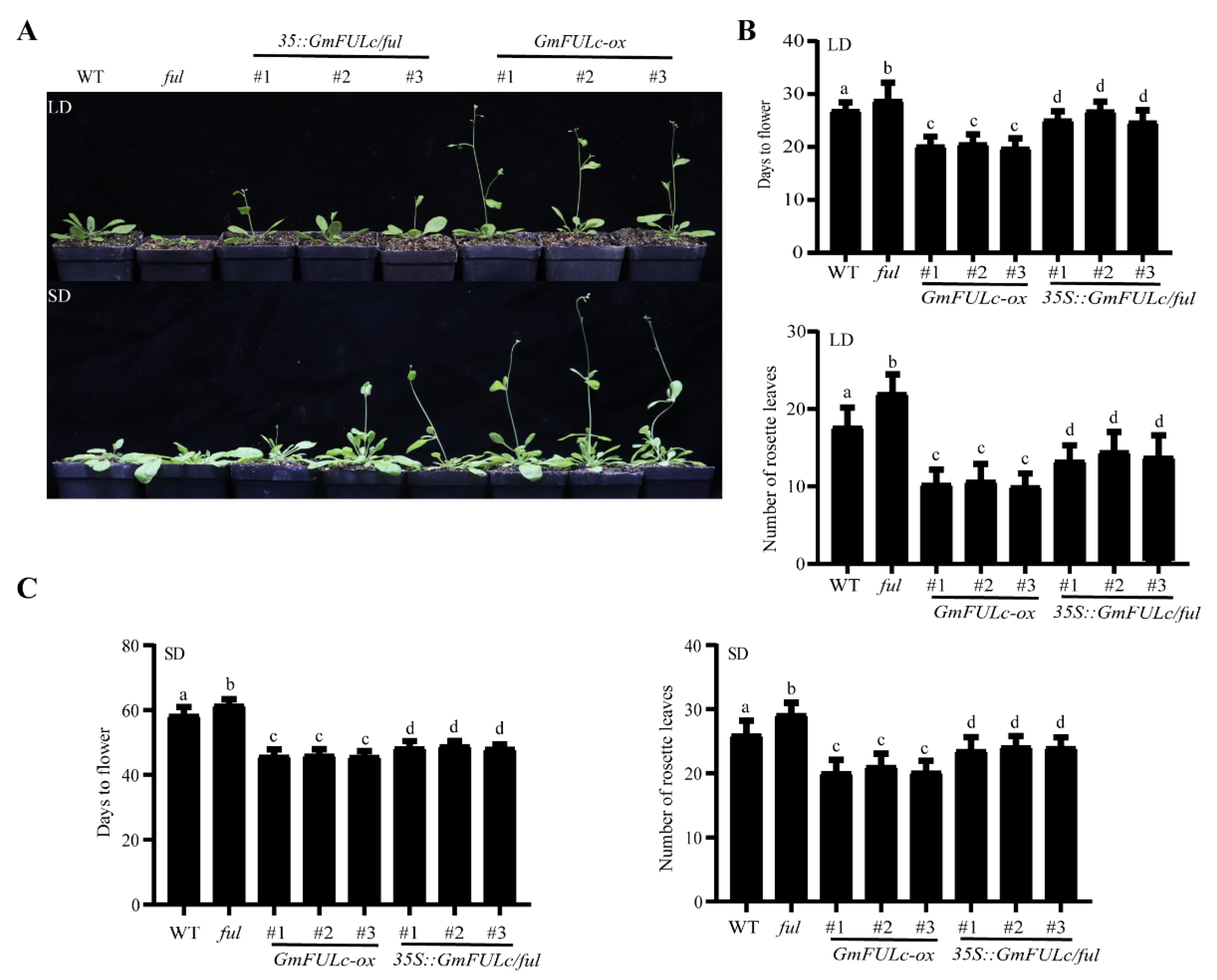
2.4. Overexpression of GmFULc May Accelerate Flowering in Transgenic Arabidopsis
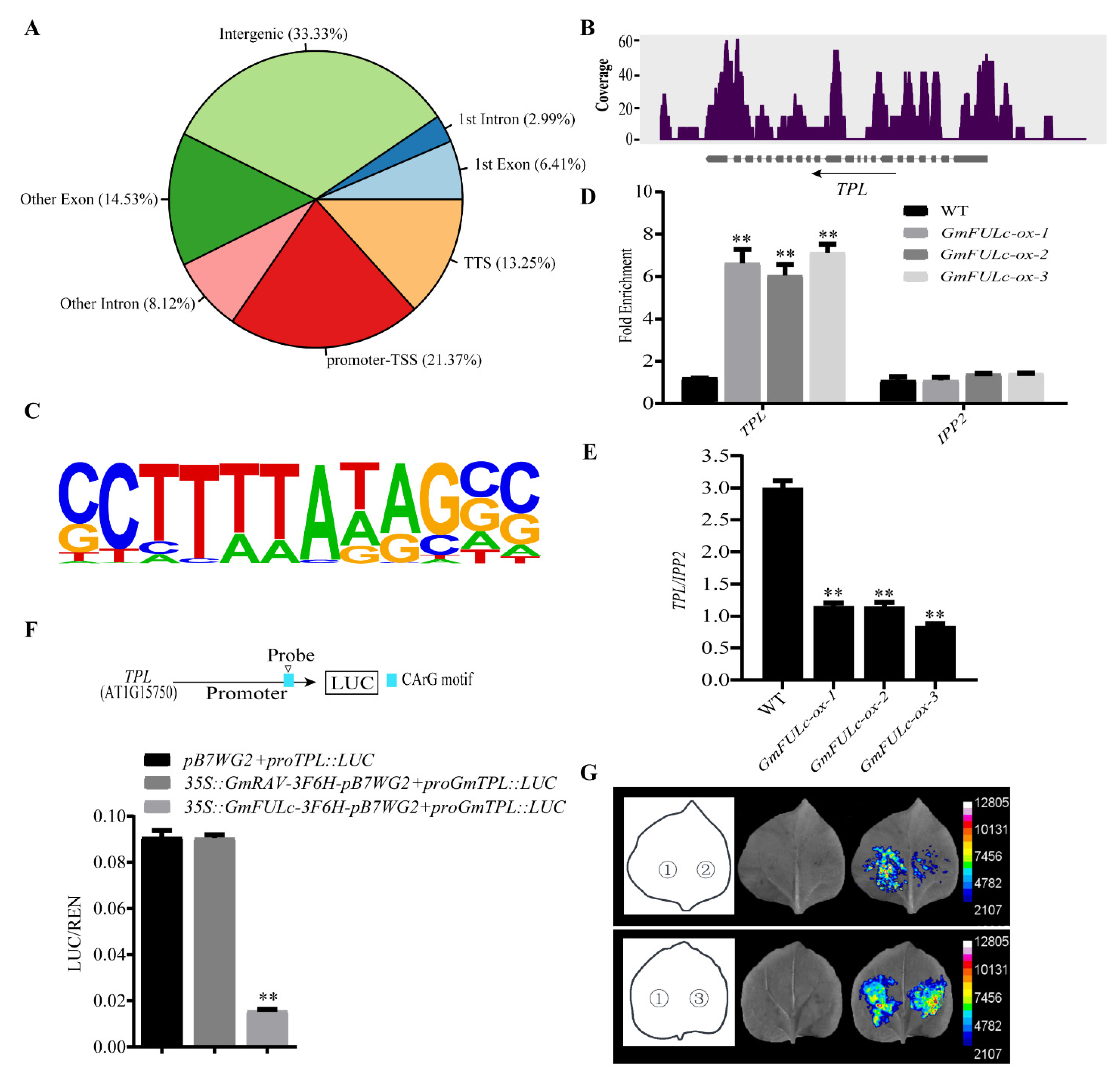
2.5. Chromatin Immunoprecipitation Sequencing Assays of GmFULc-Target Genes
2.6. GmFULc Inhibits the Transcriptional Levels of TPL
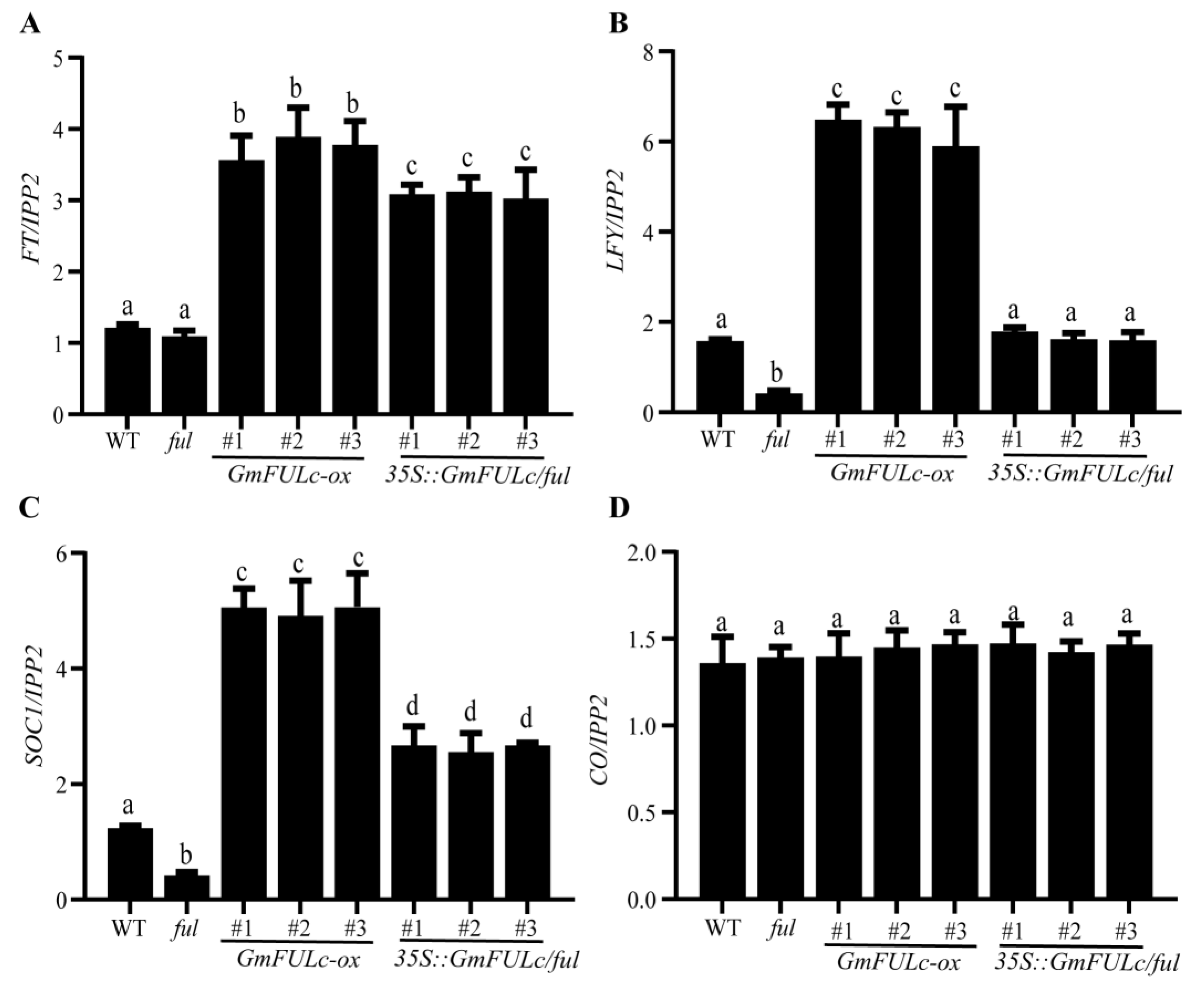
2.7. GmFULc Affects the Transcriptional Levels of Flowering Time Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Records of Data
4.2. Plasmid Construction and Generation of Transgenic Arabidopsis Plants
4.3. ChIP-Seq and ChIP-qPCR
4.4. Transient Assay of TPL Promoters Affected by GmFULc in N. Benthamiana
4.5. Quantitative RT-PCR Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kantolic, A.G.; Slafer, G.A. Development and seed number in indeterminate soybean as affected by timing and duration of exposure to long photoperiods after flowering. Ann. Bot. 2007, 99, 925–933. [Google Scholar] [CrossRef] [Green Version]
- Cober, E.R.; Morrison, M.J. Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2010, 120, 1005–1012. [Google Scholar] [CrossRef]
- Xu, M.; Xu, Z.; Liu, B.; Kong, F.; Tsubokura, Y.; Watanabe, S.; Xia, Z.; Harada, K.; Kanazawa, A.; Yamada, T.; et al. Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 2013, 13, 91. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gu, Y.; Gao, H.; Qiu, L.; Chang, R.; Chen, S.; He, C. Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 2016, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Gramzow, T. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, AR 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Coenen, H.; Ruelens, P.; Hazarika, R.R.; Al Hindi, T.; Oguis, G.K.; Vandeperre, A.; van Noort, V.; Geuten, K. Resurrected Protein Interaction Networks Reveal the Innovation Potential of Ancient Whole-Genome Duplication. Plant Cell 2018, 30, 2741–2760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Tang, D.; Lin, X.; Ding, M.; Tong, Z. Genome-wide identification of MADS-box family genes in moso bamboo (Phyllostachys edulis) and a functional analysis of PeMADS5 in flowering. BMC Plant Biol. 2018, 18, 176. [Google Scholar] [CrossRef]
- Mandel, M.A. The Arabidopsis AGL8 MADS Box Gene Is Expressed in Inflorescence Meristems and Is Negatively Regulated by APETALA1. Plant Cell Online 1995, 7, 1763–1771. [Google Scholar] [CrossRef] [Green Version]
- Litt, A.; Irish, V.F. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 2003, 165, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [Green Version]
- Torti, S.; Fornara, F.; Vincent, C.; Andres, F.; Nordstrom, K.; Gobel, U.; Knoll, D.; Schoof, H.; Coupland, G. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 2012, 24, 444–462. [Google Scholar] [CrossRef] [Green Version]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [Green Version]
- de Folter, S.; Immink, R.G.; Kieffer, M.; Parenicova, L.; Henz, S.R.; Weigel, D.; Busscher, M.; Kooiker, M.; Colombo, L.; Kater, M.M.; et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 2005, 17, 1424–1433. [Google Scholar] [CrossRef] [Green Version]
- Balanza, V.; Martinez-Fernandez, I.; Ferrandiz, C. Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J. Exp. Bot. 2014, 65, 1193–1203. [Google Scholar] [CrossRef] [Green Version]
- de Folter, S.; Angenent, G.C. trans meets cis in MADS science. Trends Plant Sci. 2006, 11, 224–231. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bemer, M.; Karlova, R.; Ballester, A.R.; Tikunov, Y.M.; Bovy, A.G.; Wolters-Arts, M.; Rossetto Pde, B.; Angenent, G.C.; de Maagd, R.A. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 2012, 24, 4437–4451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisawa, M.; Shima, Y.; Nakagawa, H.; Kitagawa, M.; Kimbara, J.; Nakano, T.; Kasumi, T.; Ito, Y. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 2014, 26, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Shima, Y.; Fujisawa, M.; Kitagawa, M.; Nakano, T.; Kimbara, J.; Nakamura, N.; Shiina, T.; Sugiyama, J.; Nakamura, T.; Kasumi, T.; et al. Tomato FRUITFULL homologs regulate fruit ripening via ethylene biosynthesis. Biosci. Biotechnol. Biochem. 2014, 78, 231–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S.; et al. A functional allele of CsFUL1 regulates fruit length through inhibiting CsSUP and auxin transport in cucumber. Plant Cell 2019. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, T.; Xu, G.; Yang, H.; Zeng, X.; Shen, Y.; Yu, D.; Huang, F. GmAGL1, a MADS-Box Gene from Soybean, Is Involved in Floral Organ Identity and Fruit Dehiscence. Front. Plant Sci. 2017, 8, 175. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, H.; Du, H.; Wang, S.; Yang, W.; Chi, Y.; Wang, J.; Huang, F.; Yu, D. Soybean MADS-box gene GmAGL1 promotes flowering via the photoperiod pathway. BMC Genom. 2018, 19, 51. [Google Scholar] [CrossRef] [Green Version]
- Lyu, J.; Cai, Z.; Li, Y.; Suo, H.; Yi, R.; Zhang, S.; Nian, H. The Floral Repressor GmFLC-like Is Involved in Regulating Flowering Time Mediated by Low Temperature in Soybean. Int. J. Mol. Sci. 2020, 21, 1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Li, M.; Xu, C.; Yang, X.; Li, D.; Zhao, X.; Wang, K.; Li, Y.; Zhang, X.; Liu, L.; et al. Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J. Cell Mol. Biol. 2018, 96, 147–162. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Li, X.; Shan, J.; Li, Y.; Zhang, Y.; Wang, Y.; Li, W.; Zhao, L. Overexpression of GmGAMYB Accelerates the Transition to Flowering and Increases Plant Height in Soybean. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Karmarkar, V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 2008, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goralogia, G.S.; Liu, T.K.; Zhao, L.; Panipinto, P.M.; Groover, E.D.; Bains, Y.S.; Imaizumi, T. CYCLING DOF FACTOR 1 represses transcription through the TOPLESS co-repressor to control photoperiodic flowering in Arabidopsis. Plant J. Cell Mol. Biol. 2017, 92, 244–262. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bemer, M.; van Mourik, H.; Muino, J.M.; Ferrandiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Luo, Q.; Yang, C.; Han, Y.; Li, W. A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 2008, 227, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Ferrario, S.; Busscher, J.; Franken, J.; Gerats, T.; Vandenbussche, M.; Angenent, G.C.; Immink, R.G. Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 2004, 16, 1490–1505. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Duan, X.; Zhang, R.; Fu, X.; Ye, L.; Kong, H.; Xu, G.; Shan, H. Prevalent Exon-Intron Structural Changes in the APETALA1/FRUITFULL, SEPALLATA, AGAMOUS-LIKE6, and FLOWERING LOCUS C MADS-Box Gene Subfamilies Provide New Insights into Their Evolution. Front. Plant Sci. 2016, 7, 598. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Jiang, B.; Gao, X.; Yue, Y.; Fei, Z.; Sun, H.; Wu, C.; Sun, S.; Hou, W.; Han, T. GmFULa, a FRUITFULL homolog, functions in the flowering and maturation of soybean. Plant Cell Rep. 2015, 34, 121–132. [Google Scholar] [CrossRef]
- Teper-Bamnolker, P.; Samach, A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 2005, 17, 2661–2675. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Tao, Q.; Guo, D.; Wei, B.; Zhang, F.; Pang, C.; Jiang, H.; Zhang, J.; Wei, T.; Gu, H.; Qu, L.J.; et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 2013, 25, 421–437. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.; Tan, W.; Yang, H.; Zhang, L.; Li, T.; Liu, B.; Zhang, D.; Lin, H. Regulation of anthocyanin accumulation via MYB75/HAT1/TPL-mediated transcriptional repression. PLoS Genet. 2019, 15, e1007993. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Zhang, Y.; Shan, J.; Sun, J.; Li, D.; Zhang, X.; Li, W.; Zhao, L. GmIDD Is Induced by Short Days in Soybean and May Accelerate Flowering When Overexpressed in Arabidopsis via Inhibiting AGAMOUS-LIKE 18. Front. Plant Sci. 2021, 12, 629069. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.; Bent, A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, N.; Winter, C.M.; Wu, M.F.; Kwon, C.S.; William, D.A.; Wagner, D. PROTOCOLS: Chromatin Immunoprecipitation from Arabidopsis Tissues. Arab. Book 2014, 12, e0170. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Wang, Z.; Lu, Q.; Wang, P.; Li, Y.; Lv, Q.; Song, X.; Li, D.; Gu, Y.; Liu, L.; et al. Overexpression of a GmGBP1 ortholog of soybean enhances the responses to flowering, stem elongation and heat tolerance in transgenic tobaccos. Plant Mol. Biol. 2013, 82, 279–299. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP Protein Mediating Signals from the Floral Pathway Integrator FT at the Shoot Apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Wang, M.; Zhao, C.; Liu, T.; Liu, Z.; Fan, Y.; Xue, Y.; Li, W.; Zhang, X.; Zhao, L. GmFULc Is Induced by Short Days in Soybean and May Accelerate Flowering in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 10333. https://doi.org/10.3390/ijms221910333
Sun J, Wang M, Zhao C, Liu T, Liu Z, Fan Y, Xue Y, Li W, Zhang X, Zhao L. GmFULc Is Induced by Short Days in Soybean and May Accelerate Flowering in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(19):10333. https://doi.org/10.3390/ijms221910333
Chicago/Turabian StyleSun, Jingzhe, Mengyuan Wang, Chuanlin Zhao, Tianmeng Liu, Zhengya Liu, Yuhuan Fan, Yongguo Xue, Wenbin Li, Xiaoming Zhang, and Lin Zhao. 2021. "GmFULc Is Induced by Short Days in Soybean and May Accelerate Flowering in Transgenic Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 19: 10333. https://doi.org/10.3390/ijms221910333
APA StyleSun, J., Wang, M., Zhao, C., Liu, T., Liu, Z., Fan, Y., Xue, Y., Li, W., Zhang, X., & Zhao, L. (2021). GmFULc Is Induced by Short Days in Soybean and May Accelerate Flowering in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences, 22(19), 10333. https://doi.org/10.3390/ijms221910333




