Two Paralogous Gb3/CD77 Synthases in Birds Show Different Preferences for Their Glycoprotein and Glycosphingolipid Substrates
Abstract
:1. Introduction
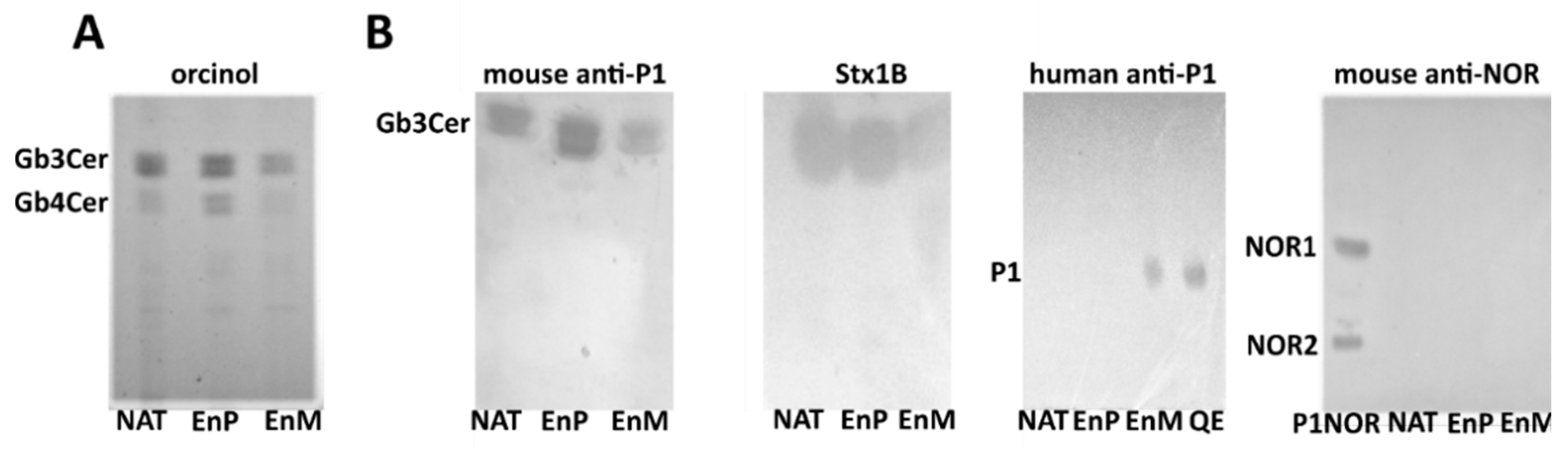
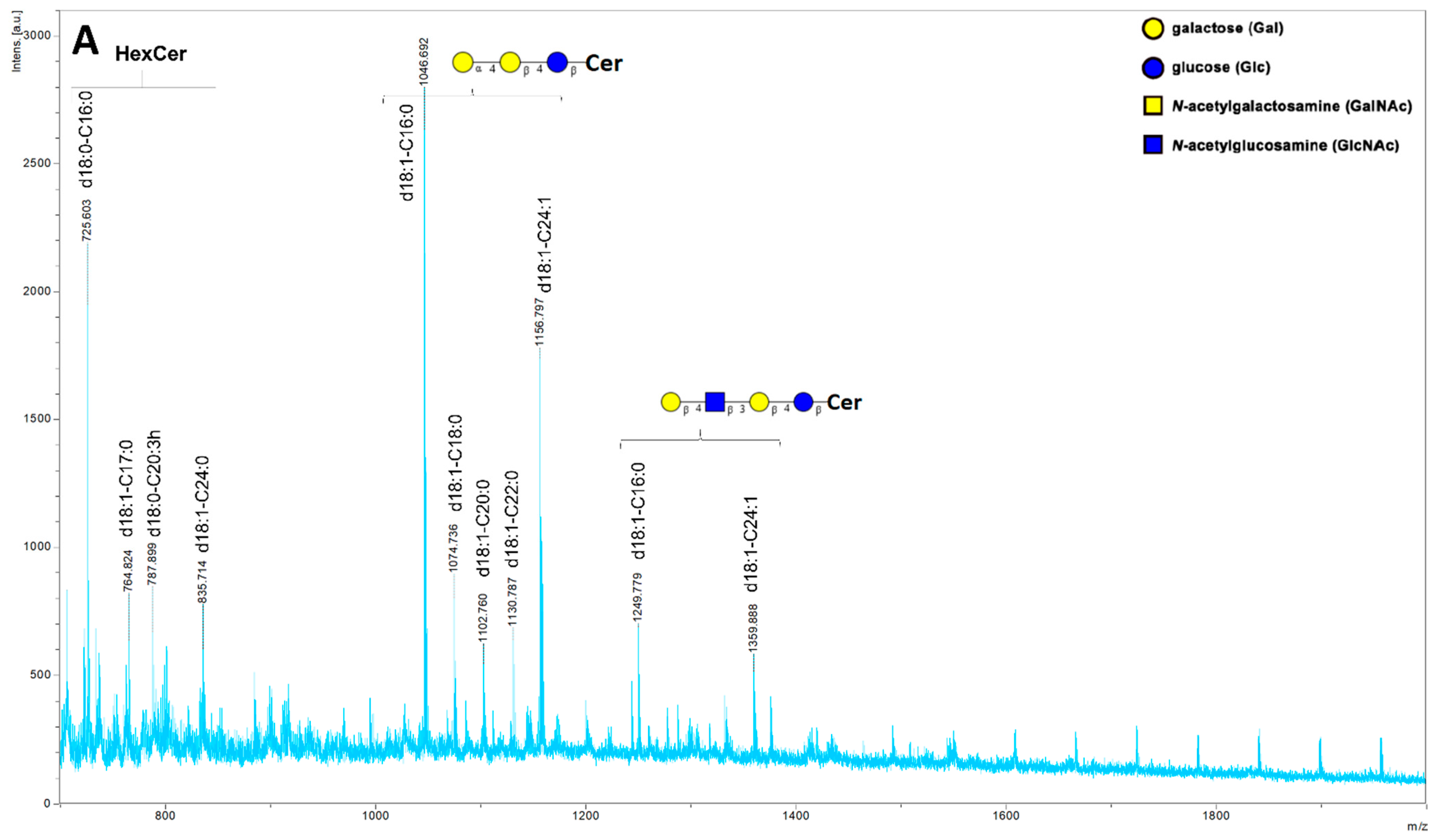
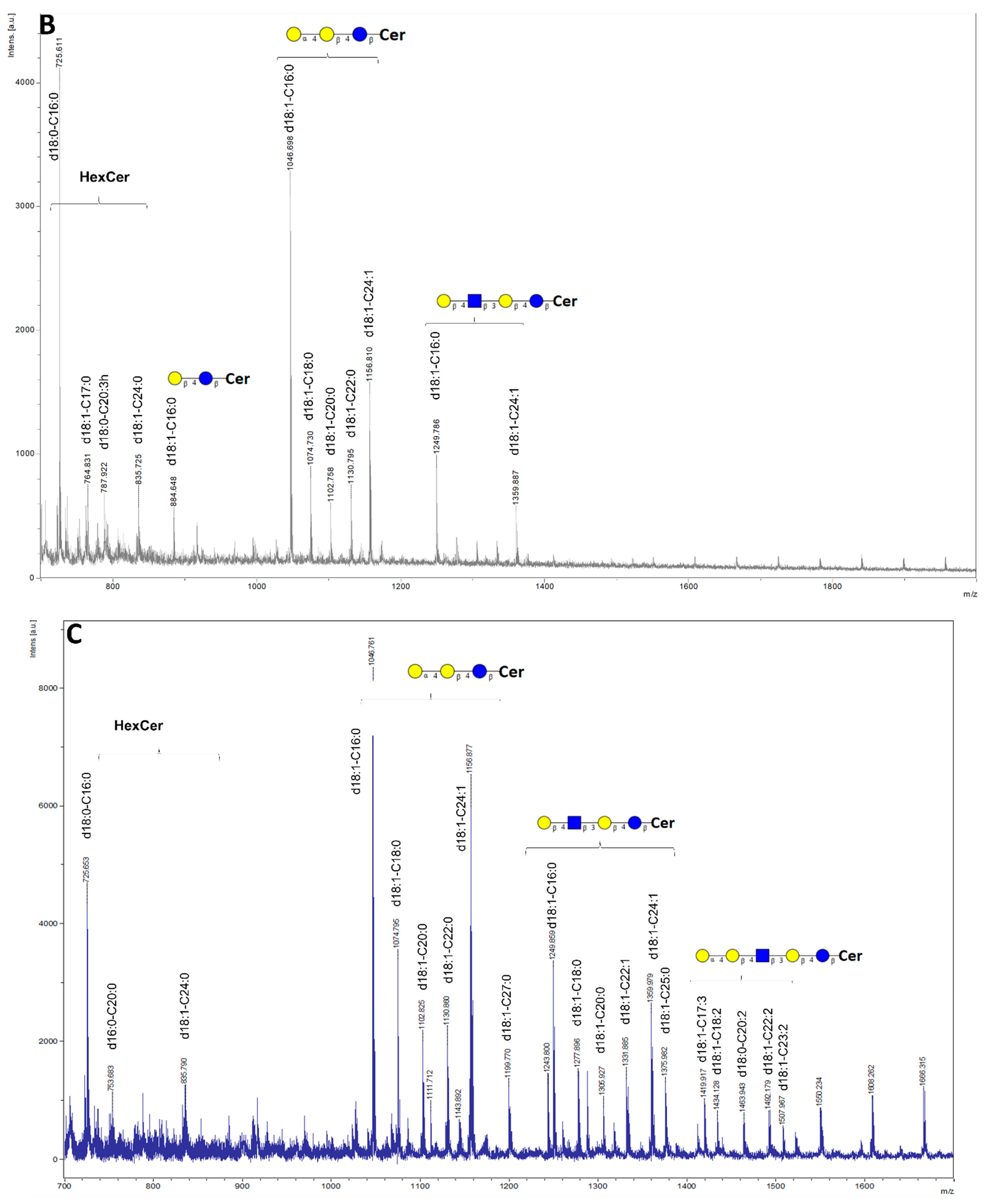
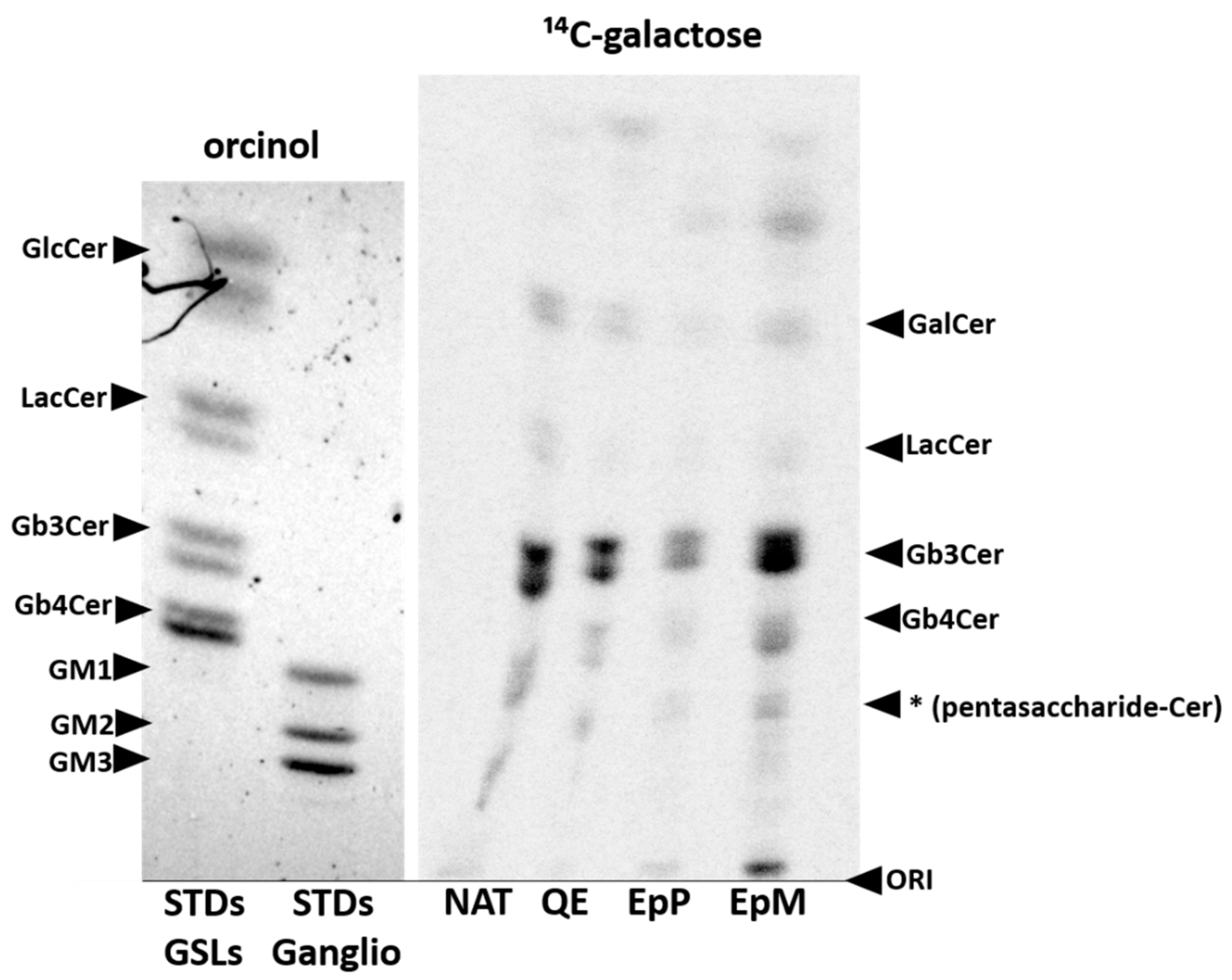
2. Results
3. Discussion
4. Materials and Methods
4.1. Isolation of Genomic DNA from Pigeon Red Blood Cells, Amplification and Cloning
4.2. Cell Culture and Transfection
4.3. Flow Cytofluorometry
4.4. Protein Lysates
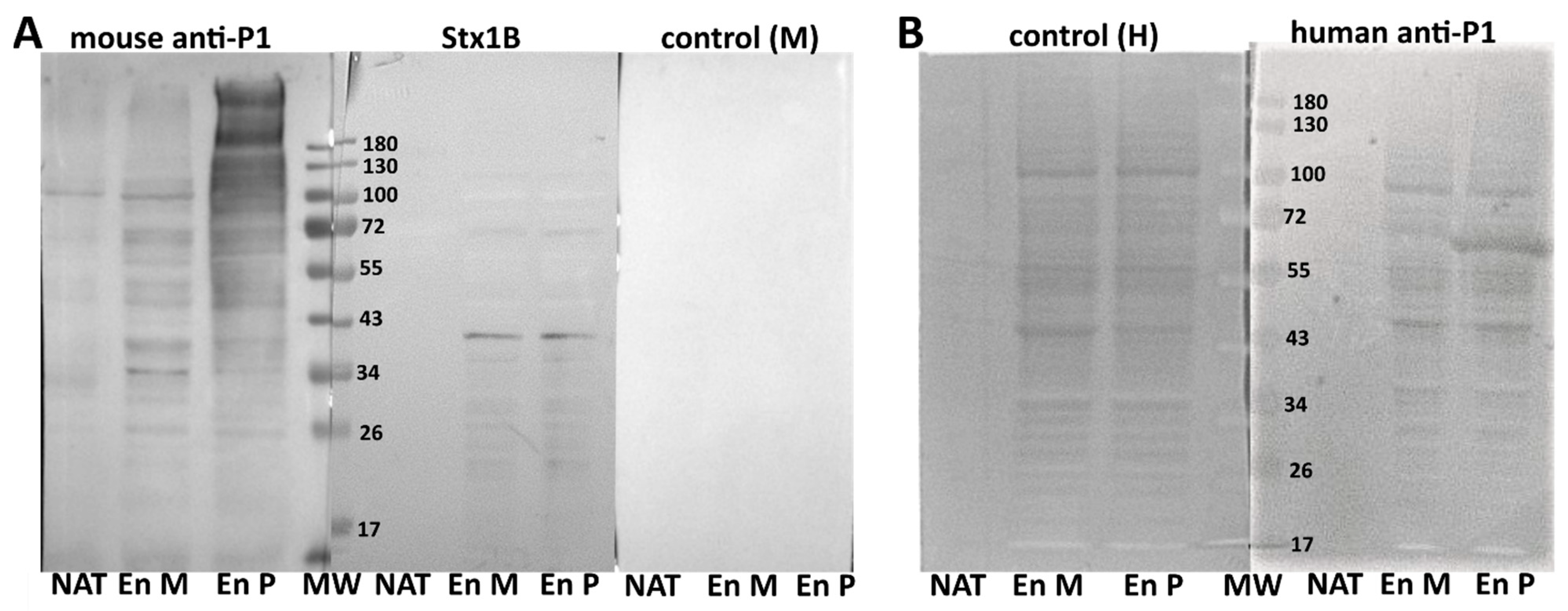
4.5. SDS-PAGE and Western Blotting
4.6. Extraction and Purification of Glycosphingolipids
4.7. Mass Spectrometry Analysis of Glycosphingolipids
4.8. Metabolic Labelling of Glycosphingolipids and TLC Analysis
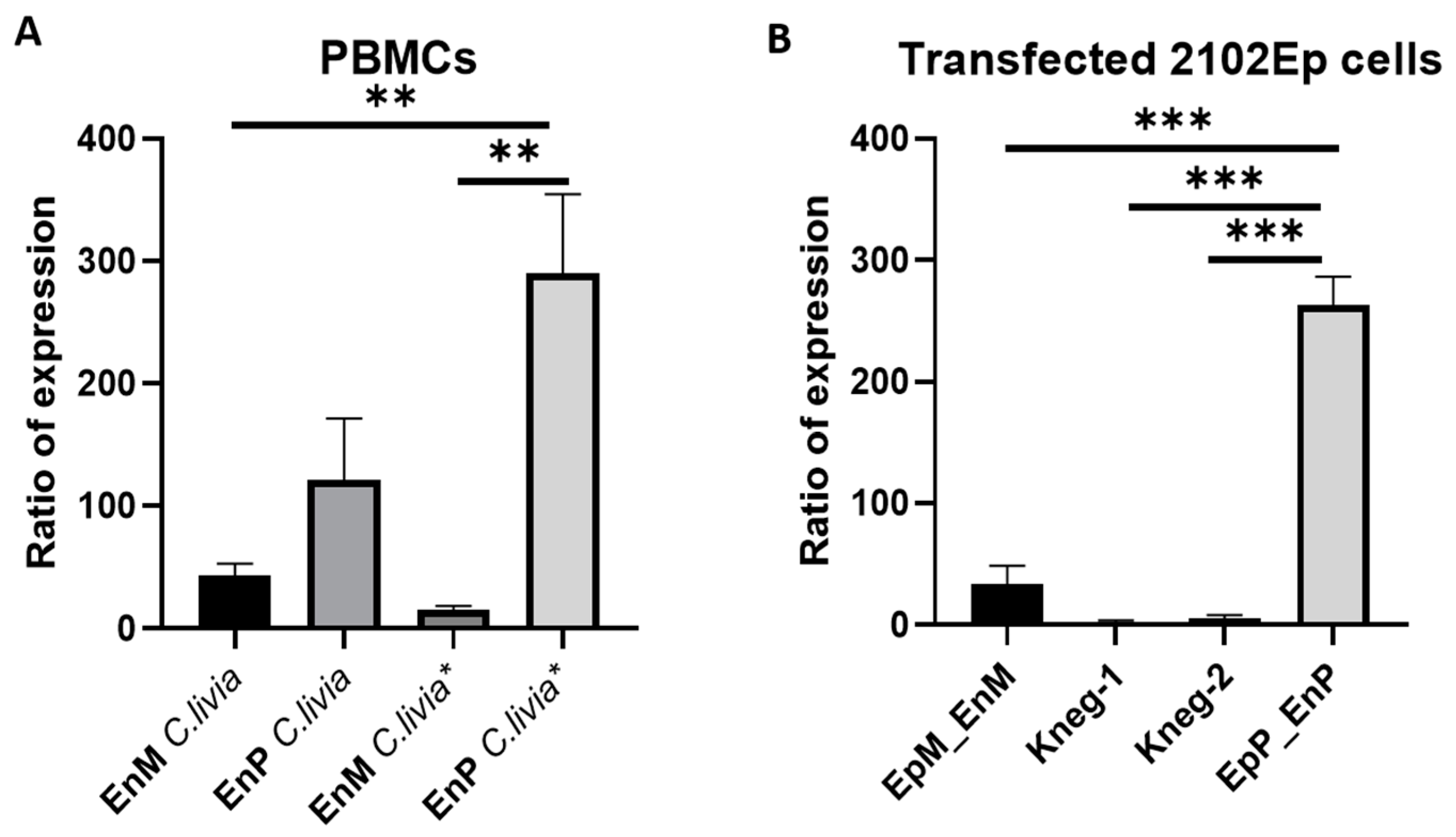
4.9. Quantitative Analysis of Avian A4GALT Transcripts
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [Green Version]
- Breton, C.; Šnajdrová, L.; Jeanneau, C.; Koča, J.; Imberty, A. Structures and mechanisms of glycosyltransferases. Glycobiology 2005, 16, 29R–37R. [Google Scholar] [CrossRef]
- Furukawa, K.; Kondo, Y.; Furukawa, K. UDP-gal: Lactosylceramide alpha 1,4-galactosyltransferase (A4GALT). In Handbook of Glycosyltransferases and Related Genes, 2nd ed.; Springer Publishing: New York, NY, USA, 2014; ISBN 9784431542407. [Google Scholar]
- Kaczmarek, R.; Buczkowska, A.; Mikołajewicz, K.; Krotkiewski, H.; Czerwinski, M. P1PK, GLOB, and FORS Blood Group Systems and GLOB Collection: Biochemical and Clinical Aspects. Do We Understand It All Yet? Transfus. Med. Rev. 2014, 28, 126–136. [Google Scholar] [CrossRef]
- Mourad, R.; Morelle, W.; Neveu, A.; Strecker, G. Diversity of O-linked glycosylation patterns between species. JBIC J. Biol. Inorg. Chem. 2001, 268, 1990–2003. [Google Scholar] [CrossRef]
- Suchanowska, A.; Kaczmarek, R.; Duk, M.; Lukasiewicz, J.; Smolarek, D.; Majorczyk, E.; Jaskiewicz, E.; Laskowska, A.; Waśniowska, K.; Grodecka, M.; et al. A Single Point Mutation in the Gene Encoding Gb3/CD77 Synthase Causes a Rare Inherited Polyagglutination Syndrome. J. Biol. Chem. 2012, 287, 38220–38230. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, H.; Ide, M.; Yamaji, T.; Mizutani, Y.; Niimi, Y.; Mutoh, T.; Kamiguchi, H.; Hirabayashi, Y. Galabiosylceramide is present in human cerebrospinal fluid. Biochem. Biophys. Res. Commun. 2021, 536, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Johannes, L.; Römer, W. Shiga toxins—From cell biology to biomedical applications. Nat. Rev. Genet. 2009, 8, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Bruyand, M.; Mariani-Kurkdjian, P.; Gouali, M.; de Valk, H.; King, L.; Le Hello, S.; Bonacorsi, S.; Loirat, C. Hemolytic uremic syndrome due to Shiga toxin-producing Escherichia coli infection. Médecine et Maladies Infectieuses 2018, 48, 167–174. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Scallan, E.; Jones-Bitton, A.; Sargeant, J.M.; Stapleton, J.; Angulo, F.J.; Yeung, D.H.; Kirk, M. Global Incidence of Human Shiga Toxin–ProducingEscherichia coliInfections and Deaths: A Systematic Review and Knowledge Synthesis. Foodborne Pathog. Dis. 2014, 11, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Okuda, T.; Numata, S.-I.; Ito, M.; Ohta, M.; Kawamura, K.; Wiels, J.; Urano, T.; Tajima, O.; Furukawa, K.; Furukawa, K. Targeted Disruption of Gb3/CD77 Synthase Gene Resulted in the Complete Deletion of Globo-series Glycosphingolipids and Loss of Sensitivity to Verotoxins. J. Biol. Chem. 2006, 281, 10230–10235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zumbrun, S.D.; Hanson, L.; Sinclair, J.F.; Freedy, J.; Melton-Celsa, A.R.; Rodriguez-Canales, J.; Hanson, J.C.; O’Brien, A.D. Human Intestinal Tissue and Cultured Colonic Cells Contain Globotriaosylceramide Synthase mRNA and the Alternate Shiga Toxin Receptor Globotetraosylceramide. Infect. Immun. 2010, 78, 4488–4499. [Google Scholar] [CrossRef] [Green Version]
- Szymczak-Kulus, K.; Weidler, S.; Bereznicka, A.; Mikolajczyk, K.; Kaczmarek, R.; Bednarz, B.; Zhang, T.; Urbaniak, A.; Olczak, M.; Park, E.Y.; et al. Human Gb3/CD77 synthase produces P1 glycotope-capped N-glycans, which mediate Shiga toxin 1 but not Shiga toxin 2 cell entry. J. Biol. Chem. 2021, 296, 100299. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Laskowski, M.; Lee, Y.C. Tracing the history of Galα1-4Gal on glycoproteins in modern birds. Biochim. Biophys. Acta-Gen. Subj. 2006, 1760, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Suzuki, N.; Tanida, I.; Kakuta, S.; Furuta, Y.; Uchiyama, Y.; Hanada, K.; Suzuki, Y.; Yamaji, T. Blood group P1 antigen–bearing glycoproteins are functional but less efficient receptors of Shiga toxin than conventional glycolipid-based receptors. J. Biol. Chem. 2020, 295, 9490–9501. [Google Scholar] [CrossRef]
- Suzuki, N.; Yamamoto, K. Molecular Cloning of Pigeon UDP-galactose:β-d-Galactoside α1,4-Galactosyltransferase and UDP-galactose:β-d-Galactoside β1,4-Galactosyltransferase, Two Novel Enzymes Catalyzing the Formation of Galα1–4Galβ1–4Galβ1–4GlcNAc Sequence. J. Biol. Chem. 2010, 285, 5178–5187. [Google Scholar] [CrossRef] [Green Version]
- Liew, C.-G.; Draper, J.S.; Walsh, J.; Moore, H.; Andrews, P.W. Transient and Stable Transgene Expression in Human Embryonic Stem Cells. Stem Cells 2008, 26, 1094. [Google Scholar] [CrossRef]
- Kaczmarek, R.; Szymczak-Kulus, K.; Bereznicka, A.; Mikołajczyk, K.; Duk, M.; Majorczyk, E.; Krop-Watorek, A.; Klausa, E.; Skowrońska, J.; Michalewska, B.; et al. Single nucleotide polymorphisms in A4GALT spur extra products of the human Gb3/CD77 synthase and underlie the P1PK blood group system. PLoS ONE 2018, 13, e0196627. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Laskowski, M.; Lee, Y.C. Phylogenetic expression of Galα1-4Gal on avian glycoproteins: Glycan differentiation inscribed in the early history of modern birds. Proc. Natl. Acad. Sci. USA 2004, 101, 9023–9028. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Nawa, D.; Su, T.-H.; Lin, C.-W.; Khoo, K.-H.; Yamamoto, K. Distribution of the Galβ1-4Gal Epitope among Birds: Species-Specific Loss of the Glycan Structure in Chicken and Its Relatives. PLoS ONE 2013, 8, e59291. [Google Scholar] [CrossRef]
- Nagai, K.-I.; Takahashi, N.; Niimura, Y. Novel biosynthesis of monogalactosyl-alkylacyl glycerolipid in Mop8 fibroblast cells transfected with a ceramide galactosyltransferase gene. Biomed. Res. Clin. Pr. 2016, 1, 103–108. [Google Scholar] [CrossRef]
- Tian, S.; Muneeruddin, K.; Choi, M.Y.; Tao, L.; Bhuiyan, R.H.; Ohmi, Y.; Furukawa, K.; Furukawa, K.; Boland, S.; Shaffer, S.A.; et al. Genome-wide CRISPR screens for Shiga toxins and ricin reveal Golgi proteins critical for glycosylation. PLoS Biol. 2018, 16, e2006951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacheco, A.R.; Lazarus, J.; Sit, B.; Schmieder, S.; Lencer, W.; Blondel, C.J.; Doench, J.G.; Davis, B.M.; Waldor, M.K. CRISPR Screen Reveals that EHEC’s T3SS and Shiga Toxin Rely on Shared Host Factors for Infection. mBio 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, S.K.; Wilhelm, I.; Schubert, T.; Zittlau, K.; Imberty, A.; Madl, J.; Eierhoff, T.; Thuenauer, R.; Römer, W. Gb3-binding lectins as potential carriers for transcellular drug delivery. Expert Opin. Drug Deliv. 2016, 14, 141–153. [Google Scholar] [CrossRef]
- Shapiro, M.D.; Kronenberg, Z.; Li, C.; Domyan, E.T.; Pan, H.; Campbell, M.; Tan, H.; Huff, C.D.; Hu, H.; Vickrey, A.I.; et al. Genomic Diversity and Evolution of the Head Crest in the Rock Pigeon. Science 2013, 339, 1063–1067. [Google Scholar] [CrossRef] [Green Version]
- Sanches, L.A.; Gomes, M.D.S.; Teixeira, R.H.F.; Cunha, M.P.V.; De Oliveira, M.G.X.; Vieira, M.A.M.; Gomes, T.; Knobl, T. Captive wild birds as reservoirs of enteropathogenic E. coli (EPEC) and Shiga-toxin producing E. coli (STEC). Braz. J. Microbiol. 2017, 48, 760–763. [Google Scholar] [CrossRef]
- Persad, A.K.; LeJeune, J.T. Animal Reservoirs of Shiga Toxin-Producing Escherichia coli. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Borges, C.A.; Maluta, R.P.; Beraldo, L.G.; Cardozo, M.V.; Guastalli, E.A.; Kariyawasam, S.; DebRoy, C.; Ávila, F.A. Captive and free-living urban pigeons ( Columba livia ) from Brazil as carriers of multidrug-resistant pathogenic Escherichia coli. Veter- J. 2017, 219, 65–67. [Google Scholar] [CrossRef] [Green Version]
- Fadel, H.M.; Afifi, R.; Al-Qabili, D.M. Characterization and zoonotic impact of Shiga toxin producing Escherichia coli in some wild bird species. Veter- World 2017, 10, 1118–1128. [Google Scholar] [CrossRef] [Green Version]
- Wallace, J.S.; Cheasty, T.; Jones, K. Isolation of Vero cytotoxin-producing Escherichia coli O157 from wild birds. J. Appl. Microbiol. 1997, 82, 399–404. [Google Scholar] [CrossRef]
- Pedersen, K.; Clark, L. A review of Shiga toxin Escherichia coli and Salmonella enterica in cattle and free-ranging birds: Potential association and epidemiological links. Human–Wildlife Conflicts 2007, 1, 68–77. [Google Scholar]
- Kauffman, M.D.; Lejeune, J. European Starlings (Sturnus vulgaris) challenged with Escherichia coli O157 can carry and transmit the human pathogen to cattle. Lett. Appl. Microbiol. 2011, 53, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Doane, C.A.; Pangloli, P.; Richards, H.A.; Mount, J.R.; Golden, D.A.; Draughon, F.A. Occurrence of Escherichia coli O157:H7 in Diverse Farm Environments. J. Food Prot. 2007, 70, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Stenfelt, L.; Westman, J.S.; Hellberg, Å.; Olsson, M.L. The P1 histo-blood group antigen is present on human red blood cell glycoproteins. Transfusion 2019, 59, 1108–1117. [Google Scholar] [CrossRef]
- Schöler, U.; Schöler, U. Inkscape; Free Software Foundation, Inc.: Boston, MA, USA, 2014. [Google Scholar]
- Samani, F.S.; Moore, J.K.; Khosravani, P.; Ebrahimi, M. Features of free software packages in flow cytometry: A comparison between four non-commercial software sources. Cytotechnology 2013, 66, 555–559. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [Green Version]
- Duk, M.; Lisowska, E. Presence of natural anti-Galα1-4GalNAcβ1-3Gal (anti-NOR) antibodies in animal sera. Glycoconj. J. 2006, 23, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Ravindran, B. Isolation of Human PBMCs. Bio-Protocol 2013, 3. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bereznicka, A.; Mikolajczyk, K.; Szymczak-Kulus, K.; Kapczynska, K.; Majorczyk, E.; Modlinska, A.; Piasecki, T.; Kaczmarek, R.; Czerwinski, M. Two Paralogous Gb3/CD77 Synthases in Birds Show Different Preferences for Their Glycoprotein and Glycosphingolipid Substrates. Int. J. Mol. Sci. 2021, 22, 9761. https://doi.org/10.3390/ijms22189761
Bereznicka A, Mikolajczyk K, Szymczak-Kulus K, Kapczynska K, Majorczyk E, Modlinska A, Piasecki T, Kaczmarek R, Czerwinski M. Two Paralogous Gb3/CD77 Synthases in Birds Show Different Preferences for Their Glycoprotein and Glycosphingolipid Substrates. International Journal of Molecular Sciences. 2021; 22(18):9761. https://doi.org/10.3390/ijms22189761
Chicago/Turabian StyleBereznicka, Anna, Krzysztof Mikolajczyk, Katarzyna Szymczak-Kulus, Katarzyna Kapczynska, Edyta Majorczyk, Anna Modlinska, Tomasz Piasecki, Radoslaw Kaczmarek, and Marcin Czerwinski. 2021. "Two Paralogous Gb3/CD77 Synthases in Birds Show Different Preferences for Their Glycoprotein and Glycosphingolipid Substrates" International Journal of Molecular Sciences 22, no. 18: 9761. https://doi.org/10.3390/ijms22189761
APA StyleBereznicka, A., Mikolajczyk, K., Szymczak-Kulus, K., Kapczynska, K., Majorczyk, E., Modlinska, A., Piasecki, T., Kaczmarek, R., & Czerwinski, M. (2021). Two Paralogous Gb3/CD77 Synthases in Birds Show Different Preferences for Their Glycoprotein and Glycosphingolipid Substrates. International Journal of Molecular Sciences, 22(18), 9761. https://doi.org/10.3390/ijms22189761





