Abstract
Chronic myeloid leukemia (CML), a hematopoietic neoplasm arising from the fusion of BCR (breakpoint cluster region) gene on chromosome 22 to the ABL (Abelson leukemia virus) gene on chromosome 9 (BCR-ABL1 oncogene), originates from a small population of leukemic stem cells with extensive capacity for self-renewal and an inflammatory microenvironment. Currently, CML treatment is based on tyrosine kinase inhibitors (TKIs). However, allogeneic hematopoietic stem cell transplantation (HSCT-allo) is currently the only effective treatment of CML. The difficulty of finding a compatible donor and high rates of morbidity and mortality limit transplantation treatment. Despite the safety and efficacy of TKIs, patients can develop resistance. Thus, microRNAs (miRNAs) play a prominent role as biomarkers and post-transcriptional regulators of gene expression. The aim of this study was to analyze the miRNA profile in CML patients who achieved cytogenetic remission after treatment with both HSCT-allo and TKI. Expression analyses of the 758 miRNAs were performed using reverse transcription quantitative polymerase chain reaction (RT-qPCR). Bioinformatics tools were used for data analysis. We detected miRNA profiles using their possible target genes and target pathways. MiR-125a-3p stood out among the downregulated miRNAs, showing an interaction network with 52 target genes. MiR-320b was the only upregulated miRNA, with an interaction network of 26 genes. The results are expected to aid future studies of miRNAs, residual leukemic cells, and prognosis in CML.
1. Introduction
Chronic myeloid leukemia (CML) was the first leukemia described in the literature, by Rudolf Virchow and John Hughes Bennett in 1845 [1]. Over the last 175 years, there have been a number of scientific discoveries involving CML and these have advanced the understanding and treatment of cancer. The understanding of the molecular pathogenesis of CML began with the discovery and correlation of t(9;22)(q34;q11) with the malignancy of the disease [2]. Followed by the identification of the chimeric BCR-ABL1 oncogene resulting from the translocation of the BCR (breakpoint cluster region) gene on chromosome 22 and ABL (Abelson leukemia virus) gene on chromosome 9 that encodes an elevated activity of oncoprotein tyrosine kinase [2].
CML is a clonal myeloproliferative neoplasm of hematopoietic stem cells with an incidence in adults of 1–2 cases per 100,000 inhabitants [3]. Three mechanisms are involved in CML oncogenesis, based on BCR-ABL1 activity: (a) altered cell adhesion to the bone marrow stroma and extracellular matrix, (b) constitutively active mitogenic signaling with reduced apoptosis of hematopoietic stem cells and progenitor cells, and (c) genetic instability leading to disease progression [4].
The identification of BCR-ABL1 tyrosine kinase as a therapeutic target has made possible the development of specific target drugs, administered orally, that molecularly control the tumor burden in CML. Imatinib mesylate (IM) was the first tyrosine kinase inhibitor (TKI) that was developed and approved for therapeutic use. IM acts as a competitive inhibitor of the ATP site on the BCR-ABL1 protein, blocking its tyrosine kinase activity and preventing the substrate from being phosphorylated. This block obstructs the transduction of signals necessary for cell proliferation [5]. Currently, new-generation TKIs are available for the treatment of CML; however, the only effective (curative) treatment of CML is allogeneic hematopoietic stem cell transplantation (HSCT-allo) [6].
HSCT-allo from a healthy allogeneic donor has been shown to consistently eradicate leukemic stem cells in most patients. The successful outcome of this treatment is related to the effectiveness of the leukemia graft reaction (LGR) orchestrated by allogeneic T cells [7].
In the last two decades, the advent of TKIs has changed the indication of HSCT-allo from an early and curative intervention to a salvage treatment, recommended for patients resistant to ITKs or in the advanced stages of the disease [6,8].
Given the evidence of therapeutic resistance to ITKs, recent advances in gene expression technology mechanisms have shown that microRNAs (miRNAs) have great potential both as therapeutic targets and as prognostic markers in different types of cancer [9,10,11].
MiRNAs are small non-protein coding RNAs that act as post-transcriptional regulators of gene expression [12]. Recently, several studies have demonstrated the potential of miRNAs as regulators of physiological and pathological processes [13]. MiRNAs play an important role in T cell development and regulation, as well as in immune reconstitution after HSCT-allo [14]. Furthermore, miRNAs have been shown to play a crucial role in neoplasm pathogenesis, promoting changes such as cell proliferation, angiogenesis, tumor growth, and metastasis [15,16].
From the first descriptions, in 2002, of the relationship between miRNA and cancer, it was observed that cancer patients exhibit dysregulated expression levels of miRNAs, both positively and negatively [17]. Consequently, each type of cancer has a miRNA expression profile relative to its condition [18]. Furthermore, deregulated miRNAs may have functional oncogenic or tumor suppressor roles [19]. Examples of these mechanisms are miR-30e and miR-203 that act as tumor suppressors, downregulating BCR-ABL1 expression in CML [20,21]. Meanwhile, miR-486 can elevate imatinib resistance by targeting FOXO1 and PTEN in CD34 positive CML cells [22].
The aim of this study was to identify a miRNA profile, target genes, and target pathways in CML-chronic phase patient groups treated with HCST-allo and imatinib mesylate, both in cytogenetic remission and over 12 months of treatment. Expression analyses of the 758 miRNAs were performed using RT-qPCR. Bioinformatics tools were used for data analysis. We detected miRNA profiles using their possible target genes and target pathways. MiR-125a-3p stood out among the downregulated miRNAs, showing an interaction network with 52 target genes. MiR-320b was the only upregulated miRNA, with an interaction network of 26 genes.
2. Results
2.1. Differentially Expressed MiRNAs
Expression data for the miRNAs obtained in this study are available from the NCBI GEO database (accession number GSE 164549). A total of 758 miRNAs were analyzed using RT-qPCR array in peripheral blood samples from 14 patients with CML-chronic phase treated with imatinib mesylate and 14 patients treated with HSCT-allo, both groups in cytogenetic remission. According to the cut-off criteria (fold change < 0.5, fold change > 2.0), 43 differentially expressed miRNAs were identified from this microarray dataset. Of these miRNAs, one (2.3%) was upregulated and 42 (97.7%) were downregulated (Table 1).

Table 1.
Differentially expressed MiRNAS.
2.2. MiRNAs Target Genes
The 43 differentially expressed miRNAs were analyzed using the miRWalk program (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/index.html, accessed on 27 August 2021) to identify possible miRNA target genes. Through this analysis we identified 1171 genes. These genes were verified and identified as differentially expressed using microarray data available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, accessed on 23 September 2020), accession number GSE 43225, in a study conducted on CML patients. Among the genes we found in our study, 95% were also identified among the genes in the microarray data from the Gene Expression Omnibus.
MiRNA expression in all patients treated with imatinib mesylate and HSCT-allo using heatmap is shown in Figure 1. Heatmap Clusterization was performed using Euclidean distance. Heatmap was generated and analyzed using Morpheus (https://software.broadinstitute.org/morpheus, accessed on 27 August 2021).
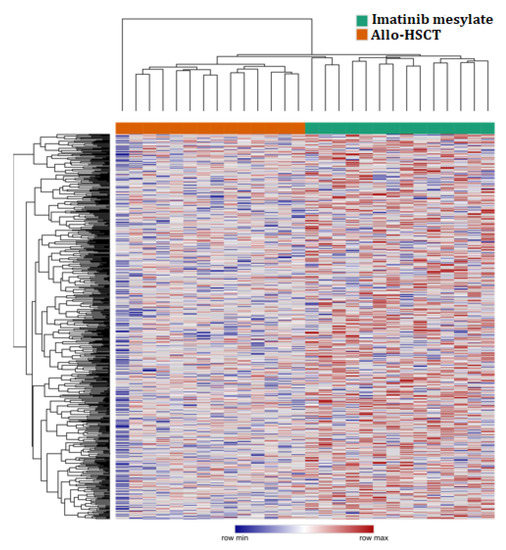
Figure 1.
Heatmap of the normalized expression values of microRNAs in allo-HSCT and imatinib mesylate groups.
2.3. Protein–Protein Interaction Network
The EnrichR tool (Mount Sinai Innovation Partners, New York, NY, USA) http://amp.pharm.mssm.edu/Enrichr/, accessed on 24 September 2020) was used to visualize the main pathways related to the target genes of the miRNAs. The Reactome program provided a biological interpretation and provided models to visualize the results. STRING (version 11.0; (String Consortium, Zurich, Switzerland), https://string-db.org/, accessed on 27 September 2020) was performed on the target genes of upregulated and downregulated miRNAs, generating data visualized through pictures created by Cytoscape software (version 3.8.0; Cytoscape Consortium, Bethesda, MA, USA), http://www.cytoscape.org, accessed on 28 September 2020) (Figure 2 and Figure 3).
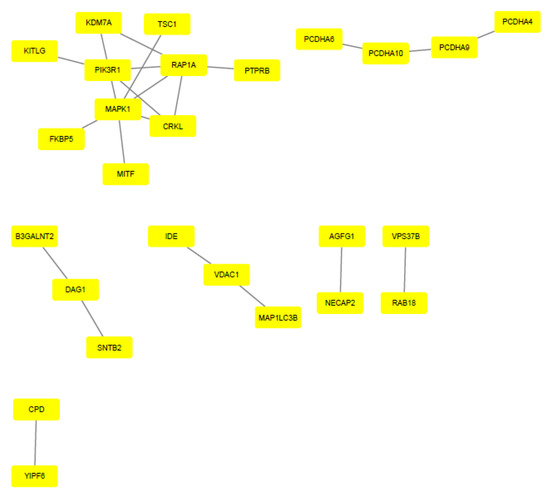
Figure 2.
Protein–protein interaction of upregulated miRNA target genes. Gene interaction was performed using STRING and Cytoscape tools. Genes are represented by yellow rectangles and lines represent protein–protein associations.
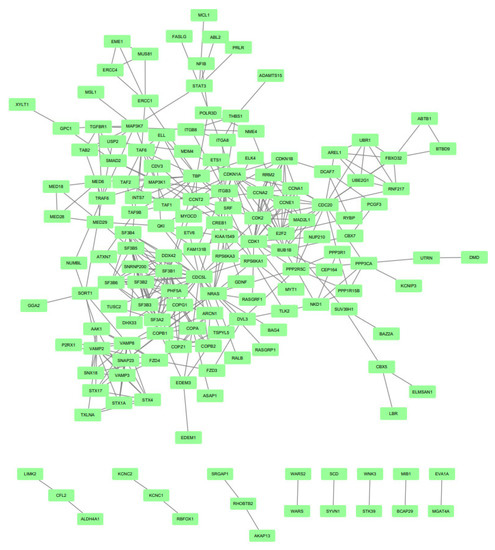
Figure 3.
Protein–protein interaction of downregulated miRNAs target genes. Gene interaction was performed using STRING and Cytoscape tools. Genes are represented by green rectangles and lines represent protein–protein associations.
2.4. Gene Ontology
Based on the network results, the genes that showed the greatest interaction with each other were selected, according to the figures generated by Cytoscape. For these genes, which were considered as the main genes, an analysis was performed using Gene Ontology (http://geneontology.org/, accessed on 24 September 2020) to identify the pathways involved with these genes. The main terms found are listed in Table 2 and Table 3.

Table 2.
Enriched terms generated by Gene Ontology for the main target genes of downregulated miRNAs.

Table 3.
Enriched terms generated by Gene Ontology for the main target genes of upregulated miRNAs.
From the profile of 43 identified miRNAs, we highlighted two miRNAs: miR-320b and miR-125a-3p. MiR-320b was the only upregulated miRNA found, and among those with low expression, miR-125a-3p was the one with the highest gene interaction, standing out among the results. MiR-125a-3p showed an interaction network with 52 target genes, and miR-320b showed an interaction network with 26 genes. The main target genes related to miR-320b and miR-125a-3p are shown in Figure 4. A summary of the methodology steps and the main results are presented in Figure 5.
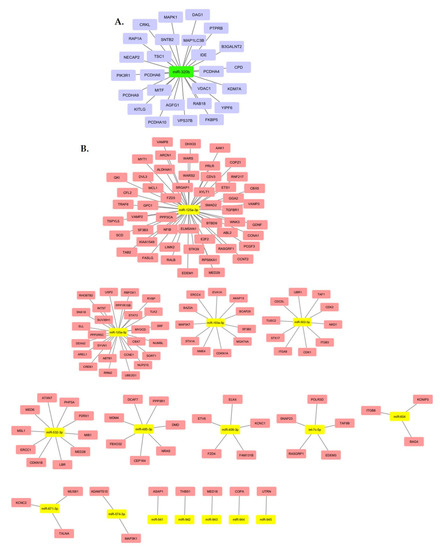
Figure 4.
Main upregulated miRNA target genes (A). Main downregulated miRNA target genes (B).
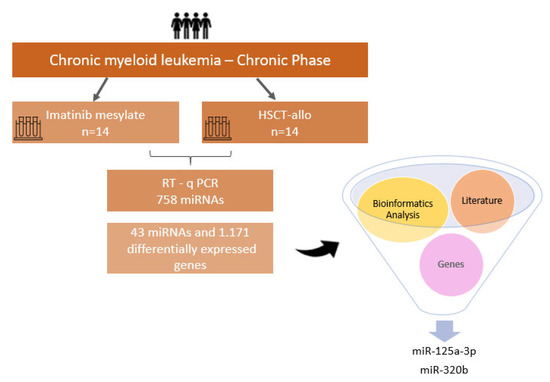
Figure 5.
Summary of methodology steps and main results.
3. Discussion
Chronic myeloid leukemia (CML) is a hematopoietic neoplasm that arises from BCR-ABL1 translocation, originating from a small population of leukemic stem cells with extensive self-renewal capacity and an inflammatory microenvironment. Currently, CML treatment is based on tyrosine kinase inhibitors and target-specific drugs that are safe and effective. However, HSCT-allo was the first treatment in CML patients to eradicate the Philadelphia chromosome. In this process, after high doses of chemotherapy, transplanted hematopoietic stem cells can reestablish the recipient’s hematopoiesis [23].
Currently, HSCT-allo is the only treatment available that promotes a CML cure; however, morbidity and mortality rates related to this treatment are high, in addition to the difficulty of finding a donor, restricting its applicability [24,25,26,27]. Therefore, after the advent of ITK, HSCT-allo began to be used for CML treatment only in patients who developed therapeutic resistance [28].
The first ITK approved for use was imatinib mesylate [29,30], a potent TKI [31,32,33]. However, 10–15% of patients in the chronic phase of CML do not show favorable changes with the use of imatinib mesylate due to resistance or intolerance to the treatment [34,35]. Despite recent advances in CML treatment, these challenges remain and prevent the development of effective therapeutic strategies [36]. With new research of miRNAs, possible new treatment mechanisms are being discovered.
In recent years, many studies have focused on the significant role of miRNAs in physiological and pathological processes [37]. These small RNAs can be identified in the peripheral blood and are used as biomarkers for several diseases [38,39]. Evidence indicates that miRNAs are directly involved in myeloid development and leukemogenesis [40].
In addition to miRNA-based studies, bioinformatics has also contributed to the understanding of processes linked to the development and progression of CML, along with the discovery of possible disease markers. Recent studies have shown increasing evidence of the interaction between genes and proteins, leading to investigations into molecular cancer mechanisms. In this context, bioinformatics is essential for integrating biological systems and computational science, helping to better understand the diagnosis processes, therapy, and prognosis of cancer and other diseases [41].
In our study, we compared two CML patient groups. One group was treated with HSCT-allo, and the other group was treated with imatinib mesylate. The clinical follow-up of transplant patients averaged between 9.6 years and 4.5 years for those treated with imatinib mesylate. In this follow-up period, we observed that five of the 14 (35.7%) patients treated with ITK achieved a deep molecular response (MR4log, MR4.5Log or undetectable) throughout the treatment and another five patients (35.7%) achieved a major molecular response (MR3Log). On the other hand, nine patients (64.3%) treated with HSCT-allo relapsed and five (35.7%) achieved major molecular response (MR3Log). By analyzing the miRNAs of these two patient groups using bioinformatics tools, it was possible to obtain a profile consisting of 43 differentially expressed miRNAs, one (2.3%) upregulated and 42 (97.7%) downregulated (Table 1).
3.1. Upregulated MiR-320b and Differentially Expressed CRKL
The treatment of cancer patients has evolved considerably; however, cell regeneration and the ability of stem cells to self-renew are significant obstacles in this process, as they can lead to tumor recurrence, metastasis, and drug resistance. MiRNAs play a fundamental role in this process, as they are unregulated in several malignant diseases and are important regulators of stem cells and cell reprogramming [42].
MiR-320, the only upregulated miRNA found in our study, was studied by Gao et al. [43], who showed that miR-320, secreted by leukemic cells, is directly related to increased cell proliferation and that leukemic cell exosomes can transport miRNAs to stromal cells, leading to reprogramming of cell niche functions.
Regarding miR-320b target genes, our data pointed to the low expression of CRKL in both groups of patients evaluated. The CRKL family is composed of five members: v-CRK, CRKI, CRKII, CRKIII, and CRL-like protein (CRKL) [44]. CRK proteins are phosphorylation substrates for the BCR-ABL1 fusion oncogene found in over 95% of CML cases. CRKL is a tyrosine-phosphorylated protein found in neutrophils from patients with CML [44]. We believe that the overexpression of miR-320b and the low expression of CRKL found in this study is a consequence of the absence of a deep molecular response in the patient groups evaluated.
3.2. MiR-125a-3p, MiR-485-3p, MiR-409-3p and MiR-574-3p Downregulated
Of the 43 differentially expressed miRNAs identified in our study, 42 (97.7%) were downregulated. Among the 42 downregulated miRNAs, we will highlight miR-125a-3p, miR-485-3p, miR-409-3p, and miR-574-3p, due to their relevance in the literature. This set of miRNAs was described by Xiong and collaborators as differentially expressed in the K562 cell line, BCR-ABL1 positive [45].
Low expression of miR-125a has been detected in bone marrow samples from patients with acute myeloid leukemia (AML) compared to the control group (healthy bone marrow donors) [38]. Previous studies have demonstrated that miR-125a plays a role in cell cycle regulation, proliferation, and apoptosis in AML [46]. In acute promyelocytic leukemia, miR-125b was found differentially expressed, promoting the proliferation of leukemic cells, and inhibiting cellular apoptosis by regulating the expression of the tumor suppressor BCL2-antagonista/killer1 (Bak1) [47].
MiR-485 has been described as poorly expressed in AML HL60 cell lines when compared to normal peripheral blood cells [48]. In a study by Li et al., miR-485-3p was one of 33 miRNAs differentially expressed in a CML patient group. These findings contribute to the understanding of the disease pathogenesis [49].
MiR-409-3p is also a miRNA described in the literature as having reduced expression in CpG-rich methylated CML cell lines [50,51]. This low expression of miRNAs has been associated with a high relapse risk in AML [52,53]. Low expression of miR-574-3p was also observed in CML patients when compared to samples from healthy controls, indicating the involvement of this miRNA in the development and progression of the disease [54].
These findings are expected to contribute to the understanding of disease pathogenesis and persistence of residual BCR-ABL1 cells observed in our patient groups after different therapeutic strategies.
3.3. MAPK and NRAS
After analyzing the target genes, we used Gene Ontology to study the enrichment of the pathways of these genes and to observe the main pathways related to these genes. Some pathways were found to be fundamental to CML development, such as regulation of protein kinase activity, processes related to cell division, and mitogen-activated-protein kinase (MAPK) activity.
The MAPK pathway is an important signaling cascade found in several types of cancer [55] and plays a central role in CML, as it is necessary for the transcription of genes involved in cell proliferation [56,57]. In our results, we found a high expression of MAP3K1 and MAP3K7 genes. These genes are members of the MAPK family [58,59]. MAP3K1 is involved in the survival and migration of tumor cells [60], and MAP3K7 is an important regulator of cell pathways associated with cell proliferation in cancer [59]. NRAS was also found to be highly expressed in our study. The NRA family is composed of three genes associated with carcinogenesis: HRAS, KRAS, and NRAS [61]. Mutations in NRAS are often found in myeloid disorders such as CML [62]. MAPK and NRAS were found to be highly expressed in our study, suggesting residual tumor activity.
In the present study, we explored through bioinformatics tools the profile of miRNAs, their target genes, and related pathways in CML patients undergoing treatment. Protein–protein interaction networks were built to relate miRNAs found with their target genes, and it was possible to obtain data that corroborate the current literature. As this is an exploratory study with real-world data and scarce samples, it was not possible to validate the results through a second round of RT-qPCR. However, we believe that this study contributes an innovative approach and useful results to the fields of oncology and bioinformatics. We stress the need for expanded studies in order to confirm the miRNA profiles discussed in this work as markers of residual disease.
4. Materials and Methods
4.1. Patients and Samples
The study was approved by the Hospital Amaral Carvalho Ethics Committee (CEPHAC-2.917.389). Patients who were followed up at Hospital Amaral Carvalho signed a form attesting to free and informed consent (the FICF form) to participate in the study. Peripheral blood samples from 28 patients with CML-chronic phase in cytogenetic remission (Philadelphia chromosome-negative) were included in the study. The patients were divided into two groups: 14 patients undergoing HSCT-allo and 14 patients treated with imatinib mesylate. A pool of leucocytes from 14 healthy blood donors was considered as the control group. The miRNA profile was determined by comparing the control group and each patients’ group to determine which miRNAs were upregulated or downregulated. The patients’ clinical data are presented in Table 4. All transplant patients received BuCy-2 as a conditioning regimen and cyclosporine and methotrexate as graft-versus-host disease prophylaxis [63,64].

Table 4.
Patients’ clinical characteristics.
4.2. RNA Extraction and Purification
Total RNA was isolated from buffy coat of fresh peripheral blood using the QIAamp® RNA Blood Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. The total RNA amount was determined by the ratios A 260 nm/A 280 nm and A 260 nm/A 230 nm (acceptable when both ratios were greater than 1.8). RNA integrity was ensured by obtaining an RNA integrity number (RIN > 8) using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany) [65,66].
4.3. Expression MiRNAs Profile and Reference Genes
The reverse transcription reaction was performed using the Taqman MicroRNA Reverse Transcription Kit in combination with Megaplex RT Primer Human Pool Set A and B (Thermo Fisher Scientific, Waltham, MA, USA). This reaction transcribed 758 miRNAs and three positive endogenous controls (U6 snRNA, RNU44, and RNU48) and one negative control [65,66].
The TaqMan® MGB strategy was used for quantitative real-time PCR (q-PCR), which is based on the specific annealing of each probe with its complementary sequence. For miRNA amplification, the Taqman® Low Density Array Human MicroRNA Arrays v2.0 (A and B) kit (ABIV®, Life Technologies, Carlsbad, CA, USA) was used according to the manufacturer´s instructions. Analyses were performed on the ViiA7 platform (ABIV®) for a total of 758 miRNAs per patient [65,66].
4.4. Bioinformatics Analysis
Bioinformatic analysis programs were selected based on the options described by Reimand et al. [67]. Expression Suite Software Version 1.1 was used to identify differentially expressed miRNAs. MiRWalk 2.0 was used to study possible target genes of differentially expressed miRNAs, which includes target prediction data generated by different algorithms (including the algorithm itself) [68]. The following algorithms were selected: miRWalk, miRDB, Micro t4, miRanda, RNAhybrid, and Targetscan. Only targets predicted by at least three of the selected algorithms were accepted. The predicted targets previously identified as differentially expressed were then verified using microarray data available from the Gene Expression Omnibus (accession number GSE 43225). Microarray data were analyzed using GEO2R script (http://www.ncbi.nlm.nih.gov/geo/geo2r/, 23 September 2020). Differentially expressed genes were considered when they showed a fold change of at least 1.5. Gene Ontology was used to search for enriched terms among differentially expressed genes using Bonferroni´s correction and accepting only terms with p ≤ 0.05. Differentially expressed genes related to upregulated and downregulated miRNAs were analyzed according to EnrichR for enrichment analysis. The Reactome was used for data analysis, and the evaluation of protein–protein interaction network (PPI), based on a list of genes, was performed using the online tool STRING version 11.0. Experiments, databases, co-expression, neighborhoods, and co-occurrence were considered. The minimum interaction score was 0.700. Cytoscape software version 3.8.0 was used to view the results.
4.5. Statistic
A comparison analysis of Ct was used to quantify miRNA expression. Differences were statistically assessed using Student’s t-test. Statistical significance was set at p < 0.05.
5. Conclusions
It was possible to determine the miRNA profiles of patients treated with HSCT-allo and imatinib mesylate, as well as their target genes and pathways. The results of our study are expected to contribute to further research in the field of CML prognosis, in particular, the identifying of new biomarkers for this disease. Bioinformatics analyzes predict, in silico, interactions that require biological validation before clinical application.
Author Contributions
The conception and coordination of the work, data analysis and manuscript preparation: P.O.M.H., N.K.H. and C.R.N.; bioinformatic analysis and manuscript preparation: J.R.B.M., L.N.M. and S.S.C.; collection and data analysis: J.C.; laboratory support and critically revision about the study: R.F.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by São Paulo Research Foundation (FAPESP 11/50629-7).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by Research Ethics Committee of Dr. Amaral Carvalho Hospital–Jau-SP-Brazil (CEPHAC—2.917.389. Approval date: 25 Septebmer 2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All datasets generated for this study are included in the article.
Acknowledgments
We thank Amaral Carvalho Hospital, Bone Marrow Transplant Service, especially its coordination, Vergílio Antônio Renzi Colturato for the support and opportunity, Anderson João Simoni, data analyst, for his availability and patients for the delivery and ongoing teachings.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Goldman, J.M. Chronic Myeloid Leukemia: A Historical Perspective. Semin. Hematol. 2010, 47, 302–311. [Google Scholar] [CrossRef]
- Spagnuolo, M.; Regazzo, G.; De Dominici, M.; Sacconi, A.; Pelosi, A.; Korita, E.; Marchesi, F.; Pisani, F.; Magenta, A.; Lulli, V.; et al. Transcriptional activation of the miR-17-92 cluster is involved in the growth-promoting effects of MYB in human Ph-positive leukemia cells. Haematologica 2018, 104, 82–92. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2018, 93, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, S. Molecular mechanisms for survival regulation of chronic myeloid leukemia stem cells. Protein Cell 2013, 4, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.G.; Antman, K.H. Imatinib Mesylate—A New Oral Targeted Therapy. N. Engl. J. Med. 2002, 346, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef]
- Zhang, G.F.; Zhou, M.; Bao, X.B.; Qui, H.Y.; Li, Z.; Xue, S.L. Imatinib mesylate versus allogeneic hematopoietic stem cell transplantation for patients with chronic myelogenous leukemia. Asian Pac. J. Cancer Prev. 2016, 17, 4477–4481. [Google Scholar] [CrossRef]
- Cardama, A.Q.; Kantarjian, H.M.; Cortes, J.E. Mechanisms of primary and secondary resistance to imatinib in chronic mye-loid leukemia. Cancer Control. 2009, 16, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Wei, D.; Li, J.; Luo, D.; Chen, G.; Dang, Y.; Cai, X. Prognostic microRNAs and their potential molecular mechanism in pancreatic cancer: A study based on The Cancer Genome Atlas and bioinformatics investigation. Mol. Med. Rep. 2017, 17, 939–951. [Google Scholar] [CrossRef]
- Keklikoglou, I.; Koerner, C.; Schmidt, C.; Zhang, J.D.; Heckmann, D.; Shavinskaya, A.; Allgayer, H.; Guckel, B.; Fehm, T.F.; Schneeweiss, A.; et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 2011, 31, 4150–4163. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Moore, B.T.; Wang, Y.; Peng, X.H.; Lappe, J.M.; Recker, R.R.; Xiao, P. MiR-422a as a potential cellular microRNA bi-omarker for postmenopausal osteoporosis. PLoS ONE 2014, 9, e97098. [Google Scholar] [CrossRef]
- Cheng, G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 2015, 81, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 1–24. [Google Scholar] [CrossRef]
- Peltier, D.; Reddy, P. Non-Coding RNA Mediated Regulation of Allogeneic T Cell Responses After Hematopoietic Transplantation. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Forterre, A.; Komuro, H.; Aminova, S.; Harada, M. A Comprehensive Review of Cancer MicroRNA Therapeutic Delivery Strategies. Cancers 2020, 12, 1852. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Klicka, K.; Włodarski, P.K. Regulators at Every Step—How microRNAs Drive Tumor Cell Invasiveness and Metastasis. Cancers 2020, 12, 3709. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Nonlinear partial differential equations and applications: Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Roth, C.; Rack, B.; Müller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12, R90. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, K.-Y.; Liu, S.-M.; Sen, S. Tumor-Associated Circulating MicroRNAs as Biomarkers of Cancer. Molecules 2014, 19, 1912–1938. [Google Scholar] [CrossRef] [PubMed]
- Hershkovitz-Rokah, O.; Modai, S.; Pasmanik-Chor, M.; Toren, A.; Shomron, N.; Raanani, P.; Shpilberg, O.; Granot, G. MiR-30e induces apoptosis and sensitizes K562 cells to imatinib treatment via regulation of the BCR–ABL protein. Cancer Lett. 2015, 356, 597–605. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, Y.; Tao, K.; Wang, X.; Xiao, Q.; Huang, Z.; Zhong, L.; Cao, W.; Wen, J.; Feng, W. Inhibition of BCR/ABL Protein Expression by miR-203 Sensitizes for Imatinib Mesylate. PLoS ONE 2013, 8, e61858. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-S.; Li, L.; Chu, S.; Shiang, K.-D.; Li, M.; Sun, H.-Y.; Xu, J.; Xiao, F.-J.; Sun, G.; Rossi, J.J.; et al. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood 2015, 125, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Pavlů, J.; Szydlo, R.M.; Goldman, J.M.; Apperley, J.F.; Schotte, R.; Rissoan, M.-C.; Bendriss-Vermare, N.; Bridon, J.-M.; Duhen, T.; Weijer, K.; et al. Three decades of transplantation for chronic myeloid leukemia: What have we learned? Blood 2011, 117, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Deininger, M.W.; Rosti, G.; Hochhaus, A.; Soverini, S.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Guilhot, F.; et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia. Blood 2013, 122, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Cortes, J.; Pane, F.; Niederwieser, D.; Saglio, G.; Apperley, J.; Cervantes, F.; Deininger, M.; Gratwohl, A.; Guilhot, F.; et al. Chronic Myeloid Leukemia: An Update of Concepts and Management Recommendations of European LeukemiaNet. J. Clin. Oncol. 2009, 27, 6041–6051. [Google Scholar] [CrossRef]
- Baccarani, M.; Saglio, G.; Goldman, J.; Hochhaus, A.; Simonsson, B.; Appelbaum, F.; Apperley, J.; Cervantes, F.; Cortes, J.; Deininger, M.; et al. Evolving concepts in the management of chronic myeloid leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 2006, 108, 1809–1820. [Google Scholar] [CrossRef]
- Gratwohl, A.; Baldomero, H.; Passweg, J.R. The role of hematopoietic stem cell transplantation in chronic myeloid leukemia. Ann. Hematol. 2015, 94, 177–186. [Google Scholar] [CrossRef]
- Soyer, N.; Uysal, A.; Tombuloglu, M.; Sahin, F.; Saydam, G.; Vural, F. Allogeneic stem cell transplantation in chronic myeloid leukemia patients: Single center experience. World J. Hematol. 2017, 6, 1–10. [Google Scholar] [CrossRef]
- O’Brien, S.G.; Guilhot, F.; Larson, R.; Gathmann, I.; Baccarani, M.; Cervantes, F.; Cornelissen, J.J.; Fischer, T.; Hochhaus, A.; Hughes, T.; et al. Imatinib Compared with Interferon and Low-Dose Cytarabine for Newly Diagnosed Chronic-Phase Chronic Myeloid Leukemia. N. Engl. J. Med. 2003, 348, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; O’Brien, S.G.; Guilhot, F.; Druker, B.; Branford, S.; Foroni, L.; Goldman, J.M.; Müller, M.C.; Radich, J.P.; Rudoltz, M.; et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia 2009, 23, 1054–1061. [Google Scholar] [CrossRef]
- Buchdunger, E.; Zimmermann, J.; Mett, H.; Meyer, T.; Müller, M.; Druker, B.; Lydon, N.B. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996, 56, 100–104. [Google Scholar]
- Druker, B.J.; Tamura, S.; Buchdunger, E.; Ohno, S.; Segal, G.M.; Fanning, S.; Zimmermann, J.; Lydon, N.B. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr–Abl positive cells. Nat. Med. 1996, 2, 561–566. [Google Scholar] [CrossRef]
- Buchdunger, E.; Cioffi, C.L.; Law, N.; Stover, D.; Ohno-Jones, S.; Druker, B.; Lydon, N.B. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J. Pharmacol. Exp. Ther. 2000, 295, 139–145. [Google Scholar]
- Rosée, P.L.; Deininger, M.W. Resistance to Imatinib: Mutations and Beyond. Semin. Hematol. 2010, 47, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Lübking, A.; Dreimane, A.; Sandin, F.; Isaksson, C.; Märkevärn, B.; Brune, M.; Ljungman, P.; Lenhoff, S.; Stenke, L.; Höglund, M.; et al. Allogeneic stem cell transplantation for chronic myeloid leukemia in the TKI era: Population-based data from the Swedish CML registry. Bone Marrow Transplant. 2019, 54, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Innes, A.J.; Milojkovic, D.; Apperley, J.F. Allogeneic transplantation for CML in the TKI era: Striking the right balance. Nat. Rev. Clin. Oncol. 2016, 13, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Zhi, F.; Cao, X.; Xie, X.; Wang, B.; Dong, W.; Gu, W.; Ling, Y.; Wang, R.; Yang, Y.; Liu, Y. Identification of Circulating MicroRNAs as Potential Biomarkers for Detecting Acute Myeloid Leukemia. PLoS ONE 2013, 8, e56718. [Google Scholar] [CrossRef]
- Fayyad-Kazan, H.; Bitar, N.; Najar, M.; Lewalle, P.; Fayyad-Kazan, M.; Badran, R.; Hamade, E.; Daher, A.; Hussein, N.; Eldirani, R.; et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 2013, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Fazi, F.; Rosa, A.; Fatica, A.; Gelmetti, V.; DE Marchis, M.L.; Nervi, C.; Bozzoni, I. A Minicircuitry Comprised of MicroRNA-223 and Transcription Factors NFI-A and C/EBPα Regulates Human Granulopoiesis. Cell 2005, 123, 819–831. [Google Scholar] [CrossRef]
- Wu, D.; Rice, C.M.; Wang, X. Cancer bioinformatics: A new approach to systems clinical medicine. BMC Bioinform. 2012, 13, 71. [Google Scholar] [CrossRef]
- Sun, D.; Luo, M.; Jeong, M.; Rodriguez, B.; Xia, Z.; Hannah, R.; Wang, H.; Le, T.; Faull, K.F.; Chen, R.; et al. Epigenomic Profiling of Young and Aged HSCs Reveals Concerted Changes during Aging that Reinforce Self-Renewal. Cell Stem Cell 2014, 14, 673–688. [Google Scholar] [CrossRef]
- Gao, C.; Zhou, C.; Zhuang, J.; Liu, L.; Wei, J.; Liu, C.; Li, H.; Sun, C. Identification of key candidate genes and miRNA-mRNA target pairs in chronic lymphocytic leukemia by integrated bioinformatics analysis. Mol. Med. Rep. 2018, 19, 362–374. [Google Scholar] [CrossRef]
- Keramatinia, A.; Ahadi, A.; Akbari, M.E.; Mohseny, M.; Jarahi, A.M.; Bahadori-Monfared, A.; Hashemi, M.; Moradi, A.; Mehrvar, N.; Kazemi, E.; et al. The roles of DNA epigenetics and clinical significance in Chronic Myeloid Leukemia: A review. Cell. Mol. Biol. 2018, 64, 58–63. [Google Scholar] [CrossRef]
- Koul, H.K.; Pal, M.; Koul, S. Role of p38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer 2013, 4, 342–359. [Google Scholar] [CrossRef]
- Ufkin, M.L.; Peterson, S.; Yang, X.; Driscoll, H.; Duarte, C.; Sathyanarayana, P. miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukemia. Leuk. Res. 2014, 38, 402–410. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, X.-Q.; Feng, D.-D.; Zhang, X.-J.; Wu, J.; Zheng, Y.-S.; Chen, X.; Xu, L.; Chen, Y.-Q. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol. Cancer 2011, 10, 108. [Google Scholar] [CrossRef]
- Valiollahi, E.; Behravan, J. Leukemogenesis associated miRNAs regulate OSKM and Tp53 genes. Biomed. Res. 2016, 2016, 376–383. [Google Scholar]
- Li, H.; Liu, L.; Zhuang, J.; Liu, C.; Zhou, C.; Yang, J.; Gao, C.; Liu, G.; Sun, C. Identification of key candidate targets and pathways for the targeted treatment of leukemia stem cells of chronic myelogenous leukemia using bioinformatics analysis. Mol. Genet. Genom. Med. 2019, 7, e851. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Karimiani, E.G.; Byers, R.J.; Rehman, S.; Westerhoff, H.V.; Day, P.J.R. Mathematical modelling of miRNA mediated BCR.ABL protein regulation in chronic myeloid leukaemia vis-a-vis therapeutic strategies. Integr. Biol. 2013, 5, 543–554. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Q.; Wu, Y.; Zhang, J.; Yi, T.; Li, W. MicroRNA-130a regulates cell malignancy by targeting RECK in chronic myeloid leu-kemia. Am. J. Transl Res. 2016, 8, 955–967. [Google Scholar]
- Liao, Q.; Wang, B.; Li, X.; Jiang, G. miRNAs in acute myeloid leukemia. Oncotarget 2017, 8, 3666–3682. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Beya, M.; Brunet, S.; Nomdedeu, J.; Tejero, R.; Diaz, T.; Pratcorona, M.; Tormo, M.; Ribera, J.M.; Escoda, L.; Duarte, R.; et al. MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leukemia 2014, 28, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, J.; Li, J.; Zhao, F.; Shen, Y.; Xing, X. Overexpression of miR-574-3p suppresses proliferation and induces apoptosis of chronic myeloid leukemia cells via targeting IL6/JAK/STAT3 pathway. Exp. Ther. Med. 2018, 16, 4296–4302. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Yang, Y.; Wang, H.; Li, J.; Wang, S.; Li, Y.; Yang, Y.; Cai, K.; Ruan, X.; Yan, J.; et al. Characterization of miRNomes in Acute and Chronic Myeloid Leukemia Cell Lines. Genom. Proteom. Bioinform. 2014, 12, 79–91. [Google Scholar] [CrossRef]
- Kim, E.K.; Choi, E.-J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2010, 1802, 396–405. [Google Scholar] [CrossRef]
- Torii, S.; Yamamoto, T.; Tsuchiya, Y.; Nishida, E. ERK MAP kinase in G1 cell cycle progression and cancer. Cancer Sci. 2006, 97, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Araujo, E.G.; Bianchi, C.; Faro, R.; Sellke, F.W.; Sato, K. Oscillation in the activities of MEK/ERK1/2 during cardiopulmonary bypass in pigs. Surgery 2001, 130, 182–191. [Google Scholar] [CrossRef]
- Zhou, L.; Dong, J.; Huang, G.; Sun, Z.; Wu, J. MicroRNA-143 inhibits cell growth by targeting ERK5 and MAP3K7 in breast cancer. Braz. J. Med. Biol. Res. 2017, 50, e5891. [Google Scholar] [CrossRef]
- Pham, T.T.; Angus, S.; Johnson, G.L. MAP3K1: Genomic Alterations in Cancer and Function in Promoting Cell Survival or Apoptosis. Genes Cancer 2013, 4, 419–426. [Google Scholar] [CrossRef]
- Rocca, S.; Carrà, G.; Poggio, P.; Morotti, A.; Brancaccio, M. Targeting few to help hundreds: JAK, MAPK and ROCK pathways as druggable targets in atypical chronic myeloid leukemia. Mol. Cancer 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.A.; Hasserjian, R.P.; Fox, P.S.; Rogers, H.J.; Geyer, J.T.; Chabot-Richards, D.; Weinzierl, E.; Hatem, J.; Jaso, J.; Kanagal-Shamanna, R.; et al. Atypical chronic myeloid leukemia is clinically distinct from unclassifiable myelodysplastic/myeloproliferative neoplasms. Blood 2014, 123, 2645–2651. [Google Scholar] [CrossRef] [PubMed]
- Tutschka, P.; Copelan, E.; Klein, J. Bone marrow transplantation for leukemia following a new busulfan and cyclophosphamide regimen. Blood 1987, 70, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Storb, R.; Deeg, H.J.; Whitehead, J.; Appelbaum, F.; Beatty, P.; Bensinger, W.; Buckner, C.D.; Clift, R.; Doney, K.; Farewell, V.; et al. Methotrexate and Cyclosporine Compared with Cyclosporine Alone for Prophylaxis of Acute Graft versus Host Disease after Marrow Transplantation for Leukemia. N. Engl. J. Med. 1986, 314, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.R.B.; De Moraes, L.N.; Cury, S.S.; Dadalto, J.; Capannacci, J.; Carvalho, R.F.; Nogueira, C.R.; Hokama, N.K.; Hokama, P.D.O.M. Comparison of microRNA Expression Profile in Chronic Myeloid Leukemia Patients Newly Diagnosed and Treated by Allogeneic Hematopoietic Stem Cell Transplantation. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.A.M.; Capannacci, J.; Hokama, N.K.; Nogueira, C.R.; Ceccarelli, M.; Cerulo, L.; D’Angelo, F.; Hokama, P.D.O.M. Circulating microRNAs expression profile in newly diagnosed and imatinib treated chronic phase—chronic myeloid leukemia. Leuk. Lymphoma 2018, 60, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).