Characterization of Mono- and Bi-Transgenic Pig-Derived Epidermal Keratinocytes Expressing Human FUT2 and GLA Genes—In Vitro Studies
Abstract
:1. Introduction
2. Results
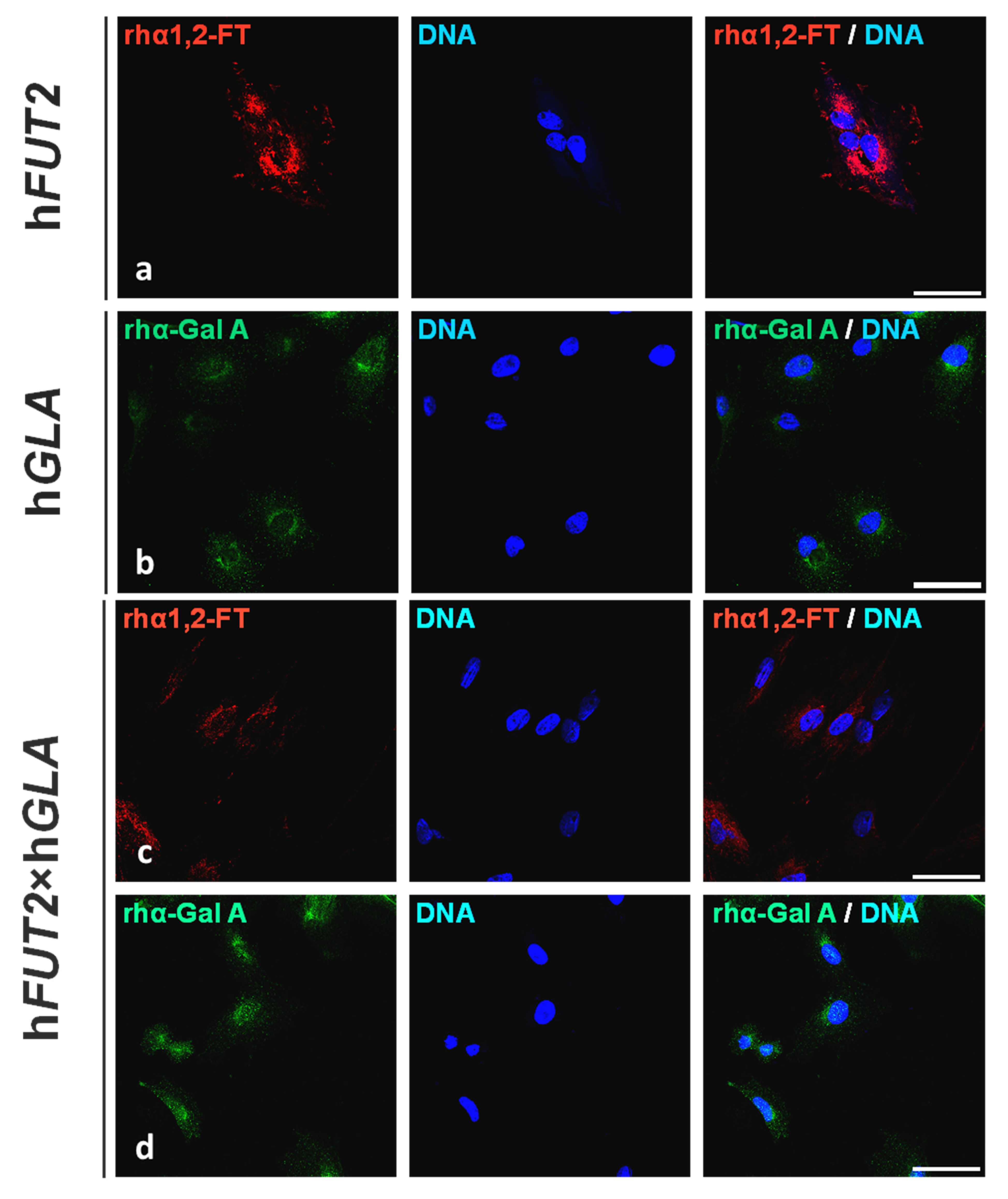
2.1. Fluorescent Immunolocalization of Recombinant Human α1,2-Fucosyltransferase (rhα1,2-FT) and α-Galactosidase A (rhα-Gal A) Enzymes in the Ex Vivo-Expanded Single- and Double-Transgenic PEKs
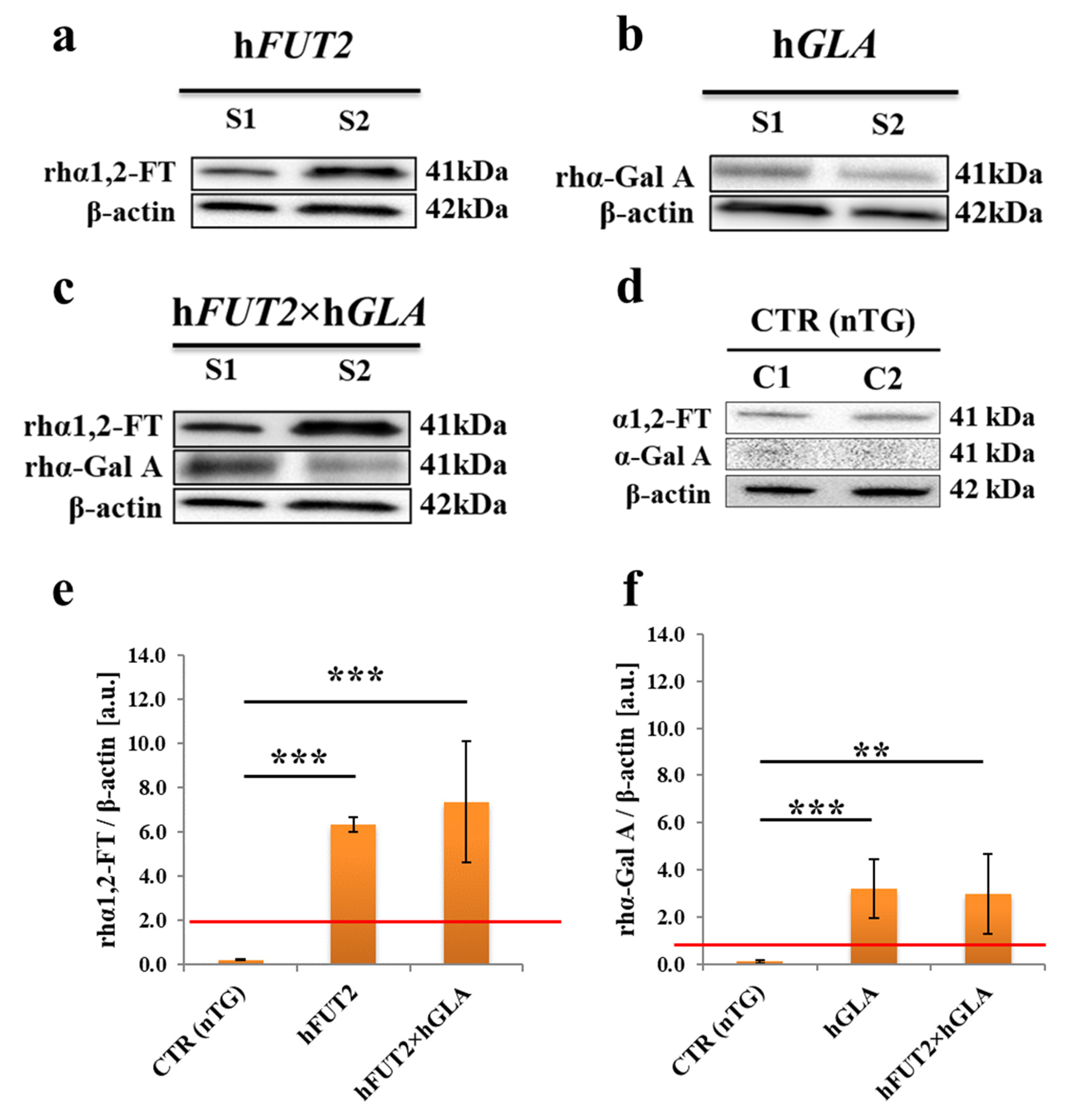
2.2. Western Blot-Mediated Determination of the Relative Expression Specific for Recombinant Human α1,2-Fucosyltransferase (rhα1,2-FT) and α-Galactosidase A (rhα-Gal A) Enzymes in the Ex Vivo-Expanded Single- and Double-Transgenic PEKs
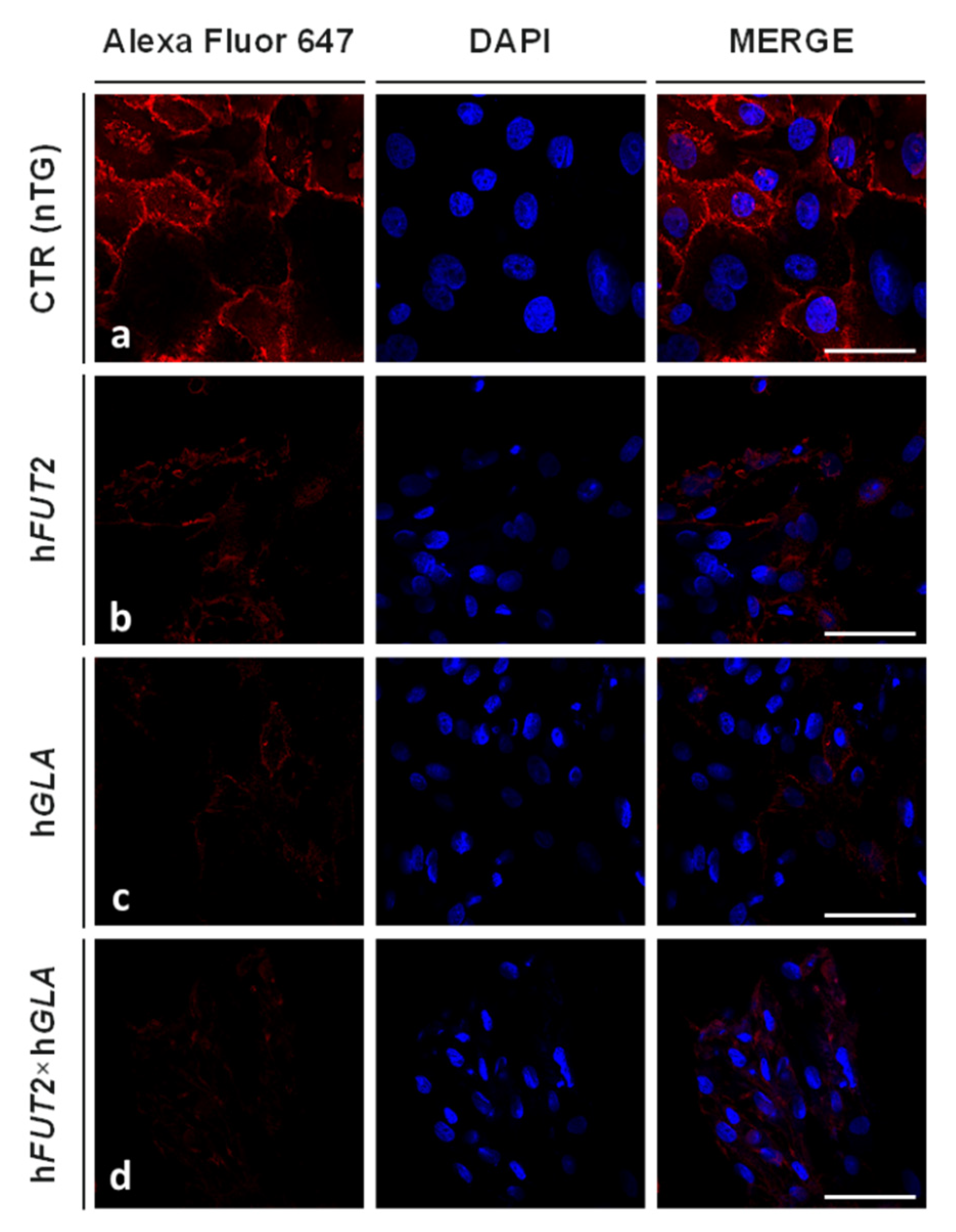
2.3. Lectin GS I-B4-Mediated Fluorocytochemically Detecting the Expression Profiles of Galα1→3 Gal Epitope in the Ex Vivo-Expanded Single-, Double- and Nontransgenic PEKs
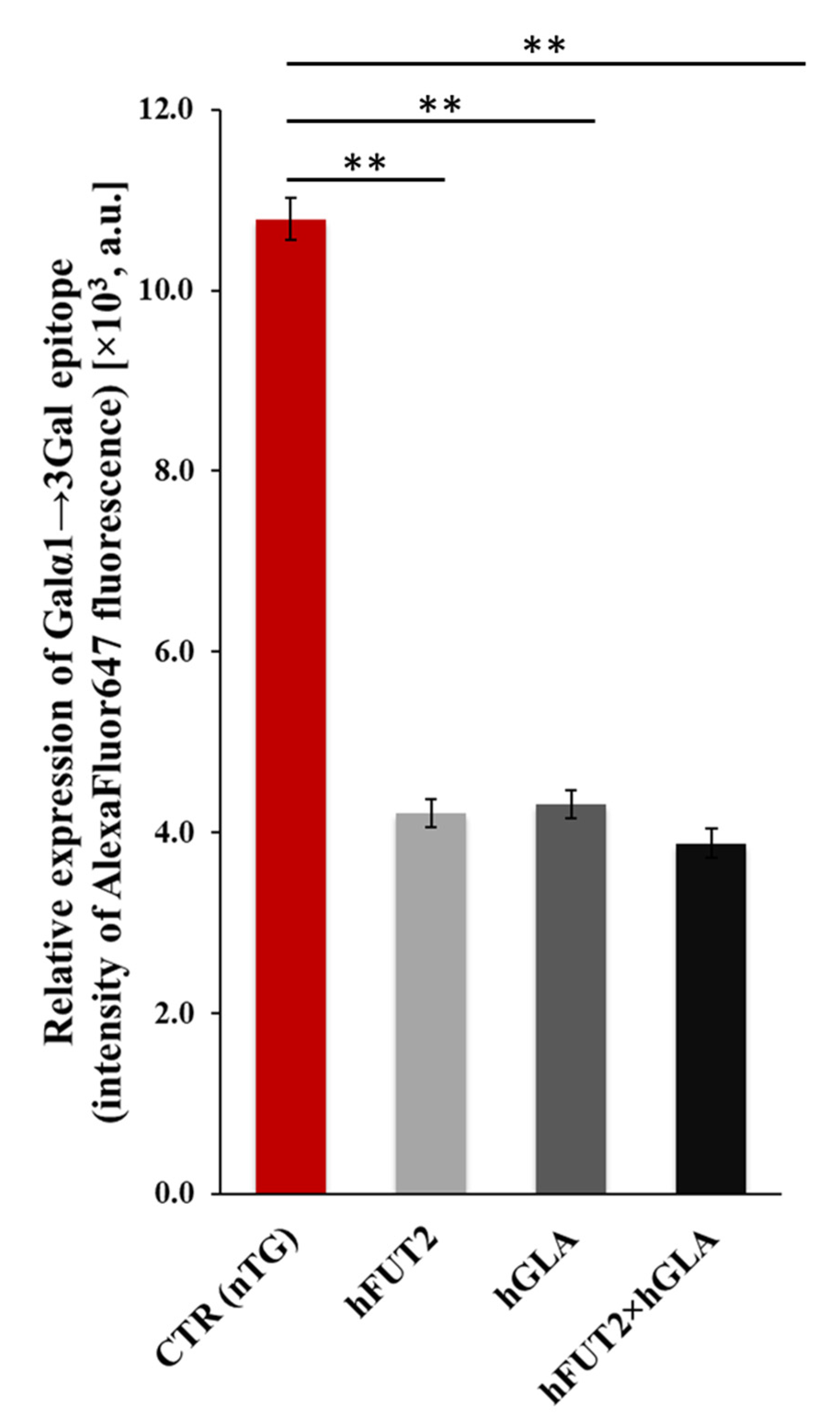
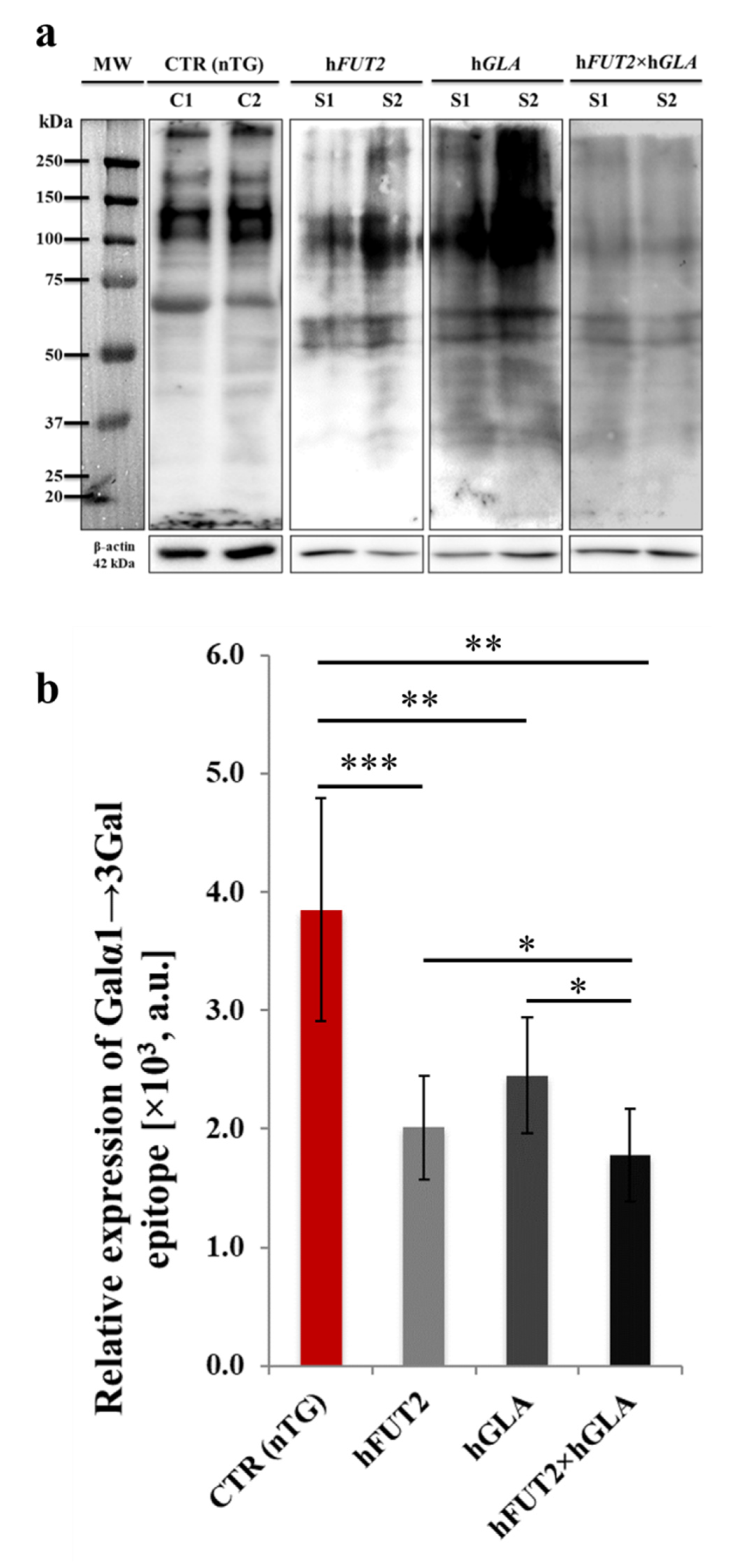
2.4. Lectin Blotting Analysis of Galα1→3Gal Epitope Expression at the Protein Level in the Ex Vivo-Expanded Single-, Double- and Nontransgenic PEKs
3. Discussion
4. Materials and Methods
4.1. Postmortem Collection of Skin Tissue Explants from Single-, Double- and Nontransgenic Pigs
4.2. Establishment of the Ex Vivo-Expanded Epidermal Keratinocytes Stemming from Skin Tissue Samples of Single-, Double- and Nontransgenic Pigs
4.3. Immunofluorescence Dyeing of the Ex Vivo-Expanded Single-, Double- and Nontransgenic PEKs
4.4. Lectin-Mediated Labelling of Galα1→3Gal Epitopes in the Ex Vivo-Expanded Single-, Double- and Nontransgenic PEKs
4.5. Confocal Microscope Analyses of the Ex Vivo-Expanded Single-, Double- and Nontransgenic PEKs
4.6. Total Protein Extraction and Western or Lectin Blot Analyses Accomplished to Determine RAs Estimated for rhα1,2-FT and rhα-Gal A Enzymes or Galα1→3Gal Epitopes at the Protein Levels in the Ex Vivo-Expanded Single-, Double- and Nontransgenic PEKs
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HAR | Hyperacute rejection |
| HRP | Horseradish peroxidase |
| LacNAc | N-Acetyllactosamine |
| NDECs | Nuclear donor epidermal cells |
| rhα1,2-FT | Recombinant human α1,2-fucosyltransferase |
| rhα-Gal A | Recombinant human α-galactosidase A |
| α1,3-GT | α1,3-Galactosyltransferase |
| PEKs | Porcine epidermal keratinocytes |
| RA | Relative abundance |
| SCNT | Somatic cell nuclear transfer |
| UDP-Gal | Uridine 5′-diphosphogalactose; Galactose-uridine-5′-diphosphate; UDP-α-D-galactose |
References
- Niemann, H.P. The production of multi-transgenic pigs: Update and perspectives for xenotransplantation. Transgenic Res. 2016, 25, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.K.; Gollackner, B.; Sachs, D.H. Will the pig solve the transplantation backlog? Annu. Rev. Med. 2002, 53, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Galili, U.; Shohet, S.B.; Kobrin, E.; Stults, C.L.; Macher, B.A. Man, apes, and Old World monkeys differ from other mammals in the expression of alpha-galactosyl epitopes on nucleated cells. J. Biol. Chem. 1988, 263, 17755–17762. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Zeyland, J.; Słomski, R.; Lipiński, D. Genetically modified pigs as organ donors for xenotransplantation. Mol. Biotechnol. 2017, 59, 435–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whyte, J.J.; Prather, R.S. Genetic modifications of pigs for medicine and agriculture. Mol. Reprod. Dev. 2011, 78, 879–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, D.K.C.; Ekser, B.; Tector, A.J. Immunobiological barriers to xenotransplantation. Int. J. Surg. 2015, 23, 211–216. [Google Scholar] [CrossRef]
- Cooper, D.K.; Dou, K.F.; Tao, K.S.; Yang, Z.X.; Tector, A.J.; Ekser, B. Pig liver xenotransplantation: A review of progress toward the clinic. Transplantation 2016, 100, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Yang, B.; Wang, R.; Qin, C. Xenotransplantation: Current status in preclinical research. Front. Immunol. 2020, 10, 3060. [Google Scholar] [CrossRef]
- Sandrin, M.S.; McKenzie, I.F. Gal alpha (1,3)Gal, the major xenoantigen(s) recognised in pigs by human natural antibodies. Immunol. Rev. 1994, 141, 169–190. [Google Scholar] [CrossRef]
- Blanken, W.M.; van den Eijnden, D.H. Biosynthesis of terminal Gal α1→3Galβ1→4GlcNAc-R oligosaccharide sequences on glycoconjugates. Purification and acceptor specificity of a UDP-Gal: N-acetyllactosaminide α1→3-galactosyltransferase from calf thymus. J. Biol. Chem. 1985, 260, 12927–12934. [Google Scholar] [CrossRef]
- Osman, N.; McKenzie, I.F.; Ostenried, K.; Ioannou, Y.A.; Desnick, R.J.; Sandrin, M.S. Combined transgenic expression of alpha-galactosidase and alpha1,2-fucosyltransferase leads to optimal reduction in the major xenoepitope Galalpha(1,3)Gal. Proc. Natl. Acad. Sci. USA 1997, 94, 14677–14682. [Google Scholar] [CrossRef] [Green Version]
- Varki, A. Factors controlling the glycosylation potential of the Golgi apparatus. Trends Cell Biol. 1998, 8, 34–40. [Google Scholar] [CrossRef]
- Hartel-Schenk, S.; Minnifield, N.; Reutter, W.; Hanski, C.; Bauer, C.; Morré, D.J. Distribution of glycosyltransferases among Golgi apparatus subfractions from liver and hepatomas of the rat. Biochim. Biophys. Acta 1991, 1115, 108–122. [Google Scholar] [CrossRef]
- Luo, Y.; Wen, J.; Luo, C.; Cummings, R.D.; Cooper, D.K.C. Pig xenogeneic antigen modification with green coffee bean α-galactosidase. Xenotransplantation 1999, 6, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Zeyland, J.; Woźniak, A.; Gawrońska, B.; Juzwa, W.; Jura, J.; Nowak, A.; Słomski, R.; Smorąg, Z.; Szalata, M.; Mazurek, U.; et al. Double transgenic pigs with combined expression of human α1,2-fucosyltransferase and α-galactosidase designed to avoid hyperacute xenograft rejection. Arch. Immunol. Ther. Exp. 2014, 62, 411–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Greco, V.; Guasch, G.; Fuchs, E.; Mombaerts, P. Mice cloned from skin cells. Proc. Natl. Acad. Sci. USA 2007, 104, 2738–2743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumi, K.; Feinberg, S.E. Skin and oral mucosal substitutes. Oral Maxillofac. Surg. Clin. N. Am. 2002, 14, 61–71. [Google Scholar] [CrossRef]
- Izumi, K.; Takacs, G.; Terashi, H.; Feinberg, S.E. Ex vivo development of a composite human oral mucosal equivalent. J. Oral Maxillofac. Surg. 1999, 57, 571–577. [Google Scholar] [CrossRef]
- Wanichpakorn, S.; Kedjarune-Laggat, U. Primary cell culture from human oral tissue: Gingival keratinocytes, gingival fibroblasts and periodontal ligament fibroblasts. Songklanakarin J. Sci. Technol. 2010, 32, 327–331. [Google Scholar]
- Xu, H.; Wan, H.; Sandor, M.; Qi, S.; Ervin, F.; Harper, J.R.; Silverman, R.P.; McQuillan, D.J. Host response to human acellular dermal matrix transplantation in a primate model of abdominal wall repair. Tissue Eng. Part A 2008, 14, 2009–2019. [Google Scholar] [CrossRef]
- Wiater, J.; Karasiński, J.; Słomski, R.; Smorąg, Z.; Wartalski, K.; Gajda, B.; Jura, J.; Romek, M. The effect of recombinant human alpha-1,2-fucosyltransferase and alpha-galactosidase A on the reduction of alpha-gal expression in the liver of transgenic pigs. Folia Biol. 2020, 68, 121–133. [Google Scholar] [CrossRef]
- Lipinski, D.; Jura, J.; Zeyland, J.; Juzwa, W.; Mały, E.; Kalak, R.; Bochenek, M.; Plawski, A.; Szalata, M.; Smorag, Z.; et al. Production of transgenic pigs expressing human a1,2-fucosyltransferase to avoid humoral xenograft rejection. Med. Weter. 2010, 66, 316–322. [Google Scholar]
- Ramirez, P.; Montoya, M.J.; Ríos, A.; García Palenciano, C.; Majado, M.; Chávez, R.; Muñoz, A.; Fernández, O.M.; Sánchez, A.; Segura, B.; et al. Prevention of hyperacute rejection in a model of orthotopic liver xenotransplantation from pig to baboon using polytransgenic pig livers (CD55, CD59, and H-transferase). Transplant. Proc. 2005, 37, 4103–4106. [Google Scholar] [CrossRef] [PubMed]
- Lutz, A.J.; Li, P.; Estrada, J.L.; Sidner, R.A.; Chihara, R.K.; Downey, S.M.; Burlak, C.; Wang, Z.Y.; Reyes, L.M.; Ivary, B.; et al. Double knockout pigs deficient in N-glycolylneuraminic acid and Galactose α-1,3-Galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation 2013, 20, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Sahara, H.; Watanabe, H.; Pomposelli, T.; Yamada, K. Lung xenotransplantation. Curr. Opin. Organ Transplant. 2017, 22, 541–548. [Google Scholar] [CrossRef]
- Klymiuk, N.; Aigner, B.; Brem, G.; Wolf, E. Genetic modification of pigs as organ donors for xenotransplantation. Mol. Reprod. Dev. 2010, 77, 209–221. [Google Scholar] [CrossRef]
- Bottino, R.; Wijkstrom, M.; van der Windt, D.J.; Hara, H.; Ezzelarab, M.; Murase, N.; Bertera, S.; He, J.; Phelps, C.; Ayares, D.; et al. Pig-to-monkey islet xenotransplantation using multi-transgenic pigs. Am. J. Transplant. 2014, 14, 2275–2287. [Google Scholar] [CrossRef] [Green Version]
- Kong, S.; Ruan, J.; Xin, L.; Fan, J.; Xia, J.; Liu, Z.; Mu, Y.; Yang, S.; Li, K. Multi-transgenic minipig models exhibiting potential for hepatic insulin resistance and pancreatic apoptosis. Mol. Med. Rep. 2016, 13, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Kwon, D.J.; Kim, D.H.; Hwang, I.S.; Kim, D.E.; Kim, H.J.; Kim, J.S.; Lee, K.; Im, G.S.; Lee, J.W.; Hwang, S. Generation of α-1,3-galactosyltransferase knocked-out transgenic cloned pigs with knocked-in five human genes. Transgenic Res. 2017, 26, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Fischer, K.; Kraner-Scheiber, S.; Petersen, B.; Rieblinger, B.; Buermann, A.; Flisikowska, T.; Flisikowski, K.; Christan, S.; Edlinger, M.; Baars, W.; et al. Efficient production of multi-modified pigs for xenotransplantation by ‘combineering’, gene stacking and gene editing. Sci. Rep. 2016, 6, 29081. [Google Scholar] [CrossRef]
- Zeyland, J.; Gawrońska, B.; Juzwa, W.; Jura, J.; Nowak, A.; Słomski, R.; Smorąg, Z.; Szalata, M.; Woźniak, A.; Lipiński, D. Transgenic pigs designed to express human α-galactosidase to avoid humoral xenograft rejection. J. Appl. Genet. 2013, 54, 293–303. [Google Scholar] [CrossRef] [Green Version]
- Haruyama, N.; Cho, A.; Kulkarni, A.B. Overview: Engineering transgenic constructs and mice. Curr. Protoc. Cell Biol. 2009, 42, 19.10.1–19.10.9. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Wu, M.; Huan, Y.; Zhang, L.; Liu, H.; Bou, G.; Luo, Y.; Mu, Y.; Liu, Z. Transgene expression is associated with copy number and cytomegalovirus promoter methylation in transgenic pigs. PLoS ONE 2009, 4, e6679. [Google Scholar] [CrossRef] [Green Version]
- Houdebine, L.M. Use of transgenic animals to improve human health and animal production. Reprod. Domest. Anim. 2005, 40, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.J.; Bissinger, P.; Bullock, D.W.; Damak, S.; Wallace, R.; Whitelaw, C.B.A.; Yull, F. Chromosomal position effects and the modulation of transgene expression. Reprod. Fertil. Dev. 1994, 6, 589–594. [Google Scholar] [CrossRef]
- Garrick, D.; Fiering, S.; Martin, D.I.K.; Whitelaw, E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998, 18, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Folger, K.R.; Wong, E.A.; Wahl, G.; Capecchi, M.R. Patterns of integration of DNA microinjected into cultured mammalian cells: Evidence for homologous recombination between injected plasmid DNA molecules. Mol. Cell. Biol. 1982, 2, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Gajda, B.; Romek, M.; Grad, I.; Krzysztofowicz, E.; Bryla, M.; Smorag, Z. Lipid content and cryotolerance of porcine embryos cultured with phenazine ethosulfate. Cryo Lett. 2011, 32, 349–357. [Google Scholar] [CrossRef]
- Romek, M.; Gajda, B.; Krzysztofowicz, E.; Kucia, M.; Uzarowska, A.; Smorag, Z. Improved quality of porcine embryos cultured with hyaluronan due to the modification of the mitochondrial membrane potential and reactive oxygen species level. Theriogenology 2017, 102, 1–9. [Google Scholar] [CrossRef]
- Wiater, J.; Samiec, M.; Skrzyszowska, M.; Lipiński, D. Trichostatin A-Assisted Epigenomic Modulation Affects the Expression Profiles of not only Recombinant Human α1,2-Fucosyltransferase and α-Galactosidase a Enzymes but also Galα1→3Gal Epitopes in Porcine Bi-Transgenic Adult Cutaneous Fibroblast Cells. Int. J. Mol. Sci. 2021, 22, 1386. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiater, J.; Samiec, M.; Wartalski, K.; Smorąg, Z.; Jura, J.; Słomski, R.; Skrzyszowska, M.; Romek, M. Characterization of Mono- and Bi-Transgenic Pig-Derived Epidermal Keratinocytes Expressing Human FUT2 and GLA Genes—In Vitro Studies. Int. J. Mol. Sci. 2021, 22, 9683. https://doi.org/10.3390/ijms22189683
Wiater J, Samiec M, Wartalski K, Smorąg Z, Jura J, Słomski R, Skrzyszowska M, Romek M. Characterization of Mono- and Bi-Transgenic Pig-Derived Epidermal Keratinocytes Expressing Human FUT2 and GLA Genes—In Vitro Studies. International Journal of Molecular Sciences. 2021; 22(18):9683. https://doi.org/10.3390/ijms22189683
Chicago/Turabian StyleWiater, Jerzy, Marcin Samiec, Kamil Wartalski, Zdzisław Smorąg, Jacek Jura, Ryszard Słomski, Maria Skrzyszowska, and Marek Romek. 2021. "Characterization of Mono- and Bi-Transgenic Pig-Derived Epidermal Keratinocytes Expressing Human FUT2 and GLA Genes—In Vitro Studies" International Journal of Molecular Sciences 22, no. 18: 9683. https://doi.org/10.3390/ijms22189683
APA StyleWiater, J., Samiec, M., Wartalski, K., Smorąg, Z., Jura, J., Słomski, R., Skrzyszowska, M., & Romek, M. (2021). Characterization of Mono- and Bi-Transgenic Pig-Derived Epidermal Keratinocytes Expressing Human FUT2 and GLA Genes—In Vitro Studies. International Journal of Molecular Sciences, 22(18), 9683. https://doi.org/10.3390/ijms22189683





