The Dynamic Network of RNP RNase P Subunits
Abstract
1. The Discovery of Different RNase P Facets
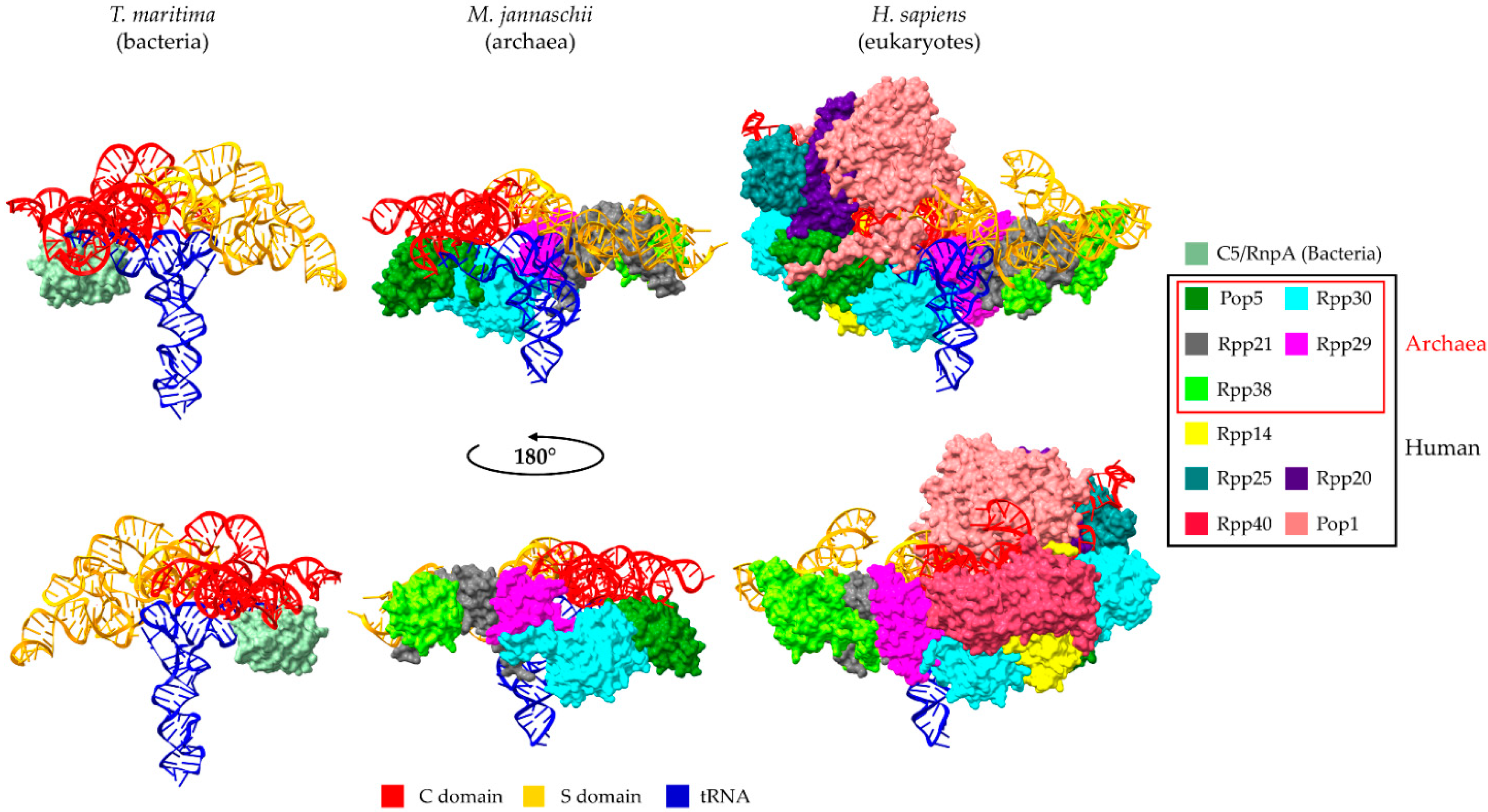
2. Universal Recognition of pre-tRNAs by RNase P Holoenzymes from a Structural Point of View
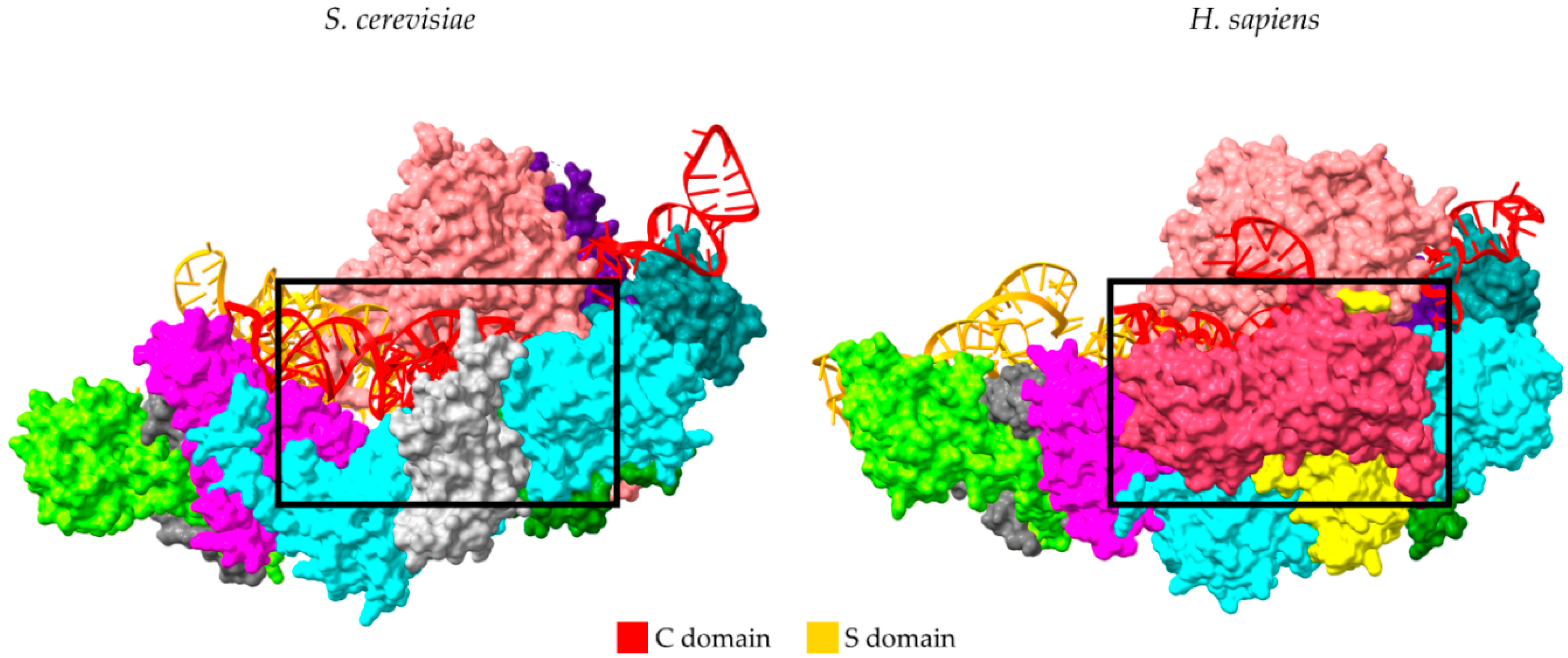
3. The Protein Subunits of Eukaryotic RNase P RNPs and Their Dynamic Networks
3.1. Rpp14
3.2. Pop5
3.3. Rpp20
3.4. Rpp21
3.5. Rpp25
3.6. Rpp29
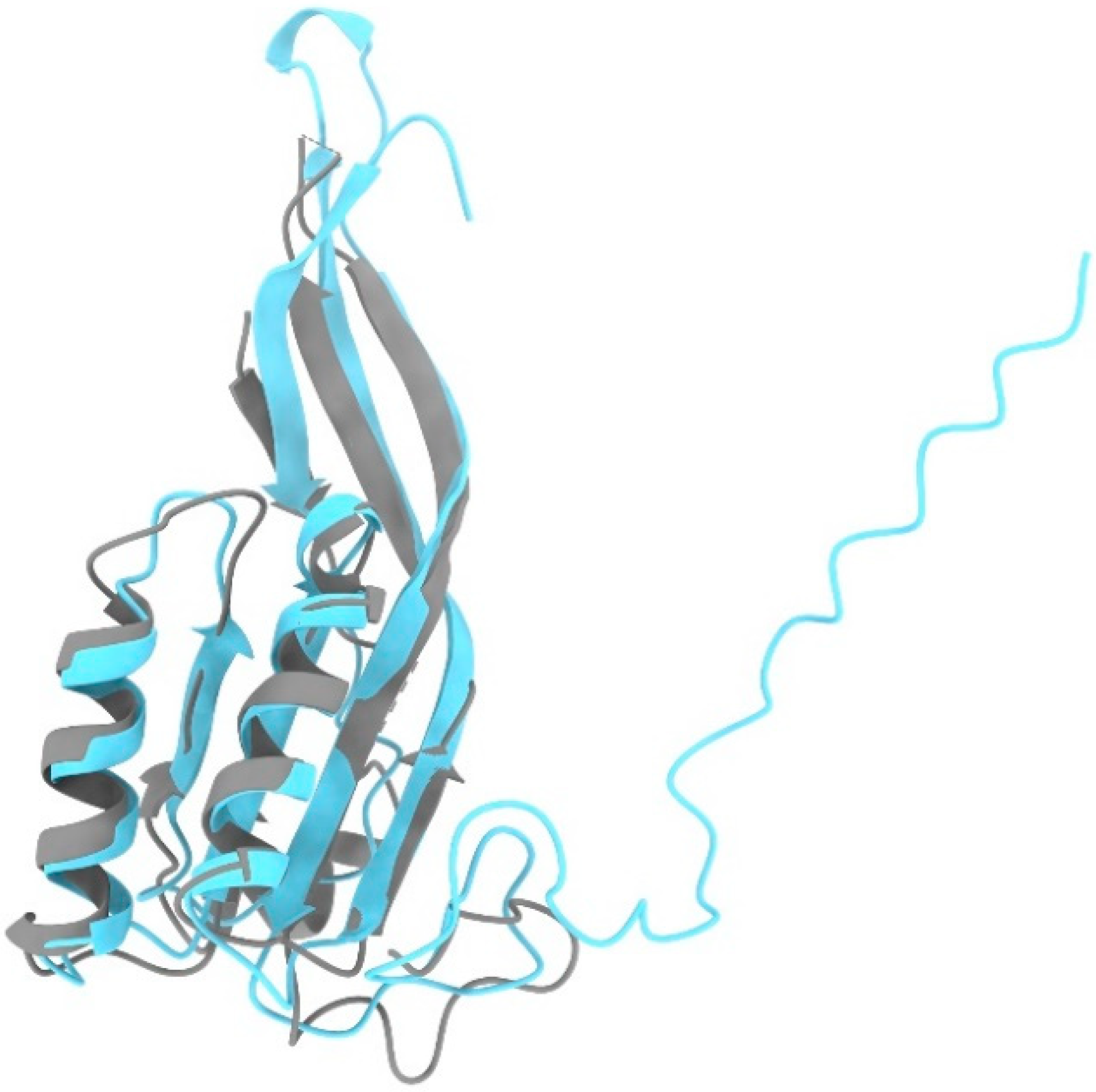
3.7. Rpp30
3.8. Rpp38
3.9. Rpp40
3.10. Pop1
4. Sub-Complexes of RNase P Protein Subunits
4.1. Rpp20 and Rpp25 Contribution to RNP Evolution
4.2. Rpp20–Rpp25–Pop1–Pop4 Couple Transcription of RPR with RNase P Holoenzyme assembly
4.3. Rpp21–Rpp29 and Their Role in RNase P Activity and Beyond
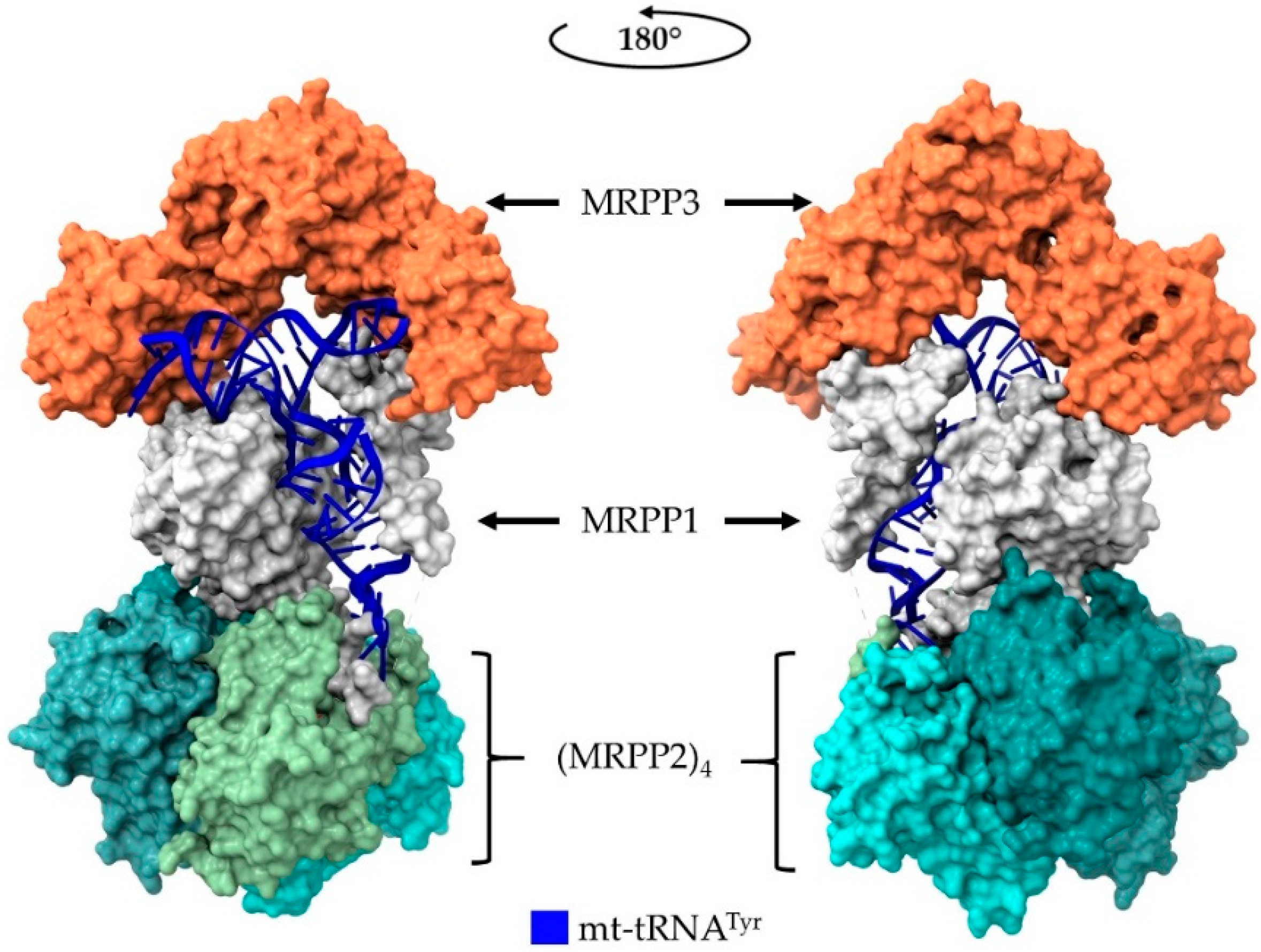
5. Human Mitochondrial RNase P
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA Moiety of Ribonuclease P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Gray, M.W.; Gopalan, V. Piece by Piece: Building a Ribozyme. J. Biol. Chem. 2020, 295, 2313–2323. [Google Scholar] [CrossRef]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragón, A. Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with TRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef]
- Wu, J.; Niu, S.; Tan, M.; Huang, C.; Li, M.; Song, Y.; Wang, Q.; Chen, J.; Shi, S.; Lan, P.; et al. Cryo-EM Structure of the Human Ribonuclease P Holoenzyme. Cell 2018, 175, 1393–1404.e11. [Google Scholar] [CrossRef]
- Wan, F.; Wang, Q.; Tan, J.; Tan, M.; Chen, J.; Shi, S.; Lan, P.; Wu, J.; Lei, M. Cryo-Electron Microscopy Structure of an Archaeal Ribonuclease P Holoenzyme. Nat. Commun. 2019, 10, 2617. [Google Scholar] [CrossRef]
- Kouzuma, Y.; Mizoguchi, M.; Takagi, H.; Fukuhara, H.; Tsukamoto, M.; Numata, T.; Kimura, M. Reconstitution of Archaeal Ribonuclease P from RNA and Four Protein Components. Biochem. Biophys. Res. Commun. 2003, 306, 666–673. [Google Scholar] [CrossRef]
- Li, K.; Williams, R.S. Cloning and Characterization of Three New Murine Genes Encoding Short Homologues of RNase P RNA. J. Biol. Chem. 1995, 270, 25281. [Google Scholar] [CrossRef]
- Wang, M.J.; Davis, N.W.; Gegenheimer, P. Novel Mechanisms for Maturation of Chloroplast Transfer RNA Precursors. EMBO J. 1988, 7, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Holzmann, J.; Frank, P.; Löffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and Functional Reconstitution of the Human Mitochondrial TRNA Processing Enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, L.; Sridhara, S.; Hällberg, B.M. The MRPP1/MRPP2 Complex Is a TRNA-Maturation Platform in Human Mitochondria. Nucleic Acids Res. 2017, 45, 12469–12480. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Gutmann, B.; Taschner, A.; Gössringer, M.; Holzmann, J.; Hartmann, R.K.; Rossmanith, W.; Giegé, P. A Single Arabidopsis Organellar Protein Has RNase P Activity. Nat. Struct. Mol. Biol. 2010, 17, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Gobert, A.; Giegé, P. PRORP Proteins Support RNase P Activity in Both Organelles and the Nucleus in Arabidopsis. Genes Dev. 2012, 26, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Pinker, F.; Bonnard, G.; Gobert, A.; Gutmann, B.; Hammani, K.; Sauter, C.; Gegenheimer, P.A.; Giegé, P. PPR Proteins Shed a New Light on RNase P Biology. RNA Biol. 2013, 10, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Rossmanith, W.; Hartmann, R.K.; Thölken, C.; Gutmann, B.; Giegé, P.; Gobert, A. Distribution of Ribonucleoprotein and Protein-Only RNase P in Eukarya. Mol. Biol. Evol. 2015, 32, 3186–3193. [Google Scholar] [CrossRef]
- Nickel, A.I.; Wäber, N.B.; Gößringer, M.; Lechner, M.; Linne, U.; Toth, U.; Rossmanith, W.; Hartmann, R.K. Minimal and RNA-Free RNase P in Aquifex Aeolicus. Proc. Natl. Acad. Sci. USA 2017, 114, 11121–11126. [Google Scholar] [CrossRef]
- Peck-Miller, K.A.; Altman, S. Kinetics of the Processing of the Precursor to 4.5 S RNA, a Naturally Occurring Substrate for RNase P from Escherichia Coli. J. Mol. Biol. 1991, 221, 1–5. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3′ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a TRNA-like Cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN/Nuclear-Retained Non-Coding RNAs Are up-Regulated upon Muscle Differentiation and Are Essential Components of Paraspeckles. Genome Res. 2008, 19, 347–359. [Google Scholar] [CrossRef]
- Reiner, R. A Role for the Catalytic Ribonucleoprotein RNase P in RNA Polymerase III Transcription. Genes Dev. 2006, 20, 1621–1635. [Google Scholar] [CrossRef]
- Jarrous, N. Roles of RNase P and Its Subunits. Trends Genet. 2017, 33, 594–603. [Google Scholar] [CrossRef]
- Zhang, J.; Ferré-DAmaré, A. Trying on TRNA for Size: RNase P and the T-Box Riboswitch as Molecular Rulers. Biomolecules 2016, 6, 18. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; McClain, W.H.; Altman, S. Cleavage of TRNA Precursors by the RNA Subunit of E. Coli Ribonuclease P (M1 RNA) Is Influenced by 3′-Proximal CCA in the Substrates. Cell 1984, 38, 219–224. [Google Scholar] [CrossRef]
- Reich, C.; Olsen, G.; Pace, B.; Pace, N. Role of the Protein Moiety of Ribonuclease P, a Ribonucleoprotein Enzyme. Science 1988, 239, 178–181. [Google Scholar] [CrossRef]
- Stathopoulos, C.; Kalpaxis, D.L.; Drainas, D. Partial Purification and Characterization of RNase P from Dictyostelium Discoideum. Eur. J. Biochem. 1995, 228, 976–980. [Google Scholar] [CrossRef]
- Kurz, J.C.; Niranjanakumari, S.; Fierke, C.A. Protein Component of Bacillus subtilis RNase P Specifically Enhances the Affinity for Precursor-TRNA Asp. Biochemistry 1998, 37, 2393–2400. [Google Scholar] [CrossRef]
- Pannucci, J.A.; Haas, E.S.; Hall, T.A.; Harris, J.K.; Brown, J.W. RNase P RNAs from Some Archaea Are Catalytically Active. Proc. Natl. Acad. Sci. USA 1999, 96, 7803–7808. [Google Scholar] [CrossRef] [PubMed]
- Kikovska, E.; Svard, S.G.; Kirsebom, L.A. Eukaryotic RNase P RNA Mediates Cleavage in the Absence of Protein. Proc. Natl. Acad. Sci. USA 2007, 104, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Blewett, N.H.; Maraia, R.J. La Involvement in TRNA and Other RNA Processing Events Including Differences among Yeast and Other Eukaryotes. Biochim. Biophys. Acta BBA—Gene Regul. Mech. 2018, 1861, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.J.; Hall, T.A.; Brown, J.W. Characterization of RNase P Holoenzymes from Methanococcus Jannaschii and Methanothermobacter Thermoautotrophicus. Biol. Chem. 2001, 382, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.K.; Haas, E.S.; Williams, D.; Frank, D.N.; Brown, J.W. New Insight into RNase P RNA Structure from Comparative Analysis of the Archaeal RNA. RNA 2001, 7, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.B.; Chan, P.P.; Cozen, A.E.; Bernick, D.L.; Brown, J.W.; Gopalan, V.; Lowe, T.M. Discovery of a Minimal Form of RNase P in Pyrobaculum. Proc. Natl. Acad. Sci. USA 2010, 107, 22493–22498. [Google Scholar] [CrossRef]
- Tsai, H.-Y.; Pulukkunat, D.K.; Woznick, W.K.; Gopalan, V. Functional Reconstitution and Characterization of Pyrococcus Furiosus RNase P. Proc. Natl. Acad. Sci. USA 2006, 103, 16147–16152. [Google Scholar] [CrossRef]
- Li, S.; Su, Z.; Lehmann, J.; Stamatopoulou, V.; Giarimoglou, N.; Henderson, F.E.; Fan, L.; Pintilie, G.D.; Zhang, K.; Chen, M.; et al. Structural Basis of Amino Acid Surveillance by Higher-Order TRNA-MRNA Interactions. Nat. Struct. Mol. Biol. 2019, 26, 1094–1105. [Google Scholar] [CrossRef]
- Phan, H.-D.; Lai, L.B.; Zahurancik, W.J.; Gopalan, V. The Many Faces of RNA-Based RNase P, an RNA-World Relic. Trends Biochem. Sci. 2021, S0968-0004(21)00162-6. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. Publ. Protein Soc. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Howard, M.J.; Lim, W.H.; Fierke, C.A.; Koutmos, M. Mitochondrial Ribonuclease P Structure Provides Insight into the Evolution of Catalytic Strategies for Precursor-TRNA 5′ Processing. Proc. Natl. Acad. Sci. USA 2012, 109, 16149–16154. [Google Scholar] [CrossRef] [PubMed]
- Karasik, A.; Shanmuganathan, A.; Howard, M.J.; Fierke, C.A.; Koutmos, M. Nuclear Protein-Only Ribonuclease P2 Structure and Biochemical Characterization Provide Insight into the Conserved Properties of TRNA 5′ End Processing Enzymes. J. Mol. Biol. 2016, 428, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Pinker, F.; Schelcher, C.; Fernandez-Millan, P.; Gobert, A.; Birck, C.; Thureau, A.; Roblin, P.; Giegé, P.; Sauter, C. Biophysical Analysis of Arabidopsis Protein-Only RNase P Alone and in Complex with TRNA Provides a Refined Model of TRNA Binding. J. Biol. Chem. 2017, 292, 13904–13913. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Pinker, F.; Fuchsbauer, O.; Gutmann, B.; Boutin, R.; Roblin, P.; Sauter, C.; Giegé, P. Structural Insights into Protein-Only RNase P Complexed with TRNA. Nat. Commun. 2013, 4, 1353. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, T.; Kaitany, K.J.; Kakuta, Y.; Kimura, M.; Fierke, C.A.; Hall, T.M.T. Pentatricopeptide Repeats of Protein-Only RNase P Use a Distinct Mode to Recognize Conserved Bases and Structural Elements of Pre-TRNA. Nucleic Acids Res. 2020, 48, 11815–11826. [Google Scholar] [CrossRef] [PubMed]
- Klemm, B.P.; Karasik, A.; Kaitany, K.J.; Shanmuganathan, A.; Henley, M.J.; Thelen, A.Z.; Dewar, A.J.L.; Jackson, N.D.; Koutmos, M.; Fierke, C.A. Molecular Recognition of Pre-TRNA by Arabidopsis Protein-Only Ribonuclease P. RNA 2017, 23, 1860–1873. [Google Scholar] [CrossRef]
- Feyh, R.; Waeber, N.B.; Prinz, S.; Giammarinaro, P.I.; Bange, G.; Hochberg, G.; Hartmann, R.K.; Altegoer, F. Structure and Mechanistic Features of the Prokaryotic Minimal RNase P. eLife 2021, 10, 70160. [Google Scholar] [CrossRef]
- Teramoto, T.; Koyasu, T.; Adachi, N.; Kawasaki, M.; Moriya, T.; Numata, T.; Senda, T.; Kakuta, Y. Minimal Protein-Only RNase P Structure Reveals Insights into TRNA Precursor Recognition and Catalysis. J. Biol. Chem. 2021, 297, 101028. [Google Scholar] [CrossRef]
- Bhatta, A.; Dienemann, C.; Cramer, P.; Hillen, H.S. Structural Basis of RNA Processing by Human Mitochondrial RNase P. Nat. Struct. Mol. Biol. 2021, 28, 713–723. [Google Scholar] [CrossRef]
- Reinhard, L.; Sridhara, S.; Hallberg, B.M. Structure of the Nuclease Subunit of Human Mitochondrial RNase P. Nucleic Acids Res. 2015, 43, 5664–5672. [Google Scholar] [CrossRef]
- Jiang, T.; Altman, S. A Protein Subunit of Human RNase P, Rpp14, and Its Interacting Partner, OIP2, Have 3′→5′ Exoribonuclease Activity. Proc. Natl. Acad. Sci. USA 2002, 99, 5295–5300. [Google Scholar] [CrossRef]
- Li, Y.; Altman, S. A Subunit of Human Nuclear RNase P Has ATPase Activity. Proc. Natl. Acad. Sci. USA 2001, 98, 441–444. [Google Scholar] [CrossRef]
- Van Eenennaam, H.; Vogelzangs, J.H.P.; Lugtenberg, D.; Van Den Hoogen, F.H.J.; Van Venrooij, W.J.; Pruijn, G.J.M. Identity of the RNase MRP- and RNase P-Associated Th/To Autoantigen. Arthritis Rheum. 2002, 46, 3266–3272. [Google Scholar] [CrossRef] [PubMed]
- Reiner, R.; Krasnov-Yoeli, N.; Dehtiar, Y.; Jarrous, N. Function and Assembly of a Chromatin-Associated RNase P That Is Required for Efficient Transcription by RNA Polymerase I. PLoS ONE 2008, 3, e4072. [Google Scholar] [CrossRef] [PubMed]
- Abu-Zhayia, E.R.; Khoury-Haddad, H.; Guttmann-Raviv, N.; Serruya, R.; Jarrous, N.; Ayoub, N. A Role of Human RNase P Subunits, Rpp29 and Rpp21, in Homology Directed-Repair of Double-Strand Breaks. Sci. Rep. 2017, 7, 1002. [Google Scholar] [CrossRef] [PubMed]
- Newhart, A.; Powers, S.L.; Shastrula, P.K.; Sierra, I.; Joo, L.M.; Hayden, J.E.; Cohen, A.R.; Janicki, S.M. RNase P Protein Subunit Rpp29 Represses Histone H3.3 Nucleosome Deposition. Mol. Biol. Cell 2016, 27, 1154–1169. [Google Scholar] [CrossRef]
- Shastrula, P.K.; Lund, P.J.; Garcia, B.A.; Janicki, S.M. Rpp29 Regulates Histone H3.3 Chromatin Assembly through Transcriptional Mechanisms. J. Biol. Chem. 2018, 293, 12360–12377. [Google Scholar] [CrossRef]
- Jagut, M.; Mihaila-Bodart, L.; Molla-Herman, A.; Alin, M.-F.; Lepesant, J.-A.; Huynh, J.-R. A Mosaic Genetic Screen for Genes Involved in the Early Steps of Drosophila Oogenesis. G3 Genes Genomes Genet. 2013, 3, 409–425. [Google Scholar] [CrossRef]
- Molla-Herman, A.; Vallés, A.M.; Ganem-Elbaz, C.; Antoniewski, C.; Huynh, J. TRNA Processing Defects Induce Replication Stress and Chk2-dependent Disruption of PiRNA Transcription. EMBO J. 2015, 34, 3009–3027. [Google Scholar] [CrossRef]
- Molla-Herman, A.; Angelova, M.T.; Ginestet, M.; Carré, C.; Antoniewski, C.; Huynh, J.-R. TRNA Fragments Populations Analysis in Mutants Affecting TRNAs Processing and TRNA Methylation. Front. Genet. 2020, 11, 518949. [Google Scholar] [CrossRef] [PubMed]
- Koenig, M.; Bentow, C.; Satoh, M.; Fritzler, M.J.; Senécal, J.-L.; Mahler, M. Autoantibodies to a Novel Rpp38 (Th/To) Derived B-Cell Epitope Are Specific for Systemic Sclerosis and Associate with a Distinct Clinical Phenotype. Rheumatology 2019, 58, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Eder, P.S.; Wesolowski, D.; Altman, S. Rpp14 and Rpp29, Two Protein Subunits of Human Ribonuclease P. RNA 1999, 5, 153–157. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jarrous, N.; Wolenski, J.S.; Wesolowski, D.; Lee, C.; Altman, S. Localization in the Nucleolus and Coiled Bodies of Protein Subunits of the Ribonucleoprotein Ribonuclease P. J. Cell Biol. 1999, 146, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Tan, M.; Zhang, Y.; Niu, S.; Chen, J.; Shi, S.; Qiu, S.; Wang, X.; Peng, X.; Cai, G.; et al. Structural Insight into Precursor TRNA Processing by Yeast Ribonuclease P. Science 2018, 362, eaat6678. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Altman, S. Protein-Protein Interactions with Subunits of Human Nuclear RNase P. Proc. Natl. Acad. Sci. USA 2001, 98, 920–925. [Google Scholar] [CrossRef]
- Anderson, J.R. Sequence-Specific RNA Binding Mediated by the RNase PH Domain of Components of the Exosome. RNA 2006, 12, 1810–1816. [Google Scholar] [CrossRef]
- Bai, S.; Herrera-Abreu, M.; Rohn, J.L.; Racine, V.; Tajadura, V.; Suryavanshi, N.; Bechtel, S.; Wiemann, S.; Baum, B.; Ridley, A.J. Identification and Characterization of a Set of Conserved and New Regulators of Cytoskeletal Organization, Cell Morphology and Migration. BMC Biol. 2011, 9, 54. [Google Scholar] [CrossRef]
- Rosenblad, M.A.; López, M.D.; Piccinelli, P.; Samuelsson, T. Inventory and Analysis of the Protein Subunits of the Ribonucleases P and MRP Provides Further Evidence of Homology between the Yeast and Human Enzymes. Nucleic Acids Res. 2006, 34, 5145–5156. [Google Scholar] [CrossRef] [PubMed]
- van Eenennaam, H.; Lugtenberg, D.; Vogelzangs, J.H.P.; van Venrooij, W.J.; Pruijn, G.J.M. HPop5, a Protein Subunit of the Human RNase MRP and RNase P Endoribonucleases. J. Biol. Chem. 2001, 276, 31635–31641. [Google Scholar] [CrossRef] [PubMed]
- Welting, T.J.M.; Peters, F.M.A.; Hensen, S.M.M.; van Doorn, N.L.; Kikkert, B.J.; Raats, J.M.H.; van Venrooij, W.J.; Pruijn, G.J.M. Heterodimerization Regulates RNase MRP/RNase P Association, Localization, and Expression of Rpp20 and Rpp25. RNA 2006, 13, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Stolc, V.; Katz, A.; Altman, S. Rpp2, an Essential Protein Subunit of Nuclear RNase P, Is Required for Processing of Precursor TRNAs and 35S Precursor RRNA in Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 1998, 95, 6716–6721. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Zhou, J. Rpp20 Interacts with SMN and Is Re-Distributed into SMN Granules in Response to Stress. Biochem. Biophys. Res. Commun. 2004, 314, 268–276. [Google Scholar] [CrossRef]
- Alderson, T.R.; Roche, J.; Gastall, H.Y.; Dias, D.M.; Pritišanac, I.; Ying, J.; Bax, A.; Benesch, J.L.P.; Baldwin, A.J. Local Unfolding of the HSP27 Monomer Regulates Chaperone Activity. Nat. Commun. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N.; Reiner, R.; Wesolowski, D.; Mann, H.; Guerrier-Takada, C.; Altman, S. Function and Subnuclear Distribution of Rpp21, a Protein Subunit of the Human Ribonucleoprotein Ribonuclease P. RNA 2001, 7, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Serruya, R.; Orlovetskie, N.; Reiner, R.; Dehtiar-Zilber, Y.; Wesolowski, D.; Altman, S.; Jarrous, N. Human RNase P Ribonucleoprotein Is Required for Formation of Initiation Complexes of RNA Polymerase III. Nucleic Acids Res. 2015, 43, 5442–5450. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Armache, A.; Yang, S.; Martínez de Paz, A.; Robbins, L.E.; Durmaz, C.; Cheong, J.Q.; Ravishankar, A.; Daman, A.W.; Ahimovic, D.J.; Klevorn, T.; et al. Histone H3.3 Phosphorylation Amplifies Stimulation-Induced Transcription. Nature 2020, 583, 852–857. [Google Scholar] [CrossRef]
- Jarrous, N. Human Ribonuclease P: Subunits, Function, and Intranuclear Localization. RNA 2002, 8, 1–7. [Google Scholar] [CrossRef][Green Version]
- Stolc, V.; Altman, S. Rpp1, an Essential Protein Subunit of Nuclear RNase P Required for Processing of Precursor TRNA and 35S Precursor RRNA in Saccharomyces Cerevisiae. Genes Dev. 1997, 11, 2926–2937. [Google Scholar] [CrossRef]
- Wang, S.-Q.; Shi, D.-Q.; Long, Y.-P.; Liu, J.; Yang, W.-C. GAMETOPHYTE DEFECTIVE 1, a Putative Subunit of RNases P/MRP, Is Essential for Female Gametogenesis and Male Competence in Arabidopsis. PLoS ONE 2012, 7, e33595. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K. TRNA Transfers to the Limelight. Genes Dev. 2003, 17, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Oshima, K.; Gao, X.; Hayashi, S.; Ueda, T.; Nakashima, T.; Kimura, M. Crystal Structures of the Archaeal RNase P Protein Rpp38 in Complex with RNA Fragments Containing a K-Turn Motif. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kovrigina, E.; Wesolowski, D.; Altman, S. Coordinate Inhibition of Expression of Several Genes for Protein Subunits of Human Nuclear RNase P. Proc. Natl. Acad. Sci. USA 2003, 100, 1598–1602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Altman, S. Inhibition of the Expression of the Human RNase P Protein Subunits Rpp21, Rpp25, Rpp29 by External Guide Sequences (EGSs) and SiRNA. J. Mol. Biol. 2004, 342, 1077–1083. [Google Scholar] [CrossRef]
- Lygerou, Z.; Pluk, H.; van Venrooij, W.J.; Séraphin, B. HPop1: An Autoantigenic Protein Subunit Shared by the Human RNase P and RNase MRP Ribonucleoproteins. EMBO J. 1996, 15, 5936–5948. [Google Scholar] [CrossRef]
- Lygerou, Z.; Mitchell, P.; Petfalski, E.; Seraphin, B.; Tollervey, D. The POP1 Gene Encodes a Protein Component Common to the RNase MRP and RNase P Ribonucleoproteins. Genes Dev. 1994, 8, 1423–1433. [Google Scholar] [CrossRef]
- Garcia, P.D.; Zakian, V.A. A New Role for Proteins Subunits of RNase P: Stabilization of the Telomerase Holoenzyme. Microb. Cell 2020, 7, 250–254. [Google Scholar] [CrossRef]
- Garcia, P.D.; Leach, R.W.; Wadsworth, G.M.; Choudhary, K.; Li, H.; Aviran, S.; Kim, H.D.; Zakian, V.A. Stability and Nuclear Localization of Yeast Telomerase Depend on Protein Components of RNase P/MRP. Nat. Commun. 2020, 11, 2173. [Google Scholar] [CrossRef]
- Abdulhadi-Atwan, M.; Klopstock, T.; Sharaf, M.; Weinberg-Shukron, A.; Renbaum, P.; Levy-Lahad, E.; Zangen, D. The Novel R211Q POP1 Homozygous Mutation Causes Different Pathogenesis and Skeletal Changes from Those of Previously Reported POP1-associated Anauxetic Dysplasia. Am. J. Med. Genet. A. 2020, 182, 1268–1272. [Google Scholar] [CrossRef]
- Goyal, M.; Banerjee, C.; Nag, S.; Bandyopadhyay, U. The Alba Protein Family: Structure and Function. Biochim. Biophys. Acta BBA Proteins Proteom. 2016, 1864, 570–583. [Google Scholar] [CrossRef]
- Lemieux, B.; Laterreur, N.; Perederina, A.; Noël, J.-F.; Dubois, M.-L.; Krasilnikov, A.S.; Wellinger, R.J. Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell 2016, 165, 1171–1181. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Bai, G.; Zhang, Y.; Huang, J. Crystal Structure of Human RPP20-RPP25 Proteins in Complex with the P3 Domain of LncRNA RMRP. J. Struct. Biol. 2021, 213, 107704. [Google Scholar] [CrossRef] [PubMed]
- Perederina, A.; Krasilnikov, A.S. The P3 Domain of Eukaryotic RNases P/MRP: Making a Protein-Rich RNA-Based Enzyme. RNA Biol. 2010, 7, 534–539. [Google Scholar] [CrossRef][Green Version]
- Perederina, A.; Esakova, O.; Quan, C.; Khanova, E.; Krasilnikov, A.S. Eukaryotic Ribonucleases P/MRP: The Crystal Structure of the P3 Domain. EMBO J. 2010, 29, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, L.; Fretz, S.; Epps, N.; Zengel, J.M. Functional Equivalence of Hairpins in the RNA Subunits of RNase MRP and RNase P in Saccharomyces Cerevisiae. RNA 2000, 6, 653–658. [Google Scholar] [CrossRef]
- Ziehler, W.A.; Morris, J.; Scott, F.H.; Millikin, C.; Engelke, D.R. An Essential Protein-Binding Domain of Nuclear RNase P RNA. RNA 2001, 7, 565–575. [Google Scholar] [CrossRef]
- Li, X. Identification of a Functional Core in the RNA Component of RNase MRP of Budding Yeasts. Nucleic Acids Res. 2004, 32, 3703–3711. [Google Scholar] [CrossRef][Green Version]
- Palsule, G.; Gopalan, V.; Simcox, A. Biogenesis of RNase P RNA from an Intron Requires Co-Assembly with Cognate Protein Subunits. Nucleic Acids Res. 2019, 47, 8746–8754. [Google Scholar] [CrossRef]
- Mann, H.; Ben-Asouli, Y.; Schein, A.; Moussa, S.; Jarrous, N. Eukaryotic RNase P: Role of RNA and Protein Subunits of a Primordial Catalytic Ribonucleoprotein in RNA-Based Catalysis. Mol. Cell 2003, 12, 925–935. [Google Scholar] [CrossRef]
- Sharin, E. RNase P: Role of Distinct Protein Cofactors in TRNA Substrate Recognition and RNA-Based Catalysis. Nucleic Acids Res. 2005, 33, 5120–5132. [Google Scholar] [CrossRef] [PubMed]
- Rossmanith, W.; Karwan, R.M. Characterization of Human Mitochondrial RNase P: Novel Aspects in TRNA Processing. Biochem. Biophys. Res. Commun. 1998, 247, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Hartig, A.; Hartmann, R.K.; Rossmanith, W. Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms. PLoS Genet. 2014, 10, e1004506. [Google Scholar] [CrossRef] [PubMed]
- Lopez Sanchez, M.I.G.; Mercer, T.R.; Davies, S.M.K.; Shearwood, A.-M.J.; Nygård, K.K.A.; Richman, T.R.; Mattick, J.S.; Rackham, O.; Filipovska, A. RNA Processing in Human Mitochondria. Cell Cycle 2011, 10, 2904–2916. [Google Scholar] [CrossRef]
- Yang, S.-Y.; He, X.-Y.; Schulz, H. Multiple Functions of Type 10 17β-Hydroxysteroid Dehydrogenase. Trends Endocrinol. Metab. 2005, 16, 167–175. [Google Scholar] [CrossRef]
- Rackham, O.; Shearwood, A.-M.J.; Mercer, T.R.; Davies, S.M.K.; Mattick, J.S.; Filipovska, A. Long Noncoding RNAs Are Generated from the Mitochondrial Genome and Regulated by Nuclear-Encoded Proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Kissinger, C.R.; Rejto, P.A.; Pelletier, L.A.; Thomson, J.A.; Showalter, R.E.; Abreo, M.A.; Agree, C.S.; Margosiak, S.; Meng, J.J.; Aust, R.M.; et al. Crystal Structure of Human ABAD/HSD10 with a Bound Inhibitor: Implications for Design of Alzheimer’s Disease Therapeutics. J. Mol. Biol. 2004, 342, 943–952. [Google Scholar] [CrossRef]
- Oerum, S.; Roovers, M.; Rambo, R.P.; Kopec, J.; Bailey, H.J.; Fitzpatrick, F.; Newman, J.A.; Newman, W.G.; Amberger, A.; Zschocke, J.; et al. Structural Insight into the Human Mitochondrial TRNA Purine N1-Methyltransferase and Ribonuclease P Complexes. J. Biol. Chem. 2018, 293, 12862–12876. [Google Scholar] [CrossRef]
- Oerum, S.; Dégut, C.; Barraud, P.; Tisné, C. M1A Post-Transcriptional Modification in TRNAs. Biomolecules 2017, 7, 20. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. TRNA Punctuation Model of RNA Processing in Human Mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Deutschmann, A.J.; Amberger, A.; Zavadil, C.; Steinbeisser, H.; Mayr, J.A.; Feichtinger, R.G.; Oerum, S.; Yue, W.W.; Zschocke, J. Mutation or Knock-down of 17β-Hydroxysteroid Dehydrogenase Type 10 Cause Loss of MRPP1 and Impaired Processing of Mitochondrial Heavy Strand Transcripts. Hum. Mol. Genet. 2014, 23, 3618–3628. [Google Scholar] [CrossRef]
- Chatfield, K.C.; Coughlin, C.R., 2nd; Friederich, M.W.; Gallagher, R.C.; Hesselberth, J.R.; Lovell, M.A.; Ofman, R.; Swanson, M.A.; Thomas, J.A.; Wanders, R.J.A.; et al. Mitochondrial Energy Failure in HSD10 Disease Is Due to Defective MtDNA Transcript Processing. Mitochondrion 2015, 21, 5. [Google Scholar] [CrossRef]
- Metodiev, M.D.; Thompson, K.; Alston, C.L.; Morris, A.A.M.; He, L.; Assouline, Z.; Rio, M.; Bahi-Buisson, N.; Pyle, A.; Griffin, H.; et al. Recessive Mutations in TRMT10C Cause Defects in Mitochondrial RNA Processing and Multiple Respiratory Chain Deficiencies. Am. J. Hum. Genet. 2016, 98, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.; Busch, J.D.; Matic, S.; Siira, S.J.; Kuznetsova, I.; Atanassov, I.; Ermer, J.A.; Shearwood, A.-M.J.; Richman, T.R.; Stewart, J.B.; et al. Hierarchical RNA Processing Is Required for Mitochondrial Ribosome Assembly. Cell Rep. 2016, 16, 1874–1890. [Google Scholar] [CrossRef]
- Saoji, M.; Sen, A.; Cox, R.T. Loss of Individual Mitochondrial Ribonuclease P Complex Proteins Differentially Affects Mitochondrial TRNA Processing In Vivo. Int. J. Mol. Sci. 2021, 22, 6066. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulos, G.N.; Klapa, M.I.; Moschonas, N.K. PICKLE 3.0: Enriching the Human Meta-Database with the Mouse Protein Interactome Extended via Mouse–Human Orthology. Bioinformatics 2021, 37, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-Scale Gene Function Analysis with the PANTHER Classification System. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Gill, T.; Cai, T.; Aulds, J.; Wierzbicki, S.; Schmitt, M.E. RNase MRP Cleaves the CLB2 MRNA To Promote Cell Cycle Progression: Novel Method of MRNA Degradation. Mol. Cell. Biol. 2004, 24, 945–953. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of M6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507. [Google Scholar] [CrossRef]
| RNase P | Bacteria (E. coli) | Archaea (M. jannaschii) | Yeast (Nuclear) | Human (Nuclear) |
|---|---|---|---|---|
| RNA subunit | M1 RNA or P RNA (377 nt) | RPR (252 nt) | RPR1 (369 nt) | RPPH1 (341 nt) |
| Protein Subunits * | RnpA/ C5/P protein (13.8 kDa) | |||
| Pop5 (15.9 kDa) | Pop5 (19.6 kDa) | Pop5 (18.8 kDa) | ||
| Rpp29 (10.9 kDa) | Pop4 (32.9 kDa) | Rpp29 (25.4 kDa) | ||
| Rpp30 (27.3 kDa) | Rpp1 (32.2 kDa) | Rpp30 (29.3 kDa) | ||
| Rpp21 (15.6 kDa) | Rpr2 (16.3 kDa) | Rpp21 (17.6 kDa) | ||
| L7Ae/Rpp38 (12.7 kDa) | Pop3 (22.6 kDa) | Rpp38 (31.8 kDa) | ||
| Pop1 (100.4 kDa) | Pop1 (114.7 kDa) | |||
| Pop6 (18.2 kDa) | Rpp20 (15.7 kDa) | |||
| Pop7/Rpp2 (15.8 kDa) | Rpp25 (20.6 kDa) | |||
| Rpp14 (13.7 kDa) | ||||
| Rpp40 (41.8 kDa) | ||||
| Pop8 (15.5 kDa) |
| Subunits | Structure | Additional Roles | References |
|---|---|---|---|
| Rpp14 |  | Exhibits 3′→5′ exoribonucleases activity | [46] |
| Pop5 |  | Unknown | |
| Rpp20 |  | Exhibits ATPase activity, promotes transcription by RNA Pol I and III, binds to chromatin of rDNA genes, Th/To autoantigen | [19,47,48,49] |
| Rpp21 |  | Promotes DNA repair through HDR | [50] |
| Rpp25 |  | Promotes transcription by RNA Pol I and III, binds to chromatin of rDNA genes, Th/To autoantigen | [19,48,49] |
| Rpp29 |  | Promotes DNA repair through HDR, nucleosome remodeling | [50,51,52] |
| Rpp30 |  | Affects piRNAs transcription and tRF biogenesis | [53,54,55] |
| Rpp38 | 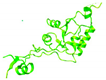 | Th/To autoantigen | [56] |
| Rpp40 |  | Unknown | |
| Pop1 |  | Transcription, Th/To autoantigen | [48,51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaukat, A.-N.; Kaliatsi, E.G.; Skeparnias, I.; Stathopoulos, C. The Dynamic Network of RNP RNase P Subunits. Int. J. Mol. Sci. 2021, 22, 10307. https://doi.org/10.3390/ijms221910307
Shaukat A-N, Kaliatsi EG, Skeparnias I, Stathopoulos C. The Dynamic Network of RNP RNase P Subunits. International Journal of Molecular Sciences. 2021; 22(19):10307. https://doi.org/10.3390/ijms221910307
Chicago/Turabian StyleShaukat, Athanasios-Nasir, Eleni G. Kaliatsi, Ilias Skeparnias, and Constantinos Stathopoulos. 2021. "The Dynamic Network of RNP RNase P Subunits" International Journal of Molecular Sciences 22, no. 19: 10307. https://doi.org/10.3390/ijms221910307
APA StyleShaukat, A.-N., Kaliatsi, E. G., Skeparnias, I., & Stathopoulos, C. (2021). The Dynamic Network of RNP RNase P Subunits. International Journal of Molecular Sciences, 22(19), 10307. https://doi.org/10.3390/ijms221910307







