Molecular Targeted Therapy and Immunotherapy for Myelodysplastic Syndrome
Abstract
1. Introduction
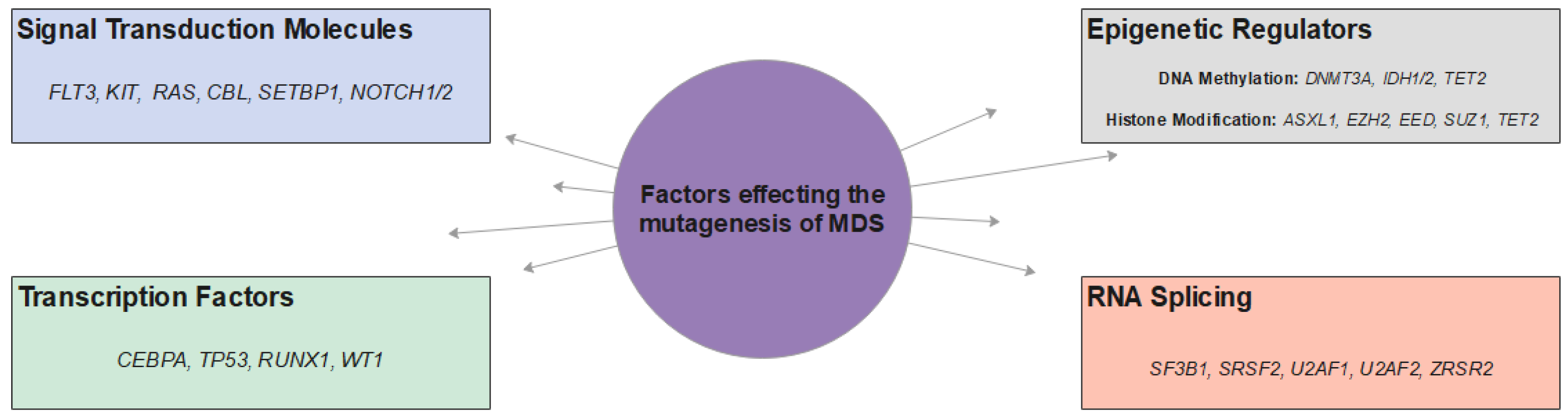
2. Molecular Pathogenesis of MDS
2.1. Signaling Molecules and Pathways
2.1.1. FLT3
2.1.2. KIT
2.1.3. RAS
2.1.4. CBL
2.1.5. SETBP1
2.1.6. NOTCH 1/2
2.1.7. Other Signal Transducers
2.2. Transcription Factors
2.2.1. TP53
2.2.2. RUNX1
2.2.3. WT1
2.2.4. CEBPA
2.2.5. Other Transcription Factors
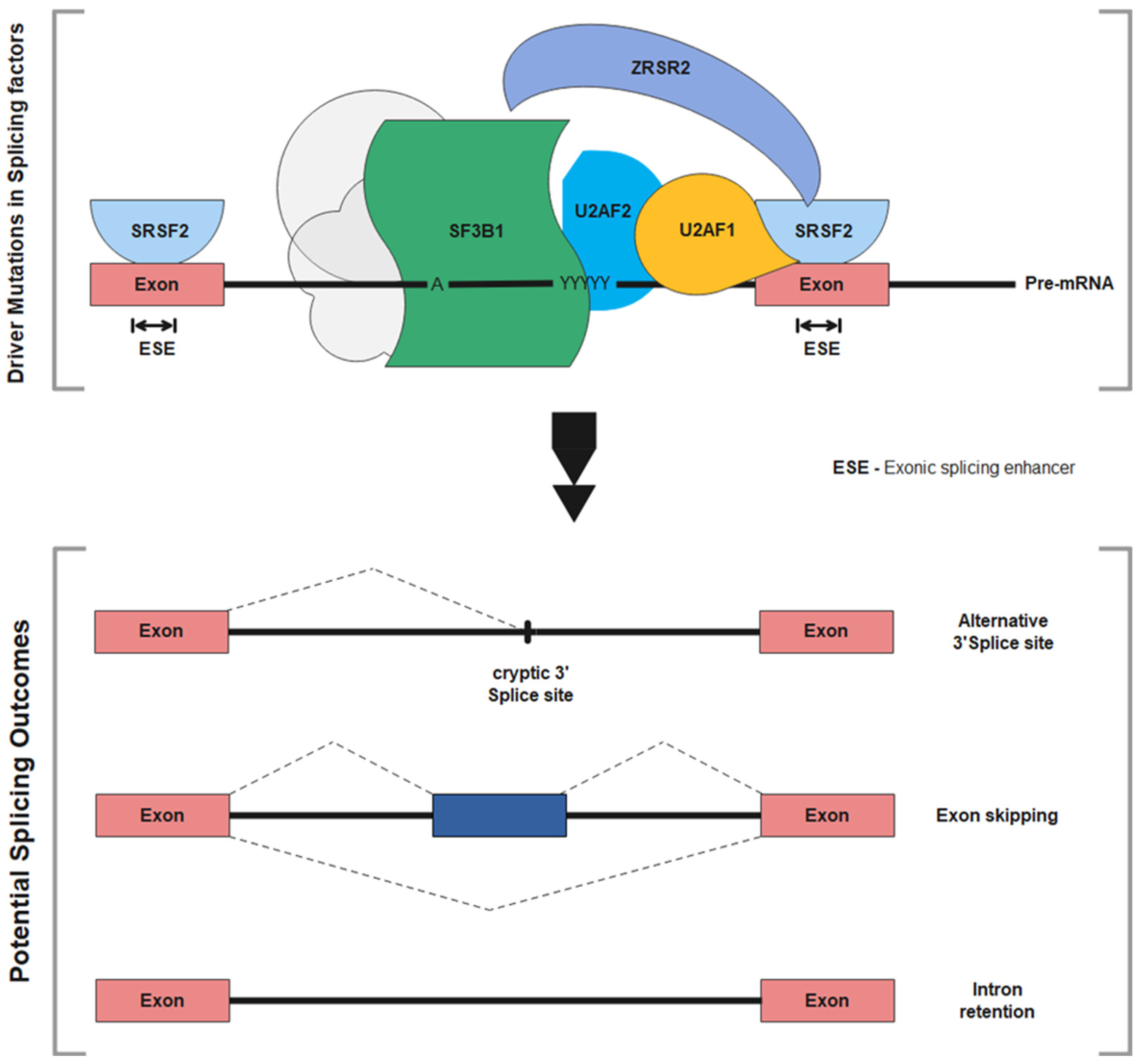
2.3. RNA Splicing
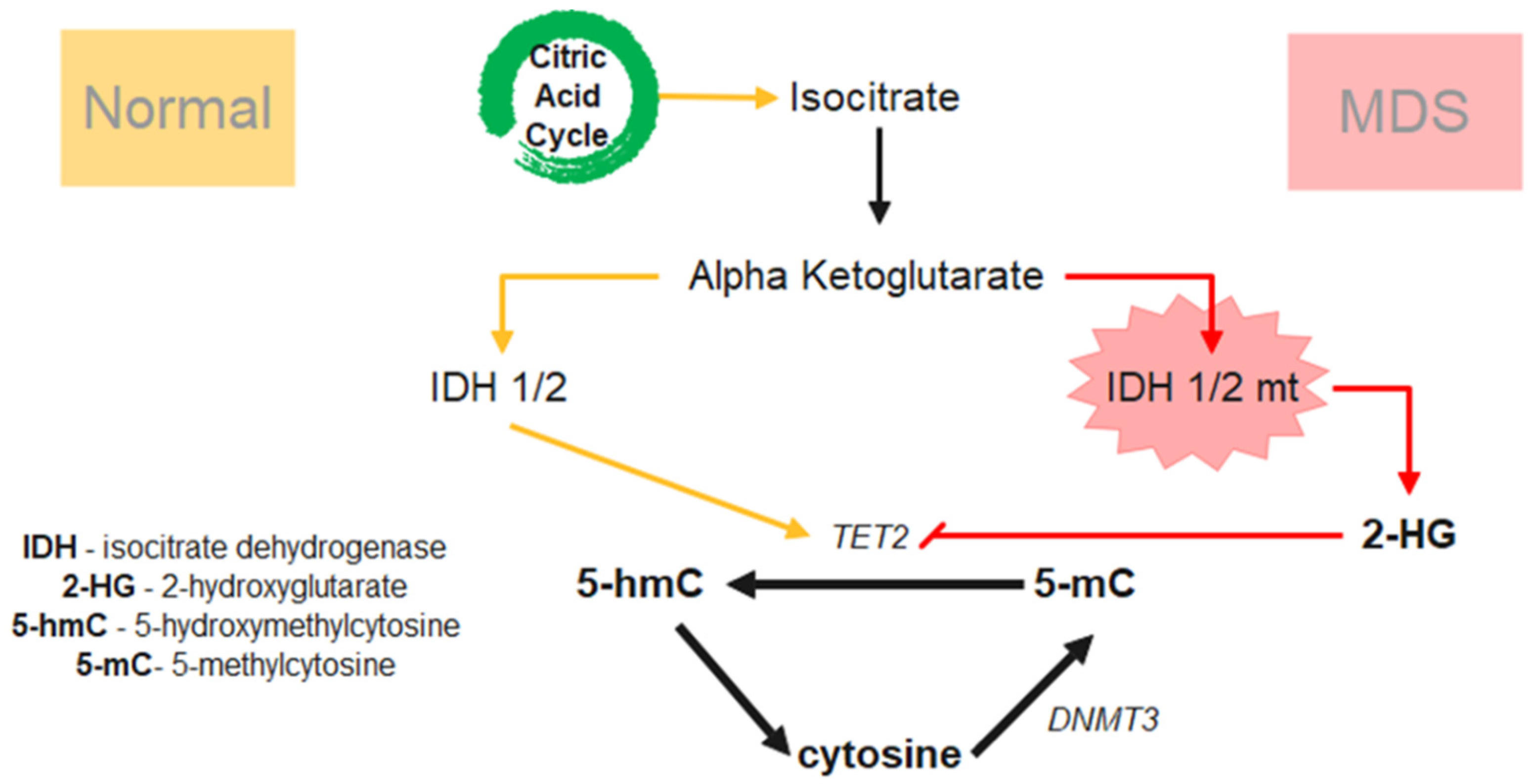
2.4. Epigenetic Dysregulation-DNA Methylation
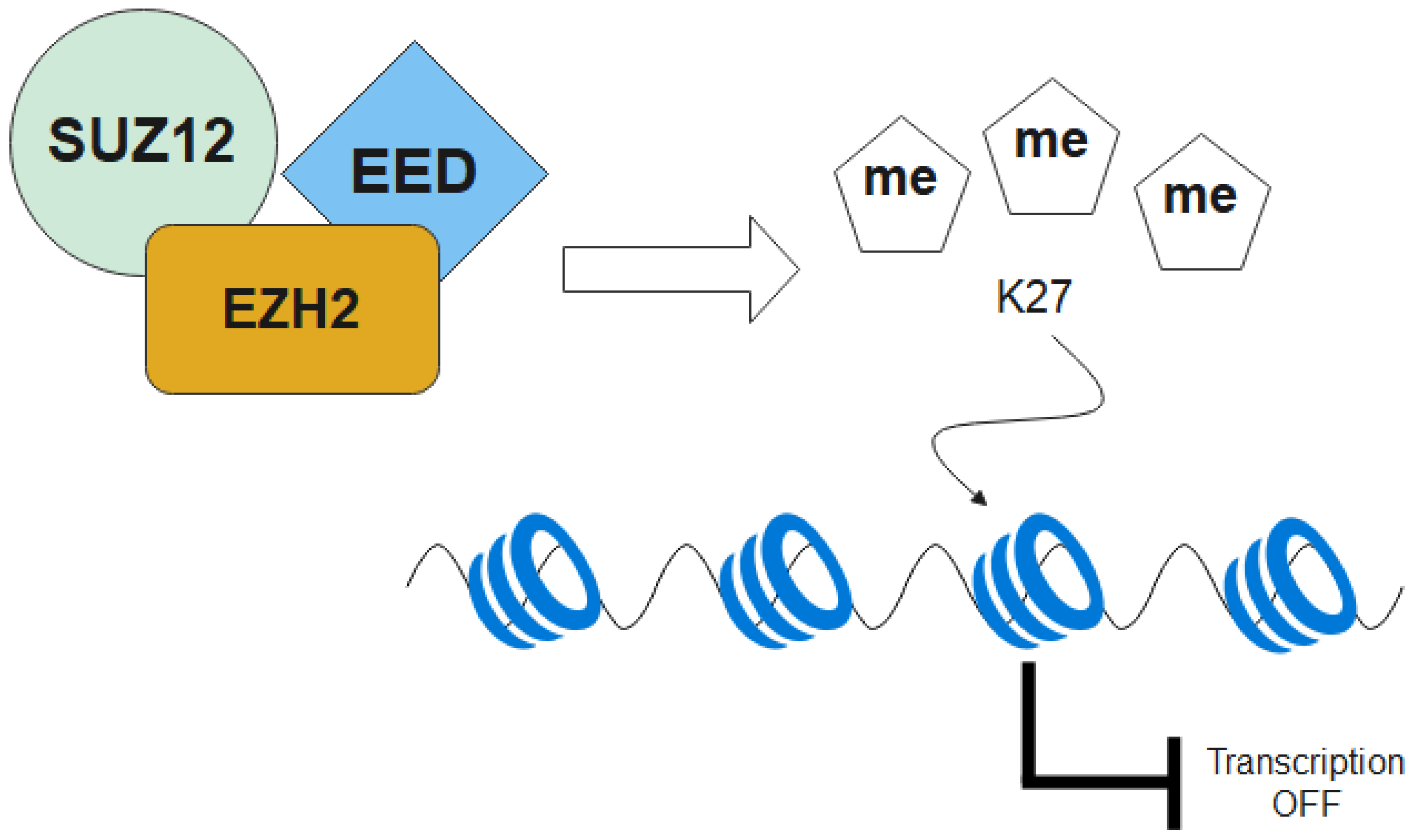
2.5. Epigenetic Dysregulation—Histone Modification
2.5.1. EZH2, EED and SUZ12
2.5.2. ASXL1 and TET2
3. Immune Dysregulation
3.1. Adaptive Immune Dysregulation
3.2. Innate Immune Dysregulation
4. Treatment of MDS
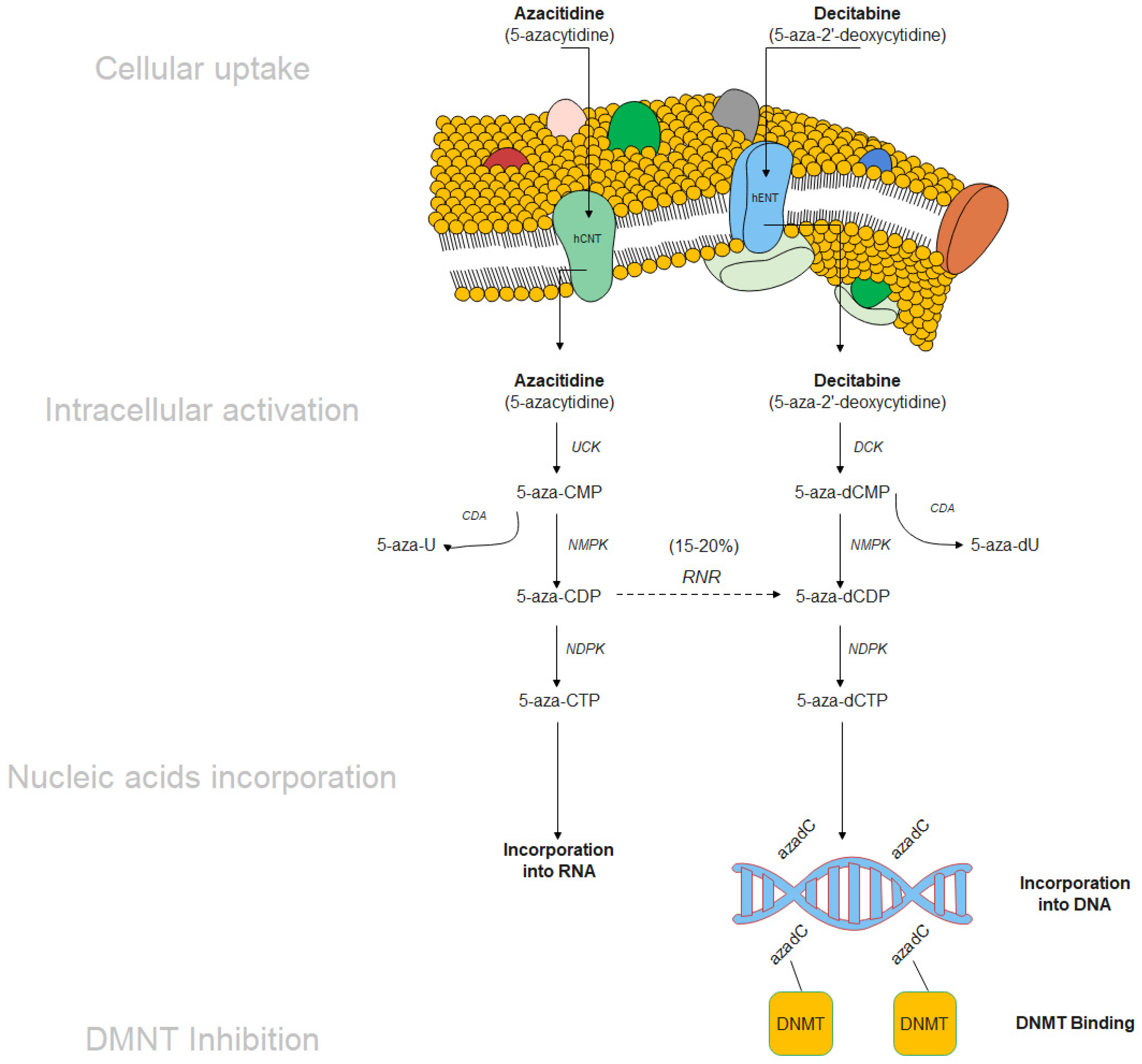
4.1. Hypomethylating Agents (HMA)
4.2. Mechanisms of Resistance to Hypomethylating Agents
4.3. Next Generation HMA—Strategies to Overcome HMA Resistance
4.3.1. Guadecitabine (SGI-110)
4.3.2. ASTX727
4.4. Targeted Therapy in Combination with Hypomethylating Agents
4.4.1. BCL-2 Inhibitors
4.4.2. IDH1/2 Inhibitors
4.4.3. FLT3 Inhibitors
4.5. Splicing Inhibition and TP53 Modulation
4.6. Immune Checkpoint Inhibition in Combination with HMA
4.6.1. Anti-PD-1, Anti-PD-L1, Anti-CTLA-4
4.6.2. Anti-TIM-3
4.6.3. Anti-CD47
4.7. Adoptive T-Cell Therapy with HMA or as Monotherapy
4.7.1. Anti-CD123
4.7.2. Anti-NKG2D
4.8. Donor-Derived Lymphocytes against Tumor-Associated Antigens
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Patnaik, M.M.; Tefferi, A. Myelodysplastic syndromes with ring sideroblasts (MDS-RS) and MDS/myeloproliferative neoplasm with RS and thrombocytosis (MDS/MPN-RS-T)—“2021 update on diagnosis, risk-stratification, and management”. Am. J. Hematol. 2021, 96, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Alessandrino, E.P.; Della Porta, M.G.; Bacigalupo, A.; Van Lint, M.T.; Falda, M.; Onida, F.; Bernardi, M.; Iori, A.P.; Rambaldi, A.; Cerretti, R.; et al. WHO classification and WPSS predict posttransplantation outcome in patients with myelodysplastic syndrome: A study from the Gruppo Italiano Trapianto di Midollo Osseo (GITMO). Blood 2008, 112, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Stevens, B.M.; Khan, N.; D’Alessandro, A.; Nemkov, T.; Winters, A.; Jones, C.L.; Zhang, W.; Pollyea, D.A.; Jordan, C.T. Characterization and targeting of malignant stem cells in patients with advanced myelodysplastic syndromes. Nat. Commun. 2018, 9, 3694. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, Y.; Gao, S.; Chen, J.; Yu, J.; Zhang, H.; Li, M.; Zhan, X.; Li, W. Immune dysregulation in myelodysplastic syndrome: Clinical features, pathogenesis and therapeutic strategies. Crit. Rev. Oncol. Hematol. 2018, 122, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Cazzola, M.; Malcovati, L.; Invernizzi, R. Myelodysplastic/myeloproliferative neoplasms. Hematol. Am. Soc. Hematol. Educ. Program. 2011, 2011, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Corrêa de Souza, D.; de Souza Fernandez, C.; Camargo, A.; Apa, A.G.; Sobral da Costa, E.; Bouzas, L.F.; Abdelhay, E.; de Souza Fernandez, T. Cytogenetic as an Important Tool for Diagnosis and Prognosis for Patients with Hypocellular Primary Myelodysplastic Syndrome. BioMed. Res. Int. 2014, 2014, 542395. [Google Scholar] [CrossRef]
- Bejar, R. Clinical and genetic predictors of prognosis in myelodysplastic syndromes. Haematologica 2014, 99, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Y.; Xu, Q.; Chen, Y.; Lv, N.; Jing, Y.; Dou, L.; Bo, J.; Hou, G.; Guo, J.; et al. Implications of mutational spectrum in myelodysplastic syndromes based on targeted next-generation sequencing. Oncotarget 2017, 8, 82475–82490. [Google Scholar] [CrossRef]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef]
- Langemeijer, S.M.; Kuiper, R.P.; Berends, M.; Knops, R.; Aslanyan, M.G.; Massop, M.; Stevens-Linders, E.; van Hoogen, P.; van Kessel, A.G.; Raymakers, R.A.; et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009, 41, 838–842. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Galili, N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat. Rev. Cancer 2012, 12, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Nazha, A.; Narkhede, M.; Radivoyevitch, T.; Seastone, D.J.; Patel, B.J.; Gerds, A.T.; Mukherjee, S.; Kalaycio, M.; Advani, A.; Przychodzen, B.; et al. Incorporation of molecular data into the Revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Leukemia 2016, 30, 2214–2220. [Google Scholar] [CrossRef]
- Gu, S.; Xia, J.; Tian, Y.; Zi, J.; Ge, Z. A novel scoring system integrating molecular abnormalities with IPSS-R can improve the risk stratification in patients with MDS. BMC Cancer 2021, 21, 134. [Google Scholar] [CrossRef]
- Bersanelli, M.; Travaglino, E.; Meggendorfer, M.; Matteuzzi, T.; Sala, C.; Mosca, E.; Chiereghin, C.; Di Nanni, N.; Gnocchi, M.; Zampini, M.; et al. Classification and Personalized Prognostic Assessment on the Basis of Clinical and Genomic Features in Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1223–1233. [Google Scholar] [CrossRef]
- Gangat, N.; Mudireddy, M.; Lasho, T.L.; Finke, C.M.; Nicolosi, M.; Szuber, N.; Patnaik, M.M.; Pardanani, A.; Hanson, C.A.; Ketterling, R.P.; et al. Mutations and prognosis in myelodysplastic syndromes: Karyotype-adjusted analysis of targeted sequencing in 300 consecutive cases and development of a genetic risk model. Am. J. Hematol. 2018, 93, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Lasho, T.L.; Patnaik, M.M.; Saeed, L.; Mudireddy, M.; Idossa, D.; Finke, C.; Ketterling, R.P.; Pardanani, A.; Gangat, N. Targeted next-generation sequencing in myelodysplastic syndromes and prognostic interaction between mutations and IPSS-R. Am. J. Hematol. 2017, 92, 1311–1317. [Google Scholar] [CrossRef]
- Gao, J.; Swaminathan, S.; Pai, N.; Johnson, Z.; Chen, Y.H.; Peterson, L.; Goolsby, C. Flow cytometric detection of altered signaling in myelodysplastic syndrome and cytopenia. Leuk. Res. 2015, 39, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Frohling, S.; Schlenk, R.F.; Breitruck, J.; Benner, A.; Kreitmeier, S.; Tobis, K.; Dohner, H.; Dohner, K. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm. Blood 2002, 100, 4372–4380. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.; Leung, A.Y.; Kwong, Y.L. Molecular and Cellular Mechanisms of Myelodysplastic Syndrome: Implications on Targeted Therapy. Int. J. Mol. Sci. 2016, 17, 440. [Google Scholar] [CrossRef] [PubMed]
- Rocnik, J.L.; Okabe, R.; Yu, J.C.; Lee, B.H.; Giese, N.; Schenkein, D.P.; Gilliland, D.G. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood 2006, 108, 1339–1345. [Google Scholar] [CrossRef]
- Georgiou, G.; Karali, V.; Zouvelou, C.; Kyriakou, E.; Dimou, M.; Chrisochoou, S.; Greka, P.; Dufexis, D.; Vervesou, E.; Dimitriadou, E.; et al. Serial determination of FLT3 mutations in myelodysplastic syndrome patients at diagnosis, follow up or acute myeloid leukaemia transformation: Incidence and their prognostic significance. Br. J. Haematol. 2006, 134, 302–306. [Google Scholar] [CrossRef]
- Daver, N.; Strati, P.; Jabbour, E.; Kadia, T.; Luthra, R.; Wang, S.; Patel, K.; Ravandi, F.; Cortes, J.; Qin Dong, X.; et al. FLT3 mutations in myelodysplastic syndrome and chronic myelomonocytic leukemia. Am. J. Hematol. 2013, 88, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Tyndel, M.S.; Kim, H.J.; Ahn, J.S.; Choi, S.H.; Park, H.J.; Kim, Y.K.; Yang, D.H.; Lee, J.J.; Jung, S.H.; et al. The clonal origins of leukemic progression of myelodysplasia. Leukemia 2017, 31, 1928–1935. [Google Scholar] [CrossRef] [PubMed]
- Zhan, D.; Park, C.Y. Stem Cells in the Myelodysplastic Syndromes. Front. Aging 2021, 2. [Google Scholar] [CrossRef]
- Rouault-Pierre, K.; Smith, A.E.; Mian, S.A.; Pizzitola, I.; Kulasekararaj, A.G.; Mufti, G.J.; Bonnet, D. Myelodysplastic syndrome can propagate from the multipotent progenitor compartment. Haematologica 2017, 102, e7–e10. [Google Scholar] [CrossRef]
- Orfao, A.; Garcia-Montero, A.C.; Sanchez, L.; Escribano, L. Recent advances in the understanding of mastocytosis: The role of KIT mutations. Br. J. Haematol. 2007, 138, 12–30. [Google Scholar] [CrossRef]
- Lorenzo, F.; Nishii, K.; Monma, F.; Kuwagata, S.; Usui, E.; Shiku, H. Mutational analysis of the KIT gene in myelodysplastic syndrome (MDS) and MDS-derived leukemia. Leuk. Res. 2006, 30, 1235–1239. [Google Scholar] [CrossRef]
- Craig, J.W.; Hasserjian, R.P.; Kim, A.S.; Aster, J.C.; Pinkus, G.S.; Hornick, J.L.; Steensma, D.P.; Coleman Lindsley, R.; DeAngelo, D.J.; Morgan, E.A. Detection of the KIT(D816V) mutation in myelodysplastic and/or myeloproliferative neoplasms and acute myeloid leukemia with myelodysplasia-related changes predicts concurrent systemic mastocytosis. Mod. Pathol. 2020, 33, 1135–1145. [Google Scholar] [CrossRef]
- Shao, Z.; Zhang, H.; Chen, G.; Wang, L.; Li, K.; Zhang, Y.; Li, L.; Sun, J. Expression and function of c-kit receptor in bone marrow mononuclear cells of patients with myelodysplastic syndromes. Chin. Med. J. 2001, 114, 481–485. [Google Scholar]
- Russkamp, N.F.; Myburgh, R.; Kiefer, J.D.; Neri, D.; Manz, M.G. Anti-CD117 immunotherapy to eliminate hematopoietic and leukemia stem cells. Exp. Hematol. 2021, 95, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, B.B.; Kadam, N.N. Mutations of myelodysplastic syndromes (MDS): An update. Mutat Res. Rev. Mutat Res. 2016, 769, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Al-Kali, A.; Luthra, R.; Bueso-Ramos, C.E.; Pierce, S.; Kadia, T.; Borthakur, G.; Estrov, Z.; Jabbour, E.; Faderl, S.; Ravandi, F.; et al. Impact of RAS Mutations In Myelodysplastic Syndrome (MDS). Blood 2010, 116, 2926. [Google Scholar] [CrossRef]
- Paquette, R.L.; Landaw, E.M.; Pierre, R.V.; Kahan, J.; Lubbert, M.; Lazcano, O.; Isaac, G.; McCormick, F.; Koeffler, H.P. N-ras mutations are associated with poor prognosis and increased risk of leukemia in myelodysplastic syndrome. Blood 1993, 82, 590–599. [Google Scholar] [CrossRef]
- Hirai, H.; Okada, M.; Mizoguchi, H.; Mano, H.; Kobayashi, Y.; Nishida, J.; Takaku, F. Relationship between an activated N-ras oncogene and chromosomal abnormality during leukemic progression from myelodysplastic syndrome. Blood 1988, 71, 256–258. [Google Scholar] [CrossRef] [PubMed]
- Sanada, M.; Suzuki, T.; Shih, L.Y.; Otsu, M.; Kato, M.; Yamazaki, S.; Tamura, A.; Honda, H.; Sakata-Yanagimoto, M.; Kumano, K.; et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 2009, 460, 904–908. [Google Scholar] [CrossRef] [PubMed]
- Katzav, S.; Schmitz, M.L. Mutations of c-Cbl in myeloid malignancies. Oncotarget 2015, 6, 10689–10696. [Google Scholar] [CrossRef]
- Reindl, C.; Quentmeier, H.; Petropoulos, K.; Greif, P.A.; Benthaus, T.; Argiropoulos, B.; Mellert, G.; Vempati, S.; Duyster, J.; Buske, C.; et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin. Cancer Res. 2009, 15, 2238–2247. [Google Scholar] [CrossRef]
- Schwaab, J.; Ernst, T.; Erben, P.; Rinke, J.; Schnittger, S.; Strobel, P.; Metzgeroth, G.; Mossner, M.; Haferlach, T.; Cross, N.C.; et al. Activating CBL mutations are associated with a distinct MDS/MPN phenotype. Ann. Hematol. 2012, 91, 1713–1720. [Google Scholar] [CrossRef]
- Minakuchi, M.; Kakazu, N.; Gorrin-Rivas, M.J.; Abe, T.; Copeland, T.D.; Ueda, K.; Adachi, Y. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur. J. Biochem. 2001, 268, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Makkinje, A.; Damuni, Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 1996, 271, 11059–11062. [Google Scholar] [CrossRef]
- Oakley, K.; Han, Y.; Vishwakarma, B.A.; Chu, S.; Bhatia, R.; Gudmundsson, K.O.; Keller, J.; Chen, X.; Vasko, V.; Jenkins, N.A.; et al. Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood 2012, 119, 6099–6108. [Google Scholar] [CrossRef]
- Makishima, H.; Yoshida, K.; Nguyen, N.; Przychodzen, B.; Sanada, M.; Okuno, Y.; Ng, K.P.; Gudmundsson, K.O.; Vishwakarma, B.A.; Jerez, A.; et al. Somatic SETBP1 mutations in myeloid malignancies. Nat. Genet. 2013, 45, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Shou, L.H.; Cao, D.; Dong, X.H.; Fang, Q.; Wu, Y.; Zhang, Y.; Fei, J.P.; Xu, B.L. Prognostic significance of SETBP1 mutations in myelodysplastic syndromes, chronic myelomonocytic leukemia, and chronic neutrophilic leukemia: A meta-analysis. PLoS ONE 2017, 12, e0171608. [Google Scholar] [CrossRef] [PubMed]
- Coccaro, N.; Tota, G.; Zagaria, A.; Anelli, L.; Specchia, G.; Albano, F. SETBP1 dysregulation in congenital disorders and myeloid neoplasms. Oncotarget 2017, 8, 51920. [Google Scholar] [CrossRef]
- Hou, H.-A.; Kuo, Y.-Y.; Tang, J.-L.; Chou, W.-C.; Yao, M.; Lai, Y.-J.; Lin, C.-C.; Chen, C.-Y.; Liu, C.-Y.; Tseng, M.-H.; et al. Clinical implications of the SETBP1 mutation in patients with primary myelodysplastic syndrome and its stability during disease progression. Am. J. Hematol. 2014, 89, 181–186. [Google Scholar] [CrossRef]
- Adema, V.; Larrayoz, M.J.; Calasanz, M.J.; Palomo, L.; Patino-Garcia, A.; Agirre, X.; Hernandez-Rivas, J.M.; Lumbreras, E.; Buno, I.; Martinez-Laperche, C.; et al. Correlation of myelodysplastic syndromes with i(17)(q10) and TP53 and SETBP1 mutations. Br. J. Haematol. 2015, 171, 137–141. [Google Scholar] [CrossRef]
- Thol, F.; Suchanek, K.J.; Koenecke, C.; Stadler, M.; Platzbecker, U.; Thiede, C.; Schroeder, T.; Kobbe, G.; Kade, S.; Loffeld, P.; et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia 2013, 27, 2072–2075. [Google Scholar] [CrossRef]
- Piazza, R.; Valletta, S.; Winkelmann, N.; Redaelli, S.; Spinelli, R.; Pirola, A.; Antolini, L.; Mologni, L.; Donadoni, C.; Papaemmanuil, E.; et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat. Genet. 2013, 45, 18–24. [Google Scholar] [CrossRef]
- Saika, M.; Inoue, D.; Nagase, R.; Sato, N.; Tsuchiya, A.; Yabushita, T.; Kitamura, T.; Goyama, S. ASXL1 and SETBP1 mutations promote leukaemogenesis by repressing TGFbeta pathway genes through histone deacetylation. Sci. Rep. 2018, 8, 15873. [Google Scholar] [CrossRef] [PubMed]
- Varnum-Finney, B.; Xu, L.; Brashem-Stein, C.; Nourigat, C.; Flowers, D.; Bakkour, S.; Pear, W.S.; Bernstein, I.D. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 2000, 6, 1278–1281. [Google Scholar] [CrossRef] [PubMed]
- Weickert, M.-T.; Hecker, J.S.; Buck, M.C.; Schreck, C.; Rivière, J.; Schiemann, M.; Schallmoser, K.; Bassermann, F.; Strunk, D.; Oostendorp, R.A.J.; et al. Bone marrow stromal cells from MDS and AML patients show increased adipogenic potential with reduced Delta-like-1 expression. Sci. Rep. 2021, 11, 5944. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.A.; Pietras, E.M.; Passegue, E. Deregulated Notch and Wnt signaling activates early-stage myeloid regeneration pathways in leukemia. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Aref, S.; Rizk, R.; El Agder, M.; Fakhry, W.; El Zafarany, M.; Sabry, M. NOTCH-1 Gene Mutations Influence Survival in Acute Myeloid Leukemia Patients. Asian Pac. J. Cancer Prev. 2020, 21, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Bravo, G.; Pierola, A.A.; Takahashi, K.; Konopleva, M.; Jabbour, E.; Borthakur, G.; Daver, N.G.; Dinardo, C.D.; Estrov, Z.; Kadia, T.M.; et al. Clinical relevance of mutations in patients with myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms with normal karyotype. J. Clin. Oncol. 2017, 35, 7053. [Google Scholar] [CrossRef]
- Fu, L.; Nara, N.; Tohda, S. Involvement of Notch signaling in myelodysplastic syndrome. Leuk. Res. 2007, 31, 1160–1161. [Google Scholar] [CrossRef]
- Beekman, R.; Touw, I.P. G-CSF and its receptor in myeloid malignancy. Blood 2010, 115, 5131–5136. [Google Scholar] [CrossRef]
- Gunawan, A.S.; McLornan, D.P.; Wilkins, B.; Waghorn, K.; Hoade, Y.; Cross, N.C.P.; Harrison, C.N. Ruxolitinib, a potent JAK1/JAK2 inhibitor, induces temporary reductions in the allelic burden of concurrent CSF3R mutations in chronic neutrophilic leukemia. Haematologica 2017, 102, e238–e240. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Wang, F.; Chen, X.; Teng, W.; Wang, M.; Zhang, Y.; Liu, H. CSF3R Mutation in Acute Leukemia and MDS Patients. Blood 2017, 130, 2669. [Google Scholar] [CrossRef]
- Fermo, E.; Zaninoni, A.; Imperiali, F.G.; Bianchi, P.; Colombi, M.; Barcellini, W.; Zanella, A. Analysis of JAK2 V167F Mutation in Myelodysplastic Syndromes. Blood 2007, 110, 4591. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627, quiz 3699. [Google Scholar] [CrossRef] [PubMed]
- Ingram, W.; Lea, N.C.; Cervera, J.; Germing, U.; Fenaux, P.; Cassinat, B.; Kiladjian, J.J.; Varkonyi, J.; Antunovic, P.; Westwood, N.B.; et al. The JAK2 V617F mutation identifies a subgroup of MDS patients with isolated deletion 5q and a proliferative bone marrow. Leukemia 2006, 20, 1319–1321. [Google Scholar] [CrossRef][Green Version]
- Cumbo, C.; Tota, G.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. TP53 in Myelodysplastic Syndromes: Recent Biological and Clinical Findings. Int. J. Mol. Sci. 2020, 21, 3432. [Google Scholar] [CrossRef]
- Platzbecker, U.; Kordasti, S. Natural born survivors: The inglorious TP53. Blood 2020, 136, 2727–2728. [Google Scholar] [CrossRef] [PubMed]
- Kulasekararaj, A.G.; Smith, A.E.; Mian, S.A.; Mohamedali, A.M.; Krishnamurthy, P.; Lea, N.C.; Gaken, J.; Pennaneach, C.; Ireland, R.; Czepulkowski, B.; et al. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br. J. Haematol. 2013, 160, 660–672. [Google Scholar] [CrossRef]
- Ebert, B.L. Deletion 5q in myelodysplastic syndrome: A paradigm for the study of hemizygous deletions in cancer. Leukemia 2009, 23, 1252–1256. [Google Scholar] [CrossRef]
- Brooks, C.L.; Gu, W. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003, 15, 164–171. [Google Scholar] [CrossRef]
- Maya, R.; Balass, M.; Kim, S.T.; Shkedy, D.; Leal, J.F.; Shifman, O.; Moas, M.; Buschmann, T.; Ronai, Z.; Shiloh, Y.; et al. ATM-dependent phosphorylation of Mdm2 on serine 395: Role in p53 activation by DNA damage. Genes Dev. 2001, 15, 1067–1077. [Google Scholar] [CrossRef]
- Da Silva-Coelho, P.; Kroeze, L.I.; Yoshida, K.; Koorenhof-Scheele, T.N.; Knops, R.; van de Locht, L.T.; de Graaf, A.O.; Massop, M.; Sandmann, S.; Dugas, M.; et al. Clonal evolution in myelodysplastic syndromes. Nat. Commun. 2017, 8, 15099. [Google Scholar] [CrossRef]
- Makishima, H.; Yoshizato, T.; Yoshida, K.; Sekeres, M.A.; Radivoyevitch, T.; Suzuki, H.; Przychodzen, B.; Nagata, Y.; Meggendorfer, M.; Sanada, M.; et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat. Genet. 2017, 49, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.; Gao, R.; Navin, N. Tumor evolution: Linear, branching, neutral or punctuated? Biochim. Biophys. Acta. Rev. Cancer 2017, 1867, 151–161. [Google Scholar] [CrossRef]
- Blyth, K.; Cameron, E.R.; Neil, J.C. The RUNX genes: Gain or loss of function in cancer. Nat. Rev. Cancer 2005, 5, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.C.; Liang, D.C.; Huang, C.F.; Shih, Y.S.; Wu, J.H.; Lin, T.L.; Shih, L.Y. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia 2009, 23, 1426–1431. [Google Scholar] [CrossRef]
- Chen, C.Y.; Lin, L.I.; Tang, J.L.; Ko, B.S.; Tsay, W.; Chou, W.C.; Yao, M.; Wu, S.J.; Tseng, M.H.; Tien, H.F. RUNX1 gene mutation in primary myelodysplastic syndrome—the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br. J. Haematol. 2007, 139, 405–414. [Google Scholar] [CrossRef]
- Niimi, H.; Harada, H.; Harada, Y.; Ding, Y.; Imagawa, J.; Inaba, T.; Kyo, T.; Kimura, A. Hyperactivation of the RAS signaling pathway in myelodysplastic syndrome with AML1/RUNX1 point mutations. Leukemia 2006, 20, 635–644. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scharnhorst, V.; van der Eb, A.J.; Jochemsen, A.G. WT1 proteins: Functions in growth and differentiation. Gene 2001, 273, 141–161. [Google Scholar] [CrossRef]
- Ellisen, L.W.; Carlesso, N.; Cheng, T.; Scadden, D.T.; Haber, D.A. The Wilms tumor suppressor WT1 directs stage-specific quiescence and differentiation of human hematopoietic progenitor cells. EMBO J. 2001, 20, 1897–1909. [Google Scholar] [CrossRef]
- Xu, F.; Wu, L.-Y.; He, Q.; Wu, D.; Zhang, Z.; Song, L.-X.; Zhao, Y.-S.; Su, J.-Y.; Zhou, L.-Y.; Guo, J.; et al. Exploration of the role of gene mutations in myelodysplastic syndromes through a sequencing design involving a small number of target genes. Sci. Rep. 2017, 7, 43113. [Google Scholar] [CrossRef]
- Huang, Q.S.; Wang, J.Z.; Qin, Y.Z.; Zeng, Q.Z.; Jiang, Q.; Jiang, H.; Lu, J.; Liu, H.X.; Liu, Y.; Wang, J.B.; et al. Overexpression of WT1 and PRAME predicts poor outcomes of patients with myelodysplastic syndromes with thrombocytopenia. Blood Adv. 2019, 3, 3406–3418. [Google Scholar] [CrossRef] [PubMed]
- Annesley, C.E.; Rabik, C.; Duffield, A.S.; Rau, R.E.; Magoon, D.; Li, L.; Huff, V.; Small, D.; Loeb, D.M.; Brown, P. Knock-in of the Wt1 R394W mutation causes MDS and cooperates with Flt3/ITD to drive aggressive myeloid neoplasms in mice. Oncotarget 2018, 9, 35313–35326. [Google Scholar] [CrossRef] [PubMed]
- Avellino, R.; Havermans, M.; Erpelinck, C.; Sanders, M.A.; Hoogenboezem, R.; van de Werken, H.J.; Rombouts, E.; van Lom, K.; van Strien, P.M.; Gebhard, C.; et al. An autonomous CEBPA enhancer specific for myeloid-lineage priming and neutrophilic differentiation. Blood 2016, 127, 2991–3003. [Google Scholar] [CrossRef] [PubMed]
- Toma, A.; Fenaux, P.; Dreyfus, F.; Cordonnier, C. Infections in myelodysplastic syndromes. Haematologica 2012, 97, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Kitaura, J.; Doki, N.; Komeno, Y.; Watanabe-Okochi, N.; Togami, K.; Nakahara, F.; Oki, T.; Enomoto, Y.; Fukuchi, Y.; et al. Two types of C/EBPα mutations play distinct but collaborative roles in leukemogenesis: Lessons from clinical data and BMT models. Blood 2011, 117, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, Y.; Li, T.; Li, Y.; Xing, H.; Sun, H.; Sun, L.; Wan, D.; Liu, Y.; Xie, X.; et al. Gene mutational analysis by NGS and its clinical significance in patients with myelodysplastic syndrome and acute myeloid leukemia. Exp. Hematol. Oncol. 2020, 9, 2. [Google Scholar] [CrossRef]
- Sportoletti, P.; Varasano, E.; Rossi, R.; Mupo, A.; Tiacci, E.; Vassiliou, G.; Martelli, M.P.; Falini, B. Mouse models of NPM1-mutated acute myeloid leukemia: Biological and clinical implications. Leukemia 2015, 29, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, M.S. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem. Res. Int. 2011, 2011, 195209. [Google Scholar] [CrossRef]
- Bains, A.; Luthra, R.; Medeiros, L.J.; Zuo, Z. FLT3 and NPM1 mutations in myelodysplastic syndromes: Frequency and potential value for predicting progression to acute myeloid leukemia. Am. J. Clin. Pathol. 2011, 135, 62–69. [Google Scholar] [CrossRef]
- Falini, B.; Sciabolacci, S.; Falini, L.; Brunetti, L.; Martelli, M.P. Diagnostic and therapeutic pitfalls in NPM1-mutated AML: Notes from the field. Leukemia 2021, 1–14. [Google Scholar] [CrossRef]
- Montalban-Bravo, G.; Kanagal-Shamanna, R.; Sasaki, K.; Patel, K.; Ganan-Gomez, I.; Jabbour, E.; Kadia, T.; Ravandi, F.; DiNardo, C.; Borthakur, G.; et al. NPM1 mutations define a specific subgroup of MDS and MDS/MPN patients with favorable outcomes with intensive chemotherapy. Blood Adv. 2019, 3, 922–933. [Google Scholar] [CrossRef]
- Pellagatti, A.; Armstrong, R.N.; Steeples, V.; Sharma, E.; Repapi, E.; Singh, S.; Sanchi, A.; Radujkovic, A.; Horn, P.; Dolatshad, H.; et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: Dysregulated genes/pathways and clinical associations. Blood 2018, 132, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Cheng, S.C. Functional roles of protein splicing factors. Biosci. Rep. 2012, 32, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.M.; Rebel, V.I. Acquired mutations that affect pre-mRNA splicing in hematologic malignancies and solid tumors. J. Natl. Cancer Inst. 2013, 105, 1540–1549. [Google Scholar] [CrossRef] [PubMed]
- Grosso, A.R.; Martins, S.; Carmo-Fonseca, M. The emerging role of splicing factors in cancer. EMBO Rep. 2008, 9, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Przychodzen, B.; Jerez, A.; Guinta, K.; Sekeres, M.A.; Padgett, R.; Maciejewski, J.P.; Makishima, H. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood 2013, 122, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.J.; Graubert, T.A. Clinical Implications of Spliceosome Mutations: Epidemiology, Clonal Hematopoiesis, and Potential Therapeutic Strategies. Blood 2016, 128, SCI-19. [Google Scholar] [CrossRef]
- Visconte, V.; Tiu, R.V.; Rogers, H.J. Pathogenesis of myelodysplastic syndromes: An overview of molecular and non-molecular aspects of the disease. Blood Res. 2014, 49, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Visconte, V.; Rogers, H.J.; Singh, J.; Barnard, J.; Bupathi, M.; Traina, F.; McMahon, J.; Makishima, H.; Szpurka, H.; Jankowska, A.; et al. SF3B1 haploinsufficiency leads to formation of ring sideroblasts in myelodysplastic syndromes. Blood 2012, 120, 3173–3186. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, D.; Guo, J.; Jin, J.; Tao, Y.; Zhang, Z.; Xu, F.; He, Q.; Li, X.; Chang, C.; et al. U2AF1 mutation promotes tumorigenicity through facilitating autophagy flux mediated by FOXO3a activation in myelodysplastic syndromes. Cell Death Dis. 2021, 12, 655. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Cazzola, M.; Boultwood, J.; Malcovati, L.; Vyas, P.; Bowen, D.; Pellagatti, A.; Wainscoat, J.S.; Hellstrom-Lindberg, E.; Gambacorti-Passerini, C.; et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011, 365, 1384–1395. [Google Scholar] [CrossRef]
- Woll, P.S.; Kjallquist, U.; Chowdhury, O.; Doolittle, H.; Wedge, D.C.; Thongjuea, S.; Erlandsson, R.; Ngara, M.; Anderson, K.; Deng, Q.; et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell 2014, 25, 794–808. [Google Scholar] [CrossRef]
- Visconte, V.; Avishai, N.; Mahfouz, R.; Tabarroki, A.; Cowen, J.; Sharghi-Moshtaghin, R.; Hitomi, M.; Rogers, H.J.; Hasrouni, E.; Phillips, J.; et al. Distinct iron architecture in SF3B1-mutant myelodysplastic syndrome patients is linked to an SLC25A37 splice variant with a retained intron. Leukemia 2015, 29, 188–195. [Google Scholar] [CrossRef]
- Mian, S.A.; Rouault-Pierre, K.; Smith, A.E.; Seidl, T.; Pizzitola, I.; Kizilors, A.; Kulasekararaj, A.G.; Bonnet, D.; Mufti, G.J. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 2015, 6, 10004. [Google Scholar] [CrossRef]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Mian, S.A.; Smith, A.E.; Kulasekararaj, A.G.; Kizilors, A.; Mohamedali, A.M.; Lea, N.C.; Mitsopoulos, K.; Ford, K.; Nasser, E.; Seidl, T.; et al. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica 2013, 98, 1058–1066. [Google Scholar] [CrossRef]
- Claus, R.; Lubbert, M. Epigenetic targets in hematopoietic malignancies. Oncogene 2003, 22, 6489–6496. [Google Scholar] [CrossRef]
- Issa, J.P. Epigenetic changes in the myelodysplastic syndrome. Hematol. Oncol. Clin. N. Am. 2010, 24, 317–330. [Google Scholar] [CrossRef]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- Khan, H.; Vale, C.; Bhagat, T.; Verma, A. Role of DNA methylation in the pathogenesis and treatment of myelodysplastic syndromes. Semin. Hematol. 2013, 50, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Luo, Y.; Zhu, S.; Maggio, D.; Yang, H.; Hu, C.; Wang, J.; Zhang, H.; Ren, Y.; Zhou, X.; et al. Isocitrate dehydrogenase 2 mutations correlate with leukemic transformation and are predicted by 2-hydroxyglutarate in myelodysplastic syndromes. J. Cancer Res. Clin. Oncol. 2018, 144, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Jabbour, E.; Ravandi, F.; Takahashi, K.; Daver, N.; Routbort, M.; Patel, K.P.; Brandt, M.; Pierce, S.; Kantarjian, H.; et al. IDH1 and IDH2 mutations in myelodysplastic syndromes and role in disease progression. Leukemia 2016, 30, 980–984. [Google Scholar] [CrossRef]
- Simard, J.-C.; Cesaro, A.; Chapeton-Montes, J.; Tardif, M.; Antoine, F.; Girard, D.; Tessier, P.A. S100A8 and S100A9 Induce Cytokine Expression and Regulate the NLRP3 Inflammasome via ROS-Dependent Activation of NF-κB1. PLoS ONE 2013, 8, e72138. [Google Scholar] [CrossRef] [PubMed]
- Basiorka, A.A.; McGraw, K.L.; Eksioglu, E.A.; Chen, X.; Johnson, J.; Zhang, L.; Zhang, Q.; Irvine, B.A.; Cluzeau, T.; Sallman, D.A.; et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 2016, 128, 2960–2975. [Google Scholar] [CrossRef]
- Maria, V.; Evaggelia, P.; Irene, M.; Maria, P.; Christos, T.; Anastasis, O.; Ioannis, I.; Pavlos, K.; Helen, A.P. Impaired clearance of apoptotic cells leads to HMGB1 release in the bone marrow of patients with myelodysplastic syndromes and induces TLR4-mediated cytokine production. Haematologica 2013, 98, 1206–1215. [Google Scholar] [CrossRef]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Maratheftis, C.I.; Andreakos, E.; Moutsopoulos, H.M.; Voulgarelis, M. Toll-like receptor-4 is up-regulated in hematopoietic progenitor cells and contributes to increased apoptosis in myelodysplastic syndromes. Clin. Cancer Res. 2007, 13, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Dimicoli, S.; Bueso-Ramos, C.; Chen, R.; Yang, H.; Neuberg, D.; Pierce, S.; Jia, Y.; Zheng, H.; Wang, H.; et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia 2013, 27, 1832–1840. [Google Scholar] [CrossRef]
- Sallman, D.A.; Cluzeau, T.; Basiorka, A.A.; List, A. Unraveling the Pathogenesis of MDS: The NLRP3 Inflammasome and Pyroptosis Drive the MDS Phenotype. Front. Oncol. 2016, 6, 151. [Google Scholar] [CrossRef]
- Sallman, D.A.; List, A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood 2019, 133, 1039–1048. [Google Scholar] [CrossRef]
- Sakhdari, A.; Class, C.; Montalban-Bravo, G.; Sasaki, K.; Bueso-Ramos, C.E.; Patel, K.P.; Khoury, J.D.; Kantarjian, H.M.; Garcia-Manero, G.; Medeiros, L.J.; et al. Loss of EZH2 Protein Expression in Myelodysplastic Syndrome Correlates with EZH2 Mutation and Portends a Worse Outcome. Blood 2019, 134, 3016. [Google Scholar] [CrossRef]
- Stasik, S.; Middeke, J.M.; Kramer, M.; Rollig, C.; Kramer, A.; Scholl, S.; Hochhaus, A.; Crysandt, M.; Brummendorf, T.H.; Naumann, R.; et al. EZH2 mutations and impact on clinical outcome: An analysis in 1,604 patients with newly diagnosed acute myeloid leukemia. Haematologica 2020, 105, e228–e231. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki-Kashio, M.; Aoyama, K.; Sashida, G.; Oshima, M.; Tomioka, T.; Muto, T.; Wang, C.; Iwama, A. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 2015, 126, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Kosmider, O.; Fenaux, P. Somatic mutations and epigenetic abnormalities in myelodysplastic syndromes. Best Pr. Res. Clin. Haematol. 2013, 26, 355–364. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Adli, M.; LaFave, L.M.; Gao, J.; Hricik, T.; Shih, A.H.; Pandey, S.; Patel, J.P.; Chung, Y.R.; Koche, R.; et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 2012, 22, 180–193. [Google Scholar] [CrossRef]
- Fisher, C.L.; Berger, J.; Randazzo, F.; Brock, H.W. A human homolog of Additional sex combs, ADDITIONAL SEX COMBS-LIKE 1, maps to chromosome 20q11. Gene 2003, 306, 115–126. [Google Scholar] [CrossRef]
- Nolte, F.; Hofmann, W.K. Myelodysplastic syndromes: Molecular pathogenesis and genomic changes. Ann. Hematol. 2008, 87, 777–795. [Google Scholar] [CrossRef] [PubMed]
- Boultwood, J.; Perry, J.; Pellagatti, A.; Fernandez-Mercado, M.; Fernandez-Santamaria, C.; Calasanz, M.J.; Larrayoz, M.J.; Garcia-Delgado, M.; Giagounidis, A.; Malcovati, L.; et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia 2010, 24, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Gelsi-Boyer, V.; Trouplin, V.; Adelaide, J.; Bonansea, J.; Cervera, N.; Carbuccia, N.; Lagarde, A.; Prebet, T.; Nezri, M.; Sainty, D.; et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 2009, 145, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Rocquain, J.; Carbuccia, N.; Trouplin, V.; Raynaud, S.; Murati, A.; Nezri, M.; Tadrist, Z.; Olschwang, S.; Vey, N.; Birnbaum, D.; et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer 2010, 10, 401. [Google Scholar] [CrossRef]
- Chen, T.C.; Hou, H.A.; Chou, W.C.; Tang, J.L.; Kuo, Y.Y.; Chen, C.Y.; Tseng, M.H.; Huang, C.F.; Lai, Y.J.; Chiang, Y.C.; et al. Dynamics of ASXL1 mutation and other associated genetic alterations during disease progression in patients with primary myelodysplastic syndrome. Blood Cancer J. 2014, 4, e177. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thol, F.; Friesen, I.; Damm, F.; Yun, H.; Weissinger, E.M.; Krauter, J.; Wagner, K.; Chaturvedi, A.; Sharma, A.; Wichmann, M.; et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 2011, 29, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T. ASXL1 mutations gain a function. Blood 2018, 131, 274–275. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.E.; Mohamedali, A.M.; Kulasekararaj, A.; Lim, Z.; Gaken, J.; Lea, N.C.; Przychodzen, B.; Mian, S.A.; Nasser, E.E.; Shooter, C.; et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 2010, 116, 3923–3932. [Google Scholar] [CrossRef] [PubMed]
- Kosmider, O.; Gelsi-Boyer, V.; Cheok, M.; Grabar, S.; Della-Valle, V.; Picard, F.; Viguie, F.; Quesnel, B.; Beyne-Rauzy, O.; Solary, E.; et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs). Blood 2009, 114, 3285–3291. [Google Scholar] [CrossRef]
- Jiang, S. Tet2 at the interface between cancer and immunity. Commun. Biol. 2020, 3, 667. [Google Scholar] [CrossRef]
- Quivoron, C.; Couronne, L.; Della Valle, V.; Lopez, C.K.; Plo, I.; Wagner-Ballon, O.; Do Cruzeiro, M.; Delhommeau, F.; Arnulf, B.; Stern, M.H.; et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011, 20, 25–38. [Google Scholar] [CrossRef]
- Grignano, E.; Jachiet, V.; Fenaux, P.; Ades, L.; Fain, O.; Mekinian, A. Autoimmune manifestations associated with myelodysplastic syndromes. Ann. Hematol. 2018, 97, 2015–2023. [Google Scholar] [CrossRef]
- Aggarwal, S.; van de Loosdrecht, A.A.; Alhan, C.; Ossenkoppele, G.J.; Westers, T.M.; Bontkes, H.J. Role of immune responses in the pathogenesis of low-risk MDS and high-risk MDS: Implications for immunotherapy. Br. J. Haematol. 2011, 153, 568–581. [Google Scholar] [CrossRef]
- Kotsianidis, I.; Bouchliou, I.; Nakou, E.; Spanoudakis, E.; Margaritis, D.; Christophoridou, A.V.; Anastasiades, A.; Tsigalou, C.; Bourikas, G.; Karadimitris, A.; et al. Kinetics, function and bone marrow trafficking of CD4+CD25+FOXP3+ regulatory T cells in myelodysplastic syndromes (MDS). Leukemia 2009, 23, 510–518. [Google Scholar] [CrossRef]
- Zou, J.X.; Rollison, D.E.; Boulware, D.; Chen, D.T.; Sloand, E.M.; Pfannes, L.V.; Goronzy, J.J.; Bai, F.; Painter, J.S.; Wei, S.; et al. Altered naive and memory CD4+ T-cell homeostasis and immunosenescence characterize younger patients with myelodysplastic syndrome. Leukemia 2009, 23, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Flies, D.B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef]
- Boddu, P.; Kantarjian, H.; Garcia-Manero, G.; Allison, J.; Sharma, P.; Daver, N. The emerging role of immune checkpoint based approaches in AML and MDS. Leuk. Lymphoma 2018, 59, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Niu, M.; Xu, L.; Luo, S.; Wu, K. Regulation of PD-L1 expression in the tumor microenvironment. J. Hematol. Oncol. 2021, 14, 10. [Google Scholar] [CrossRef]
- Yang, H.; Bueso-Ramos, C.; DiNardo, C.; Estecio, M.R.; Davanlou, M.; Geng, Q.R.; Fang, Z.; Nguyen, M.; Pierce, S.; Wei, Y.; et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia 2014, 28, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Eksioglu, E.A.; Chen, X.; Kandell, W.; Le Trinh, T.; Cen, L.; Qi, J.; Sallman, D.A.; Zhang, Y.; Tu, N.; et al. S100A9-induced overexpression of PD-1/PD-L1 contributes to ineffective hematopoiesis in myelodysplastic syndromes. Leukemia 2019, 33, 2034–2046. [Google Scholar] [CrossRef]
- Montalban-Bravo, G.; Garcia-Manero, G.; Jabbour, E. Therapeutic choices after hypomethylating agent resistance for myelodysplastic syndromes. Curr. Opin. Hematol. 2018, 25, 146–153. [Google Scholar] [CrossRef]
- Mohammadzadeh, A. Co-inhibitory receptors, transcription factors and tolerance. Int. Immunopharmacol. 2020, 84, 106572. [Google Scholar] [CrossRef] [PubMed]
- Shastri, A.; Will, B.; Steidl, U.; Verma, A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood 2017, 129, 1586–1594. [Google Scholar] [CrossRef]
- Asayama, T.; Tamura, H.; Ishibashi, M.; Kuribayashi-Hamada, Y.; Onodera-Kondo, A.; Okuyama, N.; Yamada, A.; Shimizu, M.; Moriya, K.; Takahashi, H.; et al. Functional expression of Tim-3 on blasts and clinical impact of its ligand galectin-9 in myelodysplastic syndromes. Oncotarget 2017, 8, 88904–88917. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Komrokji, R.S.; Brunner, A.M. TIM-3 pathway dysregulation and targeting in cancer. Expert Rev. Anticancer 2021, 21, 523–534. [Google Scholar] [CrossRef]
- Tao, J.; Han, D.; Gao, S.; Zhang, W.; Yu, H.; Liu, P.; Fu, R.; Li, L.; Shao, Z. CD8(+) T cells exhaustion induced by myeloid-derived suppressor cells in myelodysplastic syndromes patients might be through TIM3/Gal-9 pathway. J. Cell Mol. Med. 2020, 24, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Kikushige, Y.; Miyamoto, T.; Yuda, J.; Jabbarzadeh-Tabrizi, S.; Shima, T.; Takayanagi, S.; Niiro, H.; Yurino, A.; Miyawaki, K.; Takenaka, K.; et al. A TIM-3/Gal-9 Autocrine Stimulatory Loop Drives Self-Renewal of Human Myeloid Leukemia Stem Cells and Leukemic Progression. Cell Stem Cell 2015, 17, 341–352. [Google Scholar] [CrossRef]
- Barreyro, L.; Chlon, T.M.; Starczynowski, D.T. Chronic immune response dysregulation in MDS pathogenesis. Blood 2018, 132, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Cull, A.H.; Snetsinger, B.; Buckstein, R.; Wells, R.A.; Rauh, M.J. Tet2 restrains inflammatory gene expression in macrophages. Exp. Hematol. 2017, 55, 56–70.e13. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Shu, J.; Hu, Q.; Zhou, S.H.; Qian, Y.M.; Hu, M.H.; Hu, L.Y.; Wang, Y.G.; Zhou, Y.M.; Lu, J.H. Apoptosis in human myelodysplastic syndrome CD34+ cells is modulated by the upregulation of TLRs and histone H4 acetylation via a beta-arrestin 1 dependent mechanism. Exp. Cell Res. 2016, 340, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Bjorklund, A.T.; Carlsten, M.; Sohlberg, E.; Liu, L.L.; Clancy, T.; Karimi, M.; Cooley, S.; Miller, J.S.; Klimkowska, M.; Schaffer, M.; et al. Complete Remission with Reduction of High-Risk Clones following Haploidentical NK-Cell Therapy against MDS and AML. Clin. Cancer Res. 2018, 24, 1834–1844. [Google Scholar] [CrossRef]
- Carlsten, M.; Baumann, B.C.; Simonsson, M.; Jadersten, M.; Forsblom, A.M.; Hammarstedt, C.; Bryceson, Y.T.; Ljunggren, H.G.; Hellstrom-Lindberg, E.; Malmberg, K.J. Reduced DNAM-1 expression on bone marrow NK cells associated with impaired killing of CD34+ blasts in myelodysplastic syndrome. Leukemia 2010, 24, 1607–1616. [Google Scholar] [CrossRef]
- Carlsten, M.; Jaras, M. Natural Killer Cells in Myeloid Malignancies: Immune Surveillance, NK Cell Dysfunction, and Pharmacological Opportunities to Bolster the Endogenous NK Cells. Front. Immunol. 2019, 10, 2357. [Google Scholar] [CrossRef]
- Porzsolt, F.; Heimpel, H. Natural killer cell activity in preleukaemia. Lancet 1982, 1, 449. [Google Scholar] [CrossRef]
- Takaku, S.; Takaku, F. Natural killer cell activity and preleukaemia. Lancet 1981, 2, 1178. [Google Scholar] [CrossRef]
- Ma, L.; Delforge, M.; van Duppen, V.; Verhoef, G.; Emanuel, B.; Boogaerts, M.; Hagemeijer, A.; Vandenberghe, P. Circulating myeloid and lymphoid precursor dendritic cells are clonally involved in myelodysplastic syndromes. Leukemia 2004, 18, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Bento, L.C.; Bacal, N.S.; Rocha, F.A.; Severino, P.; Marti, L.C. Bone Marrow Monocytes and Derived Dendritic Cells from Myelodysplastic Patients Have Functional Abnormalities Associated with Defective Response to Bacterial Infection. J. Immunol. 2020, 204, 2098–2109. [Google Scholar] [CrossRef] [PubMed]
- Ganan-Gomez, I.; Wei, Y.; Starczynowski, D.T.; Colla, S.; Yang, H.; Cabrero-Calvo, M.; Bohannan, Z.S.; Verma, A.; Steidl, U.; Garcia-Manero, G. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia 2015, 29, 1458–1469. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, H.; Shao, Z. Monocyte-Derived Macrophages Are Impaired in Myelodysplastic Syndrome. J. Immunol. Res. 2016, 2016, 5479013. [Google Scholar] [CrossRef]
- Haddad, F.; Daver, N. Targeting CD47/SIRPα in Acute Myeloid Leukemia and Myelodysplastic Syndrome: Preclinical and Clinical Developments of Magrolimab. J. Immunother. Precis. Oncol. 2021, 4, 67–71. [Google Scholar] [CrossRef]
- Murata, Y.; Saito, Y.; Kotani, T.; Matozaki, T. CD47-signal regulatory protein alpha signaling system and its application to cancer immunotherapy. Cancer Sci. 2018, 109, 2349–2357. [Google Scholar] [CrossRef]
- Eladl, E.; Tremblay-LeMay, R.; Rastgoo, N.; Musani, R.; Chen, W.; Liu, A.; Chang, H. Role of CD47 in Hematological Malignancies. J. Hematol. Oncol. 2020, 13, 96. [Google Scholar] [CrossRef]
- Jiang, H.; Fu, R.; Wang, H.; Li, L.; Liu, H.; Shao, Z. CD47 is expressed abnormally on hematopoietic cells in myelodysplastic syndrome. Leuk. Res. 2013, 37, 907–910. [Google Scholar] [CrossRef]
- Soto-Pantoja, D.R.; Terabe, M.; Ghosh, A.; Ridnour, L.A.; DeGraff, W.G.; Wink, D.A.; Berzofsky, J.A.; Roberts, D.D. CD47 in the Tumor Microenvironment Limits Cooperation between Antitumor T-cell Immunity and Radiotherapy. Cancer Res. 2014, 74, 6771–6783. [Google Scholar] [CrossRef] [PubMed]
- Sorm, F.; Piskala, A.; Cihak, A.; Vesely, J. 5-Azacytidine, a new, highly effective cancerostatic. Experientia 1964, 20, 202–203. [Google Scholar] [CrossRef] [PubMed]
- Boultwood, J.; Wainscoat, J.S. Gene silencing by DNA methylation in haematological malignancies. Br. J. Haematol. 2007, 138, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Joeckel, T.E.; Lubbert, M. Clinical results with the DNA hypomethylating agent 5-aza-2’-deoxycytidine (decitabine) in patients with myelodysplastic syndromes: An update. Semin. Hematol. 2012, 49, 330–341. [Google Scholar] [CrossRef]
- Cheson, B.D.; Bennett, J.M.; Kantarjian, H.; Pinto, A.; Schiffer, C.A.; Nimer, S.D.; Lowenberg, B.; Beran, M.; de Witte, T.M.; Stone, R.M.; et al. Report of an international working group to standardize response criteria for myelodysplastic syndromes. Blood 2000, 96, 3671–3674. [Google Scholar] [PubMed]
- Steensma, D.P.; Baer, M.R.; Slack, J.L.; Buckstein, R.; Godley, L.A.; Garcia-Manero, G.; Albitar, M.; Larsen, J.S.; Arora, S.; Cullen, M.T.; et al. Multicenter study of decitabine administered daily for 5 days every 4 weeks to adults with myelodysplastic syndromes: The alternative dosing for outpatient treatment (ADOPT) trial. J. Clin. Oncol. 2009, 27, 3842–3848. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Arelin, V.; Gruenebach, F.; Krusch, M.; Schmidt, S.M.; Salih, H.R. Azacytidine impairs NK cell reactivity while decitabine augments NK cell responsiveness toward stimulation. Int. J. Cancer 2011, 128, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-R.; Kim, H.J.; Sohn, M.-J.; Lim, J.-Y.; Park, K.-S.; Lee, S.; Chung, N.-G.; Jeong, D.-C.; Min, C.-K.; Kim, Y.-J. Effects of decitabine on allogeneic immune reactions of donor lymphocyte infusion via activation of dendritic cells. Exp. Hematol. Oncol. 2020, 9, 22. [Google Scholar] [CrossRef]
- Stomper, J.; Rotondo, J.C.; Greve, G.; Lübbert, M. Hypomethylating agents (HMA) for the treatment of acute myeloid leukemia and myelodysplastic syndromes: Mechanisms of resistance and novel HMA-based therapies. Leukemia 2021, 35, 1873–1889. [Google Scholar] [CrossRef]
- Gañán-Gómez, I.; Alfonso, A.; Ogoti, Y.; Yang, H.; Montalban-Bravo, G.; Yu, A.C.; Marchesini, M.; Bueso-Ramos, C.E.; Takahashi, K.; Clise-Dwyer, K.; et al. Hematopoietic Architecture of MDS before and after Progression Reveals Two Biologically Distinct Disease Subtypes. Blood 2016, 128, 4310. [Google Scholar] [CrossRef]
- Nahas, M.R.; Stroopinsky, D.; Rosenblatt, J.; Cole, L.; Pyzer, A.R.; Anastasiadou, E.; Sergeeva, A.; Ephraim, A.; Washington, A.; Orr, S.; et al. Hypomethylating agent alters the immune microenvironment in acute myeloid leukaemia (AML) and enhances the immunogenicity of a dendritic cell/AML vaccine. Br. J. Haematol. 2019, 185, 679–690. [Google Scholar] [CrossRef]
- Xiong, H.; Mittman, S.; Rodriguez, R.; Moskalenko, M.; Pacheco-Sanchez, P.; Yang, Y.; Nickles, D.; Cubas, R. Anti-PD-L1 Treatment Results in Functional Remodeling of the Macrophage Compartment. Cancer Res. 2019, 79, 1493–1506. [Google Scholar] [CrossRef]
- Jia, X.; Yang, W.; Zhou, X.; Han, L.; Shi, J. Influence of demethylation on regulatory T and Th17 cells in myelodysplastic syndrome. Oncol. Lett. 2020, 19, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Costantini, B.; Kordasti, S.Y.; Kulasekararaj, A.G.; Jiang, J.; Seidl, T.; Abellan, P.P.; Mohamedali, A.; Thomas, N.S.; Farzaneh, F.; Mufti, G.J. The effects of 5-azacytidine on the function and number of regulatory T cells and T-effectors in myelodysplastic syndrome. Haematologica 2013, 98, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Diesch, J.; Zwick, A.; Garz, A.K.; Palau, A.; Buschbeck, M.; Gotze, K.S. A clinical-molecular update on azanucleoside-based therapy for the treatment of hematologic cancers. Clin. Epigenet. 2016, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Jelinek, J.; Si, J.; Shu, J.; Issa, J.P. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood 2009, 113, 659–667. [Google Scholar] [CrossRef]
- Duchmann, M.; Itzykson, R. Clinical update on hypomethylating agents. Int. J. Hematol. 2019, 110, 161–169. [Google Scholar] [CrossRef]
- Valencia, A.; Masala, E.; Rossi, A.; Martino, A.; Sanna, A.; Buchi, F.; Canzian, F.; Cilloni, D.; Gaidano, V.; Voso, M.T.; et al. Expression of nucleoside-metabolizing enzymes in myelodysplastic syndromes and modulation of response to azacitidine. Leukemia 2014, 28, 621–628. [Google Scholar] [CrossRef]
- Sripayap, P.; Nagai, T.; Uesawa, M.; Kobayashi, H.; Tsukahara, T.; Ohmine, K.; Muroi, K.; Ozawa, K. Mechanisms of resistance to azacitidine in human leukemia cell lines. Exp. Hematol. 2014, 42, 294–306.e292. [Google Scholar] [CrossRef]
- Gruber, E.; Franich, R.L.; Shortt, J.; Johnstone, R.W.; Kats, L.M. Distinct and overlapping mechanisms of resistance to azacytidine and guadecitabine in acute myeloid leukemia. Leukemia 2020, 34, 3388–3392. [Google Scholar] [CrossRef]
- Orskov, A.D.; Treppendahl, M.B.; Skovbo, A.; Holm, M.S.; Friis, L.S.; Hokland, M.; Gronbaek, K. Hypomethylation and up-regulation of PD-1 in T cells by azacytidine in MDS/AML patients: A rationale for combined targeting of PD-1 and DNA methylation. Oncotarget 2015, 6, 9612–9626. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manero, G.; Gore, S.D.; Cogle, C.; Ward, R.; Shi, T.; Macbeth, K.J.; Laille, E.; Giordano, H.; Sakoian, S.; Jabbour, E.; et al. Phase I study of oral azacitidine in myelodysplastic syndromes, chronic myelomonocytic leukemia, and acute myeloid leukemia. J. Clin. Oncol. 2011, 29, 2521–2527. [Google Scholar] [CrossRef] [PubMed]
- Laille, E.; Shi, T.; Garcia-Manero, G.; Cogle, C.R.; Gore, S.D.; Hetzer, J.; Kumar, K.; Skikne, B.; MacBeth, K.J. Pharmacokinetics and Pharmacodynamics with Extended Dosing of CC-486 in Patients with Hematologic Malignancies. PLoS ONE 2015, 10, e0135520. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Gore, S.D.; Kambhampati, S.; Scott, B.; Tefferi, A.; Cogle, C.R.; Edenfield, W.J.; Hetzer, J.; Kumar, K.; Laille, E.; et al. Efficacy and safety of extended dosing schedules of CC-486 (oral azacitidine) in patients with lower-risk myelodysplastic syndromes. Leukemia 2016, 30, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manero, G.; Santini, V.; Almeida, A.; Platzbecker, U.; Jonasova, A.; Silverman, L.R.; Falantes, J.; Reda, G.; Buccisano, F.; Fenaux, P.; et al. Phase III, Randomized, Placebo-Controlled Trial of CC-486 (Oral Azacitidine) in Patients With Lower-Risk Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1426–1436. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Daver, N.G.; Xu, J.; Chao, M.; Chung, T.; Tan, A.; Wang, V.; Wei, A.; Vyas, P.; Sallman, D.A. Magrolimab + azacitidine versus azacitidine + placebo in untreated higher risk (HR) myelodysplastic syndrome (MDS): The phase 3, randomized, ENHANCE study. J. Clin. Oncol. 2021, 39, TPS7055. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Roboz, G.J.; Kropf, P.L.; Yee, K.W.L.; O’Connell, C.L.; Tibes, R.; Walsh, K.J.; Podoltsev, N.A.; Griffiths, E.A.; Jabbour, E.; et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: Phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 2017, 18, 1317–1326. [Google Scholar] [CrossRef]
- Issa, J.J.; Roboz, G.; Rizzieri, D.; Jabbour, E.; Stock, W.; O’Connell, C.; Yee, K.; Tibes, R.; Griffiths, E.A.; Walsh, K.; et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 2015, 16, 1099–1110. [Google Scholar] [CrossRef]
- Sebert, M.; Renneville, A.; Bally, C.; Peterlin, P.; Beyne-Rauzy, O.; Legros, L.; Gourin, M.P.; Sanhes, L.; Wattel, E.; Gyan, E.; et al. A phase II study of guadecitabine in higher-risk myelodysplastic syndrome and low blast count acute myeloid leukemia after azacitidine failure. Haematologica 2019, 104, 1565–1571. [Google Scholar] [CrossRef]
- Astex-Pharmaceuticals; Otuka-Harmaceuticals. Astex and Otsuka Announce Results of Phase 3 ASTRAL-2 and ASTRAL-3 Studies of Guadecitabine (SGI-110) in Patients with Previously Treated Acute Myeloid Leukemia (AML) and Myelodysplastic Syndromes or Chronic Myelomonocytic Leukemia (MDS/CMML). Available online: https://astx.com/astex-and-otsuka-announce-results-of-phase-3-astral-2-and-astral-3-studies-of-guadecitabine-sgi-110-in-patients-with-previously-treated-acute-myeloid-leukemia-aml-and-myelodysplastic-syndromes-or/ (accessed on 17 September 2021).
- Savona, M.R.; Odenike, O.; Amrein, P.C.; Steensma, D.P.; DeZern, A.E.; Michaelis, L.C.; Faderl, S.; Harb, W.; Kantarjian, H.; Lowder, J.; et al. An oral fixed-dose combination of decitabine and cedazuridine in myelodysplastic syndromes: A multicentre, open-label, dose-escalation, phase 1 study. Lancet Haematol. 2019, 6, e194–e203. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Griffiths, E.A.; Steensma, D.P.; Roboz, G.J.; Wells, R.; McCloskey, J.; Odenike, O.; DeZern, A.E.; Yee, K.; Busque, L.; et al. Oral cedazuridine/decitabine for MDS and CMML: A phase 2 pharmacokinetic/pharmacodynamic randomized crossover study. Blood 2020, 136, 674–683. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; McCloskey, J.; Griffiths, E.A.; Yee, K.W.L.; Zeidan, A.M.; Al-Kali, A.; Dao, K.-H.; Deeg, H.J.; Patel, P.A.; Sabloff, M.; et al. Pharmacokinetic Exposure Equivalence and Preliminary Efficacy and Safety from a Randomized Cross over Phase 3 Study (ASCERTAIN study) of an Oral Hypomethylating Agent ASTX727 (cedazuridine/decitabine) Compared to IV Decitabine. Blood 2019, 134, 846. [Google Scholar] [CrossRef]
- Jehangir, W.; Karabachev, A.; Jahangir, T.; Umyarova, E. Myelodysplastic Syndrome with Transfusion Dependence Treated with Venetoclax. Case Rep. Hematol. 2020, 2020, 9031067. [Google Scholar] [CrossRef]
- Pagliuca, S.; Gurnari, C.; Visconte, V. Molecular Targeted Therapy in Myelodysplastic Syndromes: New Options for Tailored Treatments. Cancers 2021, 13, 784. [Google Scholar] [CrossRef]
- Gangat, N.; Tefferi, A. Venetoclax-based chemotherapy in acute and chronic myeloid neoplasms: Literature survey and practice points. Blood Cancer J. 2020, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.N.; John, A.J.; Kaufmann, S.H. Resistance to venetoclax and hypomethylating agents in acute myeloid leukemia. Cancer Drug Resist. 2021, 4, 125–142. [Google Scholar] [CrossRef] [PubMed]
- Jilg, S.; Hauch, R.T.; Kauschinger, J.; Buschhorn, L.; Odinius, T.O.; Dill, V.; Muller-Thomas, C.; Herold, T.; Prodinger, P.M.; Schmidt, B.; et al. Venetoclax with azacitidine targets refractory MDS but spares healthy hematopoiesis at tailored dose. Exp. Hematol. Oncol. 2019, 8, 9. [Google Scholar] [CrossRef]
- Wei, A.H.; Garcia, J.S.; Borate, U.; Fong, C.Y.; Baer, M.R.; Nolte, F.; Peterlin, P.; Jurcic, J.G.; Garcia-Manero, G.; Hong, W.-J.; et al. A Phase 1b Study Evaluating the Safety and Efficacy of Venetoclax in Combination with Azacitidine in Treatment-Naïve Patients with Higher-Risk Myelodysplastic Syndrome. Blood 2019, 134, 568. [Google Scholar] [CrossRef]
- Bewersdorf, J.P.; Zeidan, A.M. Management of patients with higher-risk myelodysplastic syndromes after failure of hypomethylating agents: What is on the horizon? Best Pr. Res. Clin. Haematol. 2021, 34, 101245. [Google Scholar] [CrossRef]
- Ball, B.J.; Famulare, C.A.; Stein, E.M.; Tallman, M.S.; Derkach, A.; Roshal, M.; Gill, S.I.; Manning, B.M.; Koprivnikar, J.; McCloskey, J.; et al. Venetoclax and hypomethylating agents (HMAs) induce high response rates in MDS, including patients after HMA therapy failure. Blood Adv. 2020, 4, 2866–2870. [Google Scholar] [CrossRef] [PubMed]
- Zeidan, A.M.; Garcia, J.S.; Fenaux, P.; Platzbecker, U.; Miyazaki, Y.; Xiao, Z.-J.; Zhou, Y.; Naqvi, K.; Kye, S.; Garcia-Manero, G. Phase 3 VERONA study of venetoclax with azacitidine to assess change in complete remission and overall survival in treatment-naïve higher-risk myelodysplastic syndromes. J. Clin. Oncol. 2021, 39, TPS7054. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Stevens, B.M.; Jones, C.L.; Winters, A.; Pei, S.; Minhajuddin, M.; D’Alessandro, A.; Culp-Hill, R.; Riemondy, K.A.; Gillen, A.E.; et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat. Med. 2018, 24, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.L.; Stevens, B.M.; Pollyea, D.A.; Culp-Hill, R.; Reisz, J.A.; Nemkov, T.; Gehrke, S.; Gamboni, F.; Krug, A.; Winters, A.; et al. Nicotinamide Metabolism Mediates Resistance to Venetoclax in Relapsed Acute Myeloid Leukemia Stem Cells. Cell Stem Cell 2020, 27, 748–764.e744. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Watts, J.M.; Stein, E.M.; De Botton, S.; Fathi, A.T.; Prince, G.T.; Stein, A.S.; Foran, J.M.; Stone, R.M.; Patel, P.A.; et al. Ivosidenib (AG-120) Induced Durable Remissions and Transfusion Independence in Patients with IDH1-Mutant Relapsed or Refractory Myelodysplastic Syndrome: Results from a Phase 1 Dose Escalation and Expansion Study. Blood 2018, 132, 1812. [Google Scholar] [CrossRef]
- Zeng, Z.; Konopleva, M. Concurrent inhibition of IDH and methyltransferase maximizes therapeutic efficacy in IDH mutant acute myeloid leukemia. Haematologica 2021, 106, 324–326. [Google Scholar] [CrossRef] [PubMed]
- Gil-Perez, A.; Montalban-Bravo, G. Management of myelodysplastic syndromes after failure of response to hypomethylating agents. Adv. Hematol 2019, 10, 2040620719847059. [Google Scholar] [CrossRef] [PubMed]
- Foran, J.M.; DiNardo, C.D.; Watts, J.M.; Stein, E.M.; De Botton, S.; Fathi, A.T.; Prince, G.T.; Stein, A.; Stone, R.M.; Patel, P.A.; et al. Ivosidenib (AG-120) in Patients with IDH1-Mutant Relapsed/Refractory Myelodysplastic Syndrome: Updated Enrollment of a Phase 1 Dose Escalation and Expansion Study. Blood 2019, 134, 4254. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Foran, J.M.; Watts, J.M.; Stein, E.M.; Botton, S.D.; Fathi, A.T.; Prince, G.T.; Stein, A.S.; Stone, R.M.; Patel, P.A.; et al. MDS-265: Ivosidenib (IVO) in Patients with IDH1-Mutant Relapsed/Refractory Myelodysplastic Syndrome (R/R MDS): Updated Enrollment of a Phase 1 Dose Escalation and Expansion Study. Clin. Lymphoma Myeloma Leuk. 2020, 20, S321. [Google Scholar] [CrossRef]
- Cortes, J.E.; Wang, E.S.; Watts, J.M.; Lee, S.; Baer, M.R.; Dao, K.-H.; Dinner, S.; Yang, J.; Donnellan, W.B.; Schwarer, A.P.; et al. Olutasidenib (FT-2102) Induces Rapid Remissions in Patients with IDH1-Mutant Myelodysplastic Syndrome: Results of Phase 1/2 Single Agent Treatment and Combination with Azacitidine. Blood 2019, 134, 674. [Google Scholar] [CrossRef]
- Watts, J.M.; Baer, M.R.; Lee, S.; Yang, J.; Dinner, S.N.; Prebet, T.; Schiller, G.J.; Seiter, K.; Ferrell, P.B.; Kelly, P.F.; et al. A phase 1 dose escalation study of the IDH1m inhibitor, FT-2102, in patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS). J. Clin. Oncol. 2018, 36, 7009. [Google Scholar] [CrossRef]
- Stein, E.M.; DiNardo, C.D.; Pollyea, D.A.; Fathi, A.T.; Roboz, G.J.; Altman, J.K.; Stone, R.M.; DeAngelo, D.J.; Levine, R.L.; Flinn, I.W.; et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017, 130, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Stein, E.M.; Fathi, A.T.; DiNardo, C.D.; Pollyea, D.A.; Roboz, G.J.; Collins, R.; Sekeres, M.A.; Stone, R.M.; Attar, E.C.; Frattini, M.G.; et al. Enasidenib in patients with mutant IDH2 myelodysplastic syndromes: A phase 1 subgroup analysis of the multicentre, AG221-C-001 trial. Lancet Haematol. 2020, 7, e309–e319. [Google Scholar] [CrossRef]
- Santini, V. Enasidenib: A magic bullet for myelodysplastic syndromes? Lancet Haematol 2020, 7, e275–e276. [Google Scholar] [CrossRef]
- Richard-Carpentier, G.; DeZern, A.E.; Takahashi, K.; Konopleva, M.Y.; Loghavi, S.; Masarova, L.; Alvarado, Y.; Ravandi, F.; Montalban Bravo, G.; Naqvi, K.; et al. Preliminary Results from the Phase II Study of the IDH2-Inhibitor Enasidenib in Patients with High-Risk IDH2-Mutated Myelodysplastic Syndromes (MDS). Blood 2019, 134, 678. [Google Scholar] [CrossRef]
- Venugopal, S.; Dinardo, C.D.; Takahashi, K.; Konopleva, M.; Loghavi, S.; Borthakur, G.; Dezern, A.E.; Masarova, L.; Daver, N.G.; Short, N.J.; et al. Phase II study of the IDH2-inhibitor enasidenib in patients with high-risk IDH2-mutated myelodysplastic syndromes (MDS). J. Clin. Oncol. 2021, 39, 7010. [Google Scholar] [CrossRef]
- Strati, P.; Kantarjian, H.; Ravandi, F.; Nazha, A.; Borthakur, G.; Daver, N.; Kadia, T.; Estrov, Z.; Garcia-Manero, G.; Konopleva, M.; et al. Phase I/II trial of the combination of midostaurin (PKC412) and 5-azacytidine for patients with acute myeloid leukemia and myelodysplastic syndrome. Am. J. Hematol. 2015, 90, 276–281. [Google Scholar] [CrossRef]
- Fischer, T.; Stone, R.M.; Deangelo, D.J.; Galinsky, I.; Estey, E.; Lanza, C.; Fox, E.; Ehninger, G.; Feldman, E.J.; Schiller, G.J.; et al. Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J. Clin. Oncol. 2010, 28, 4339–4345. [Google Scholar] [CrossRef]
- Rao, A.; Rizzieri, D.A.; Vanegas, E.; Moore, J.O.; Gockerman, J.P.; Diehl, L.F.; Adams, D.; Warzecho, J.; Decastro, C. Pilot Study of Sorafenib for Myelodysplastic Syndrome. Blood 2012, 120, 4948. [Google Scholar] [CrossRef]
- Macdonald, D.A.; Assouline, S.E.; Brandwein, J.; Kamel-Reid, S.; Eisenhauer, E.A.; Couban, S.; Caplan, S.; Foo, A.; Walsh, W.; Leber, B. A phase I/II study of sorafenib in combination with low dose cytarabine in elderly patients with acute myeloid leukemia or high-risk myelodysplastic syndrome from the National Cancer Institute of Canada Clinical Trials Group: Trial IND.186. Leuk. Lymphoma 2013, 54, 760–766. [Google Scholar] [CrossRef]
- Abdelall, W.; Kantarjian, H.M.; Borthakur, G.; Garcia-Manero, G.; Patel, K.P.; Jabbour, E.J.; Daver, N.G.; Kadia, T.; Gborogen, R.A.; Konopleva, M.; et al. The Combination of Quizartinib with Azacitidine or Low Dose Cytarabine Is Highly Active in Patients (Pts) with FLT3-ITD Mutated Myeloid Leukemias: Interim Report of a Phase I/II Trial. Blood 2016, 128, 1642. [Google Scholar] [CrossRef]
- Armstrong, R.N.; Steeples, V.; Singh, S.; Sanchi, A.; Boultwood, J.; Pellagatti, A. Splicing factor mutations in the myelodysplastic syndromes: Target genes and therapeutic approaches. Adv. Biol. Regul. 2018, 67, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Boultwood, J. Splicing factor gene mutations in the myelodysplastic syndromes: Impact on disease phenotype and therapeutic applications. Adv. Biol. Regul. 2017, 63, 59–70. [Google Scholar] [CrossRef]
- Taylor, J.; Lee, S.C. Mutations in spliceosome genes and therapeutic opportunities in myeloid malignancies. Genes Chromosomes Cancer 2019, 58, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Eskens, F.A.; Ramos, F.J.; Burger, H.; O’Brien, J.P.; Piera, A.; de Jonge, M.J.; Mizui, Y.; Wiemer, E.A.; Carreras, M.J.; Baselga, J.; et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res. 2013, 19, 6296–6304. [Google Scholar] [CrossRef] [PubMed]
- Brierley, C.K.; Steensma, D.P. Targeting Splicing in the Treatment of Myelodysplastic Syndromes and Other Myeloid Neoplasms. Curr. Hematol. Malig. Rep. 2016, 11, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Seiler, M.; Yoshimi, A.; Darman, R.; Chan, B.; Keaney, G.; Thomas, M.; Agrawal, A.A.; Caleb, B.; Csibi, A.; Sean, E.; et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 2018, 24, 497–504. [Google Scholar] [CrossRef]
- Steensma, D.P.; Wermke, M.; Klimek, V.M.; Greenberg, P.L.; Font, P.; Komrokji, R.S.; Yang, J.; Brunner, A.M.; Carraway, H.E.; Ades, L.; et al. Results of a Clinical Trial of H3B-8800, a Splicing Modulator, in Patients with Myelodysplastic Syndromes (MDS), Acute Myeloid Leukemia (AML) or Chronic Myelomonocytic Leukemia (CMML). Blood 2019, 134, 673. [Google Scholar] [CrossRef]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007, 445, 661–665. [Google Scholar] [CrossRef]
- Martins, C.P.; Brown-Swigart, L.; Evan, G.I. Modeling the Therapeutic Efficacy of p53 Restoration in Tumors. Cell 2006, 127, 1323–1334. [Google Scholar] [CrossRef]
- Bykov, V.J.; Zhang, Q.; Zhang, M.; Ceder, S.; Abrahmsen, L.; Wiman, K.G. Targeting of Mutant p53 and the Cellular Redox Balance by APR-246 as a Strategy for Efficient Cancer Therapy. Front. Oncol. 2016, 6, 21. [Google Scholar] [CrossRef]
- Lambert, J.M.; Gorzov, P.; Veprintsev, D.B.; Söderqvist, M.; Segerbäck, D.; Bergman, J.; Fersht, A.R.; Hainaut, P.; Wiman, K.G.; Bykov, V.J. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 2009, 15, 376–388. [Google Scholar] [CrossRef]
- Sallman, D.A.; DeZern, A.E.; Garcia-Manero, G.; Steensma, D.P.; Roboz, G.J.; Sekeres, M.A.; Cluzeau, T.; Sweet, K.L.; McLemore, A.; McGraw, K.L.; et al. Eprenetapopt (APR-246) and Azacitidine in TP53-Mutant Myelodysplastic Syndromes. J. Clin. Oncol. 2021, 39, 1584–1594. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Tallman, M.S.; Martinelli, G.; Ribrag, V.; Yang, H.; Balakumaran, A.; Chlosta, S.; Zhang, Y.; Smith, B.D. Pembrolizumab, a PD-1 Inhibitor, in Patients with Myelodysplastic Syndrome (MDS) after Failure of Hypomethylating Agent Treatment. Blood 2016, 128, 345. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Knaus, H.A.; Robinson, T.M.; Towlerton, A.M.H.; Warren, E.H.; Zeidner, J.F.; Blackford, A.L.; Duffield, A.S.; Rizzieri, D.; Frattini, M.G.; et al. A Multi-center Phase I Trial of Ipilimumab in Patients with Myelodysplastic Syndromes following Hypomethylating Agent Failure. Clin. Cancer Res. 2018, 24, 3519–3527. [Google Scholar] [CrossRef] [PubMed]
- Chien, K.S.; Borthakur, G.M.; Naqvi, K.; Daver, N.G.; Cortes, J.E.; DiNardo, C.D.; Jabbour, E.; Andreeff, M.; Alvarado, Y.; Bose, P.; et al. Updated Preliminary Results from a Phase II Study Combining Azacitidine and Pembrolizumab in Patients with Higher-Risk Myelodysplastic Syndrome. Blood 2019, 134, 4240. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Sasaki, K.; Montalban-Bravo, G.; Daver, N.G.; Jabbour, E.J.; Alvarado, Y.; DiNardo, C.D.; Ravandi, F.; Borthakur, G.; Bose, P.; et al. A Phase II Study of Nivolumab or Ipilimumab with or without Azacitidine for Patients with Myelodysplastic Syndrome (MDS). Blood 2018, 132, 465. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Daver, N.G.; Montalban-Bravo, G.; Jabbour, E.J.; DiNardo, C.D.; Kornblau, S.M.; Bose, P.; Alvarado, Y.; Ohanian, M.; Borthakur, G.; et al. A Phase II Study Evaluating the Combination of Nivolumab (Nivo) or Ipilimumab (Ipi) with Azacitidine in Pts with Previously Treated or Untreated Myelodysplastic Syndromes (MDS). Blood 2016, 128, 344. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Cavenagh, J.; Voso, M.T.; Taussig, D.; Tormo, M.; Boss, I.; Copeland, W.B.; Gray, V.E.; Previtali, A.; O’Connor, T.; et al. Efficacy and Safety of Azacitidine (AZA) in Combination with the Anti-PD-L1 Durvalumab (durva) for the Front-Line Treatment of Older Patients (pts) with Acute Myeloid Leukemia (AML) Who Are Unfit for Intensive Chemotherapy (IC) and Pts with Higher-Risk Myelodysplastic Syndromes (HR-MDS): Results from a Large, International, Randomized Phase 2 Study. Blood 2019, 134, 829. [Google Scholar] [CrossRef]
- Gerds, A.T.; Scott, B.L.; Greenberg, P.L.; Khaled, S.K.; Lin, T.L.; Pollyea, D.A.; Verma, A.; Dail, M.; Green, C.; Ma, C.; et al. PD-L1 Blockade with Atezolizumab in Higher-Risk Myelodysplastic Syndrome: An Initial Safety and Efficacy Analysis. Blood 2018, 132, 466. [Google Scholar] [CrossRef]
- Brunner, A.M.; Esteve, J.; Porkka, K.; Knapper, S.; Vey, N.; Scholl, S.; Garcia-Manero, G.; Wermke, M.; Janssen, J.; Traer, E.; et al. Efficacy and Safety of Sabatolimab (MBG453) in Combination with Hypomethylating Agents (HMAs) in Patients with Acute Myeloid Leukemia (AML) and High-Risk Myelodysplastic Syndrome (HR-MDS): Updated Results from a Phase 1b Study. Blood 2020, 136, 1–2. [Google Scholar] [CrossRef]
- Borate, U.; Esteve, J.; Porkka, K.; Knapper, S.; Vey, N.; Scholl, S.; Garcia-Manero, G.; Wermke, M.; Janssen, J.; Traer, E.; et al. Phase Ib Study of the Anti-TIM-3 Antibody MBG453 in Combination with Decitabine in Patients with High-Risk Myelodysplastic Syndrome (MDS) and Acute Myeloid Leukemia (AML). Blood 2019, 134, 570. [Google Scholar] [CrossRef]
- Chao, M.P.; Takimoto, C.H.; Feng, D.D.; McKenna, K.; Gip, P.; Liu, J.; Volkmer, J.-P.; Weissman, I.L.; Majeti, R. Therapeutic Targeting of the Macrophage Immune Checkpoint CD47 in Myeloid Malignancies. Front. Oncol. 2020, 9, 1380. [Google Scholar] [CrossRef]
- Sallman, D.A.; Asch, A.S.; Al Malki, M.M.; Lee, D.J.; Donnellan, W.B.; Marcucci, G.; Kambhampati, S.; Daver, N.G.; Garcia-Manero, G.; Komrokji, R.S.; et al. The First-in-Class Anti-CD47 Antibody Magrolimab (5F9) in Combination with Azacitidine Is Effective in MDS and AML Patients: Ongoing Phase 1b Results. Blood 2019, 134, 569. [Google Scholar] [CrossRef]
- Sallman, D.A.; Malki, M.A.; Asch, A.S.; Lee, D.J.; Kambhampati, S.; Donnellan, W.B.; Bradley, T.J.; Vyas, P.; Jeyakumar, D.; Marcucci, G.; et al. Tolerability and efficacy of the first-in-class anti-CD47 antibody magrolimab combined with azacitidine in MDS and AML patients: Phase Ib results. J. Clin. Oncol. 2020, 38, 7507. [Google Scholar] [CrossRef]
- Stevens, B.M.; Zhang, W.; Pollyea, D.A.; Winters, A.; Gutman, J.; Smith, C.; Budde, E.; Forman, S.J.; Jordan, C.T.; Purev, E. CD123 CAR T cells for the treatment of myelodysplastic syndrome. Exp. Hematol. 2019, 74, 52–63.e53. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Tao, J.L.; Fu, R.; Wang, H.Q.; Jiang, H.J.; Yue, L.Z.; Zhang, W.; Liu, H.; Shao, Z.H. Increased CD34 + CD38 -CD123 + cells in myelodysplastic syndrome displaying malignant features similar to those in AML. Int. J. Hematol. 2014, 100, 60–69. [Google Scholar] [CrossRef]
- Houchins, J.P.; Yabe, T.; McSherry, C.; Miyokawa, N.; Bach, F.H. Isolation and characterization of NK cell or NK/T cell-specific cDNA clones. J. Mol. Cell Immunol. 1990, 4, 295–304; discussion 305–296. [Google Scholar] [PubMed]
- Carapito, R.; Bahram, S. Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunol. Rev. 2015, 267, 88–116. [Google Scholar] [CrossRef] [PubMed]
- Epling-Burnette, P.K.; Bai, F.; Painter, J.S.; Rollison, D.E.; Salih, H.R.; Krusch, M.; Zou, J.; Ku, E.; Zhong, B.; Boulware, D.; et al. Reduced natural killer (NK) function associated with high-risk myelodysplastic syndrome (MDS) and reduced expression of activating NK receptors. Blood 2007, 109, 4816–4824. [Google Scholar] [CrossRef]
- Gacerez, A.T.; Arellano, B.; Sentman, C.L. How Chimeric Antigen Receptor Design Affects Adoptive T Cell Therapy. J. Cell Physiol. 2016, 231, 2590–2598. [Google Scholar] [CrossRef]
- Giuliani, M.; Poggi, A. Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies. Cells 2020, 9, 1578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Barber, A.; Sentman, C.L. Generation of antitumor responses by genetic modification of primary human T cells with a chimeric NKG2D receptor. Cancer Res. 2006, 66, 5927–5933. [Google Scholar] [CrossRef] [PubMed]
- Barber, A.; Meehan, K.R.; Sentman, C.L. Treatment of multiple myeloma with adoptively transferred chimeric NKG2D receptor-expressing T cells. Gene 2011, 18, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, S.H.; Murad, J.; Werner, L.; Daley, H.; Trebeden-Negre, H.; Gicobi, J.K.; Schmucker, A.; Reder, J.; Sentman, C.L.; Gilham, D.E.; et al. Phase I Trial of Autologous CAR T Cells Targeting NKG2D Ligands in Patients with AML/MDS and Multiple Myeloma. Cancer Immunol. Res. 2019, 7, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Sallman, D.A.; Brayer, J.; Sagatys, E.M.; Lonez, C.; Breman, E.; Agaugué, S.; Verma, B.; Gilham, D.E.; Lehmann, F.F.; Davila, M.L. NKG2D-based chimeric antigen receptor therapy induced remission in a relapsed/refractory acute myeloid leukemia patient. Haematologica 2018, 103, e424–e426. [Google Scholar] [CrossRef]
- Spear, P.; Wu, M.R.; Sentman, M.L.; Sentman, C.L. NKG2D ligands as therapeutic targets. Cancer Immun. 2013, 13, 8. [Google Scholar]
- Le Bert, N.; Gasser, S. Advances in NKG2D ligand recognition and responses by NK cells. Immunol. Cell Biol. 2014, 92, 230–236. [Google Scholar] [CrossRef]
- Hong, S.; Rybicki, L.; Corrigan, D.; Hamilton, B.K.; Sobecks, R.; Kalaycio, M.; Dean, R.M.; Hill, B.T.; Pohlman, B.; Jagadeesh, D.; et al. Targeted Treatment and Survival Following Relapse after Allogeneic Hematopoietic Cell Transplantation for Acute Leukemia and MDS in the Contemporary Era. Blood 2019, 134, 4567. [Google Scholar] [CrossRef]
- Claiborne, J.; Bandyopathyay, D.; Roberts, C.; Hawks, K.; Aziz, M.; Simmons, G.; Wiedl, C.; Chung, H.; Clark, W.; McCarty, J.; et al. Managing post allograft relapse of myeloid neoplasms: Azacitidine and donor lymphocyte infusions as salvage therapy. Leuk. Lymphoma 2019, 60, 2733–2743. [Google Scholar] [CrossRef]
- Zeiser, R.; Beelen, D.W.; Bethge, W.; Bornhauser, M.; Bug, G.; Burchert, A.; Christopeit, M.; Duyster, J.; Finke, J.; Gerbitz, A.; et al. Biology-Driven Approaches to Prevent and Treat Relapse of Myeloid Neoplasia after Allogeneic Hematopoietic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2019, 25, e128–e140. [Google Scholar] [CrossRef]
- Lulla, P.; Naik, S.; Tzannou, I.; Mukhi, S.; Kuvalekar, M.; Robertson, C.; Ramos, C.A.; Carrum, G.; Kamble, R.T.; Gee, A.P.; et al. Administering Leukemia-Directed Donor Lymphocytes to Patients with AML or MDS to Prevent or Treat Post-Allogeneic HSCT Relapse. Biol. Blood Marrow Transplant. 2019, 25, S10–S11. [Google Scholar] [CrossRef]
- Epperly, R.; Gottschalk, S.; Velasquez, M.P. A Bump in the Road: How the Hostile AML Microenvironment Affects CAR T Cell Therapy. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Warlick, E.D.; Miller, J.S. Myelodysplastic syndromes: The role of the immune system in pathogenesis. Leuk. Lymphoma 2011, 52, 2045–2049. [Google Scholar] [CrossRef] [PubMed]
| Drug | Phase | Disease Subtype | Regimen | Status | Clinical Trial Identifier | |
|---|---|---|---|---|---|---|
| Immune Checkpoint Inhibitors | ||||||
| Pembrolizumab | Anti-PD1 | 1 | Hematologic malignancies | Pembrolizumab | Completed | NCT01953692 |
| 1 | MDS | Pembrolizumab + Entinostat | Active, not recruiting | NCT02936752 | ||
| 2 | MDS | Pembrolizumab + AZA | Recruiting | NCT03094637 | ||
| 1 | ND/RR AML/MDS | Pembrolizumab + DEC | Recruiting | NCT03969446 | ||
| Nivolumab | 2 | MDS, R/R MDS | Nivolumab + AZA, Ipilimumab + AZA | Recruiting | NCT02530463 | |
| 2/3 | AML, MDS | AZA + Nivolumab/Midostaurin, DEC + Cytarabine | Active, not recruiting | NCT03092674 | ||
| Durvalumab (MEDI4736) | Anti-PD-L1 | 1 | MDS | Durvalumab, Durvalumab + Tremelimumab | Completed | NCT02117219 |
| 2 | MDS | Durvalumab + AZA | Active, not recruiting | NCT02281084 | ||
| 2 | AML, MDS | AZA, AZA + Durvalumab | Active, not recruiting | NCT02775903 | ||
| Ipilimumab | Anti-CTLA-4 | 1 | R/R MDS, AML | Ipilimumab + DEC | Recruiting | NCT02890329 |
| Sabatolimab (MBG453) | Anti-TIM-3 | 1 | AML, HR-MDS | MBG453, or in combination with PDR001/DEC/AZA | Active, not recruiting | NCT03066648 |
| 2 | HR-MDS | MBG453 + HMA | Active, not recruiting | NCT03946670 | ||
| 3 | HR-MDS, CMML-2 | MBG453 + AZA | Recruiting | NCT04266301 | ||
| 2 | HR-MDS | Sabatolimab + AZA + Venetoclax | Not yet recruiting | NCT04812548 | ||
| 2 | HR-MDS | Sabatolimab + AZA/DEC | Not yet recruiting | NCT04878432 | ||
| TTI-621 (SIRPαFc) | Anti-CD47 | 1 | Hematologic malignancies, solid tumors | TTI-621 for MDS | Recruiting | NCT02663518 |
| Magrolimab | 1 | Hematologic malignancies | Magrolimab, Magrolimab + AZA | Recruiting | NCT03248479 | |
| 3 | MDS | AZA, AZA + Magrolimab | Recruiting | NCT04313881 | ||
| AK117 | 1/2 | MDS | AK117 + AZA | Recruiting | NCT04900350 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, P.; Yim, R.; Yung, Y.; Chu, H.-T.; Yip, P.-K.; Gill, H. Molecular Targeted Therapy and Immunotherapy for Myelodysplastic Syndrome. Int. J. Mol. Sci. 2021, 22, 10232. https://doi.org/10.3390/ijms221910232
Lee P, Yim R, Yung Y, Chu H-T, Yip P-K, Gill H. Molecular Targeted Therapy and Immunotherapy for Myelodysplastic Syndrome. International Journal of Molecular Sciences. 2021; 22(19):10232. https://doi.org/10.3390/ijms221910232
Chicago/Turabian StyleLee, Paul, Rita Yim, Yammy Yung, Hiu-Tung Chu, Pui-Kwan Yip, and Harinder Gill. 2021. "Molecular Targeted Therapy and Immunotherapy for Myelodysplastic Syndrome" International Journal of Molecular Sciences 22, no. 19: 10232. https://doi.org/10.3390/ijms221910232
APA StyleLee, P., Yim, R., Yung, Y., Chu, H.-T., Yip, P.-K., & Gill, H. (2021). Molecular Targeted Therapy and Immunotherapy for Myelodysplastic Syndrome. International Journal of Molecular Sciences, 22(19), 10232. https://doi.org/10.3390/ijms221910232







