Roles of Enteric Neural Stem Cell Niche and Enteric Nervous System Development in Hirschsprung Disease
Abstract
1. Introduction
2. Hirschsprung Disease and the Emerging Regenerative Therapeutic Options
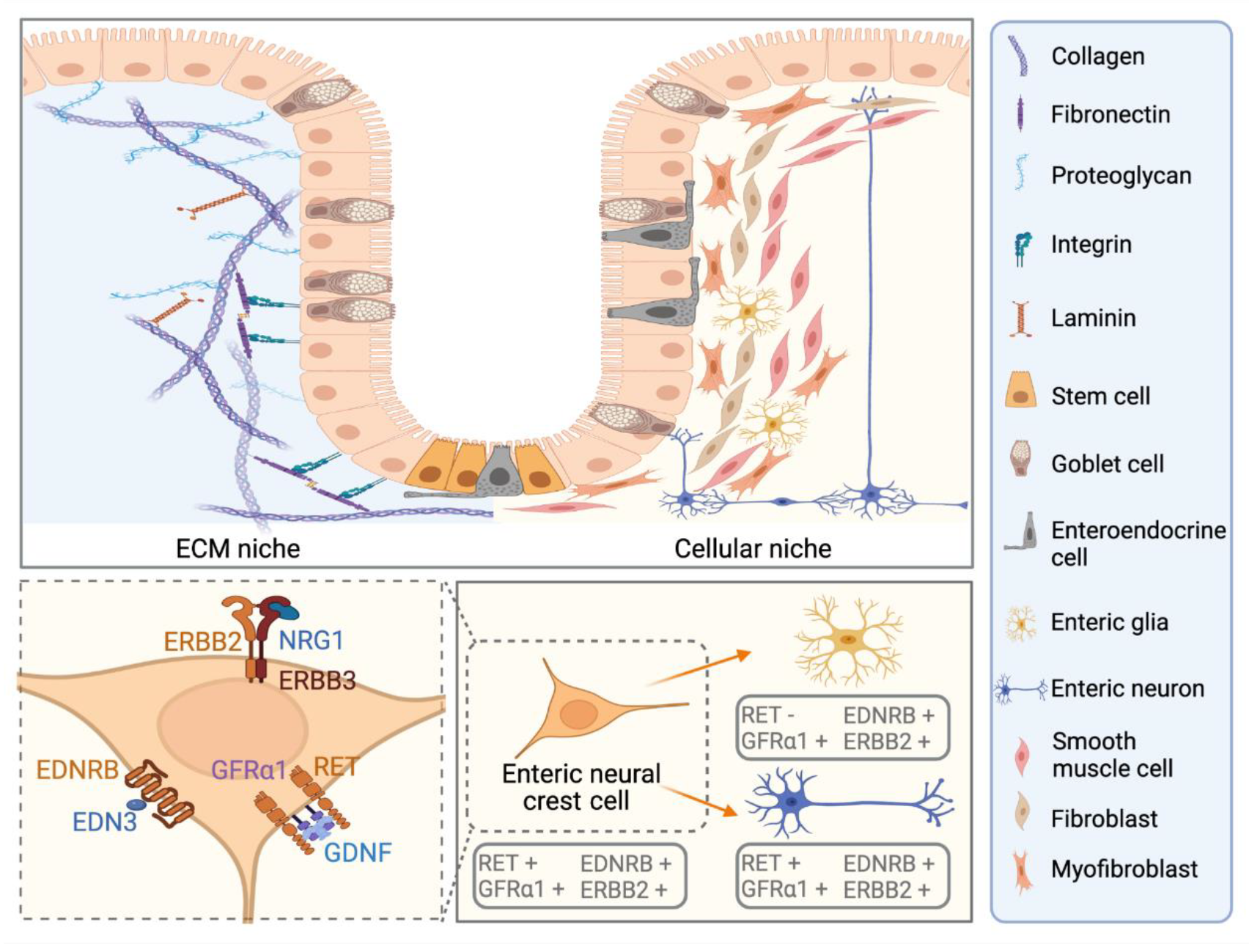
3. Known and Novel Contribution of Cellular Niche Factors to ENS Defect in HSCR
3.1. Established Roles and the Emerging Therapeutic Potentials of GDNF and EDN3 in Modulating RET and EDNRB Signalling
3.2. Neuregulins and Semaphorins as Novel Niche Factors for ENS Dysfunction in HSCR
3.3. Other Niche Factors Implicated by Genetic Defects in HSCR Patients
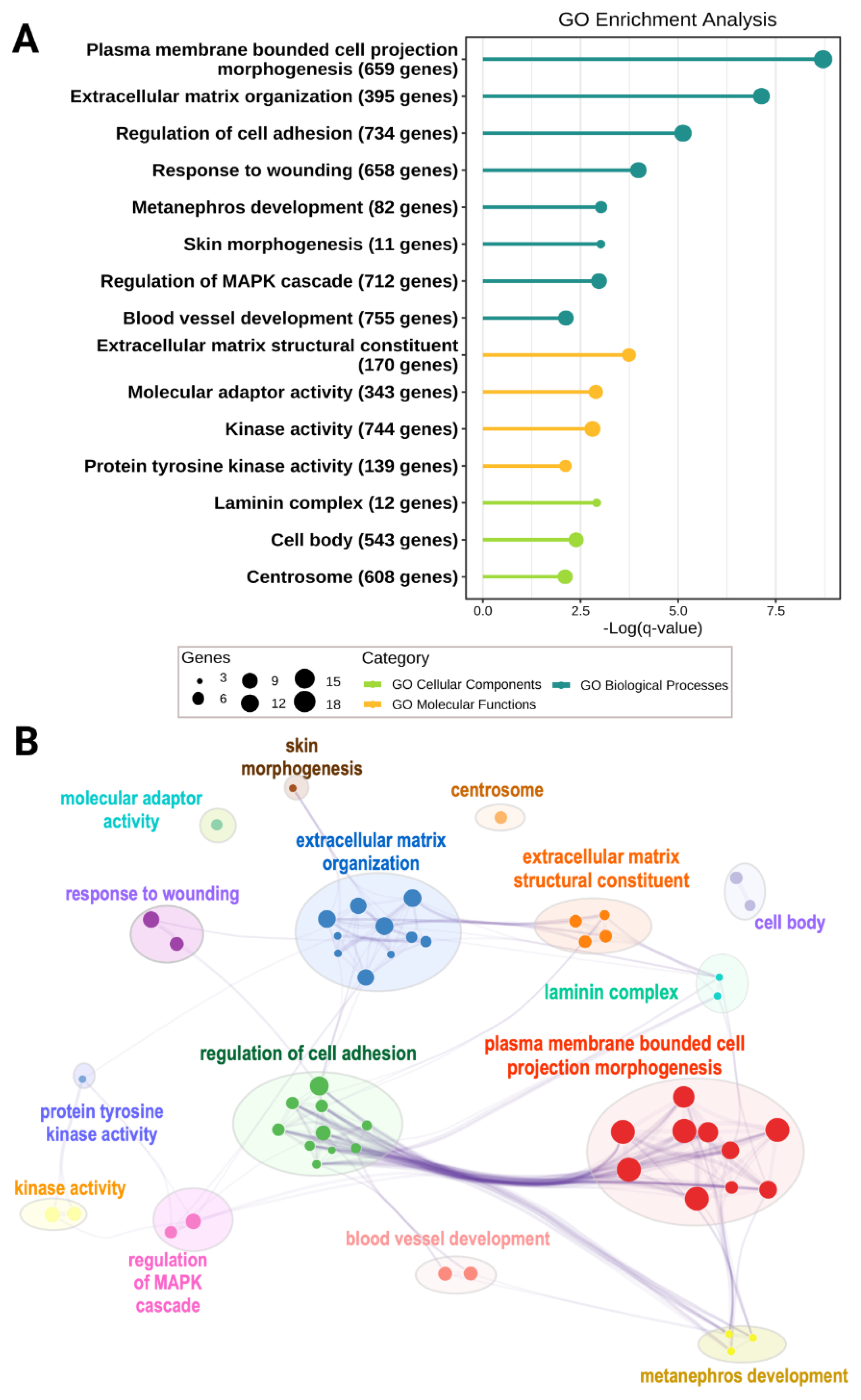
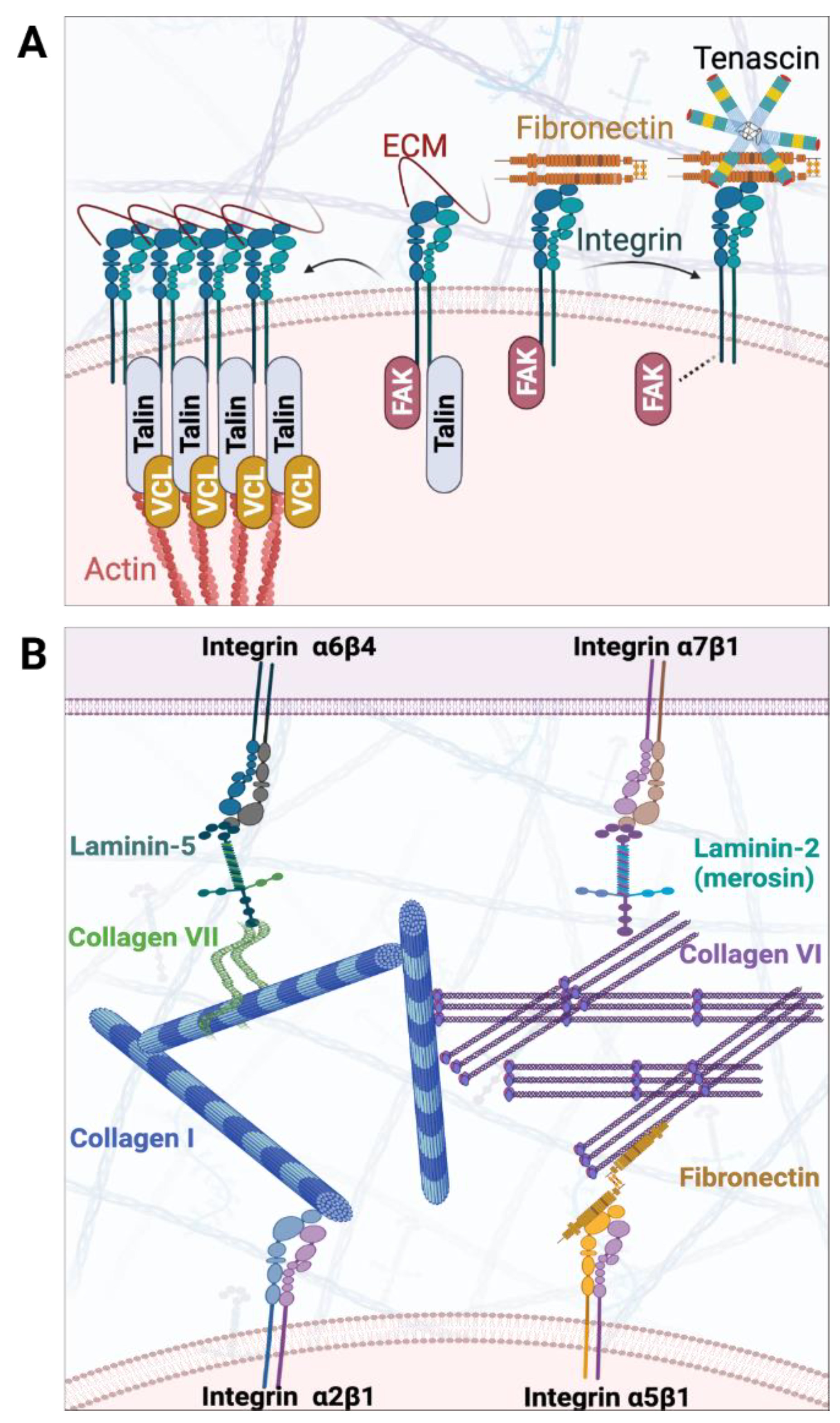
4. ECM Niche in ENS Development
4.1. Established Findings of ECM Abnormalities along the Colon of HSCR Patients
4.2. Recent Findings of ECM Abnormalities in Colon of Mutant Model Organisms
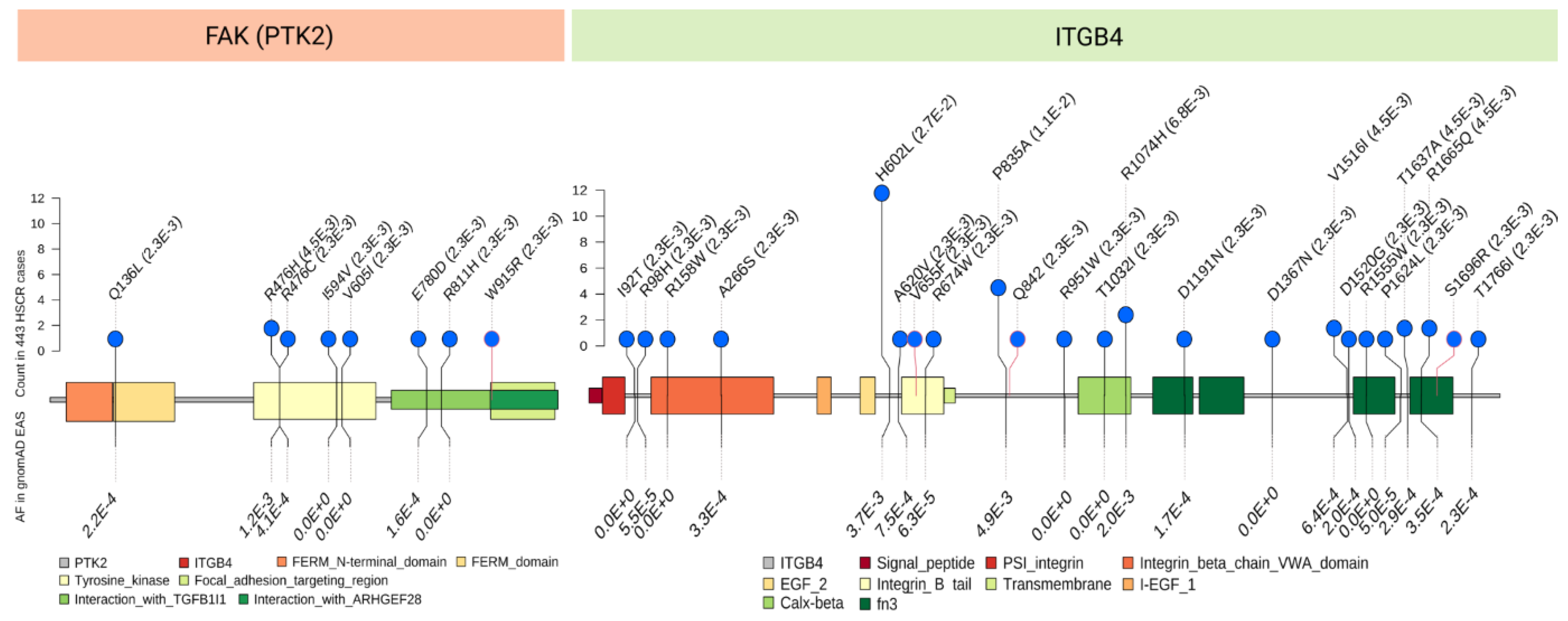
4.3. Novel Discoveries of Intrinsic Niche Defects of HSCR Patients Attributed to Genetic Aberrations from Recent Sequencing Studies
5. Conclusions and Future Perspectives on Regenerative Medicine of HSCR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rao, M.; Gershon, M.D. The bowel and beyond: The enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 517–528. [Google Scholar] [CrossRef]
- Furness, J.B.; Stebbing, M.J. The first brain: Species comparisons and evolutionary implications for the enteric and central nervous systems. Neurogastroenterol. Motil. 2018, 30, e13234. [Google Scholar] [CrossRef]
- Rao, M.; Gershon, M.D. Enteric nervous system development: What could possibly go wrong? Nat. Rev. Neurosci. 2018, 19, 552–565. [Google Scholar] [CrossRef]
- Tam, P.K.H.; Garcia-Barceló, M. Genetic basis of Hirschsprung’s disease. Pediatric Surg. Int. 2009, 25, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.K.H.; Tang, C.S.M.; Garcia-Barceló, M.-M. Genetics of Hirschsprung’s Disease. In Hirschsprung’s Disease and Allied Disorders; Puri, P., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 121–131. [Google Scholar]
- Amiel, J.; Sproat-Emison, E.; Garcia-Barcelo, M.; Lantieri, F.; Burzynski, G.; Borrego, S.; Pelet, A.; Arnold, S.; Miao, X.; Griseri, P.; et al. Hirschsprung disease, associated syndromes and genetics: A review. J. Med. Genet. 2008, 45, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.H.Y.; Wong, K.K.Y.; Tam, P.K.H.; Leung, M.W.Y.; Chao, N.S.Y.; Liu, K.K.W.; Chan, E.K.W.; Tam, Y.H.; Lee, K.H. Are all patients with short segment Hirschsprung’s disease equal? A retrospective multicenter study. Pediatr. Surg. Int. 2018, 34, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.K. Hirschsprung’s disease: A bridge for science and surgery. J. Pediatr. Surg. 2016, 51, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.J.; Goldstein, A.M.; Newgreen, D.F.; Stamp, L.; Schäfer, K.H.; Metzger, M.; Hotta, R.; Young, H.M.; Andrews, P.W.; Thapar, N.; et al. White paper on guidelines concerning enteric nervous system stem cell therapy for enteric neuropathies. Dev. Biol. 2016, 417, 229–251. [Google Scholar] [CrossRef] [PubMed]
- Stamp, L.A.; Young, H.M. Recent advances in regenerative medicine to treat enteric neuropathies: Use of human cells. Neurogastroenterol. Motil. 2017, 29, e12993. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.P.; Lau, S.T.; Wong, J.K.; Gui, H.; Wang, R.X.; Zhou, T.; Lai, W.H.; Tse, H.F.; Tam, P.K.; Garcia-Barcelo, M.M.; et al. Correction of Hirschsprung-Associated Mutations in Human Induced Pluripotent Stem Cells via Clustered Regularly Interspaced Short Palindromic Repeats/Cas9, Restores Neural Crest Cell Function. Gastroenterology 2017, 153, 139–153.e8. [Google Scholar] [CrossRef]
- Fattahi, F.; Steinbeck, J.A.; Kriks, S.; Tchieu, J.; Zimmer, B.; Kishinevsky, S.; Zeltner, N.; Mica, Y.; El-Nachef, W.; Zhao, H.; et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 2016, 531, 105–109. [Google Scholar] [CrossRef]
- Cooper, J.E.; Natarajan, D.; McCann, C.J.; Choudhury, S.; Godwin, H.; Burns, A.J.; Thapar, N. In vivo transplantation of fetal human gut-derived enteric neural crest cells. Neurogastroenterol. Motil. 2017, 29, e12900. [Google Scholar] [CrossRef]
- Frith, T.J.R.; Gogolou, A.; Hackland, J.O.S.; Hewitt, Z.A.; Moore, H.D.; Barbaric, I.; Thapar, N.; Burns, A.J.; Andrews, P.W.; Tsakiridis, A.; et al. Retinoic Acid Accelerates the Specification of Enteric Neural Progenitors from In-Vitro-Derived Neural Crest. Stem Cell Rep. 2020, 15, 557–565. [Google Scholar] [CrossRef]
- Espinosa-Medina, I.; Jevans, B.; Boismoreau, F.; Chettouh, Z.; Enomoto, H.; Müller, T.; Birchmeier, C.; Burns, A.J.; Brunet, J.-F. Dual origin of enteric neurons in vagal Schwann cell precursors and the sympathetic neural crest. Proc. Natl. Acad. Sci. USA 2017, 114, 11980–11985. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Lui, V.C.; Sham, M.H.; Cheung, A.N.; Tam, P.K. HOXB5 expression is spatially and temporarily regulated in human embryonic gut during neural crest cell colonization and differentiation of enteric neuroblasts. Dev. Dyn. 2003, 228, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Heanue, T.A.; Pachnis, V. Enteric nervous system development and Hirschsprung’s disease: Advances in genetic and stem cell studies. Nat. Rev. Neurosci. 2007, 8, 466–479. [Google Scholar] [CrossRef]
- Kawai, K.; Takahashi, M. Intracellular RET signaling pathways activated by GDNF. Cell Tissue Res. 2020, 382, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Obermayr, F.; Hotta, R.; Enomoto, H.; Young, H.M. Development and developmental disorders of the enteric nervous system. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 43–57. [Google Scholar] [CrossRef]
- Bondurand, N.; Dufour, S.; Pingault, V. News from the endothelin-3/EDNRB signaling pathway: Role during enteric nervous system development and involvement in neural crest-associated disorders. Dev. Biol. 2018, 444 (Suppl. 1), S156–S169. [Google Scholar] [CrossRef] [PubMed]
- Gianino, S.; Grider, J.R.; Cresswell, J.; Enomoto, H.; Heuckeroth, R.O. GDNF availability determines enteric neuron number by controlling precursor proliferation. Development 2003, 130, 2187–2198. [Google Scholar] [CrossRef] [PubMed]
- Uesaka, T.; Nagashimada, M.; Enomoto, H. GDNF signaling levels control migration and neuronal differentiation of enteric ganglion precursors. J. Neurosci. 2013, 33, 16372–16382. [Google Scholar] [CrossRef] [PubMed]
- Soret, R.; Schneider, S.; Bernas, G.; Christophers, B.; Souchkova, O.; Charrier, B.; Righini-Grunder, F.; Aspirot, A.; Landry, M.; Kembel, S.W.; et al. Glial Cell-Derived Neurotrophic Factor Induces Enteric Neurogenesis and Improves Colon Structure and Function in Mouse Models of Hirschsprung Disease. Gastroenterology 2020, 159, 1824–1838.e17. [Google Scholar] [CrossRef]
- Watanabe, Y.; Stanchina, L.; Lecerf, L.; Gacem, N.; Conidi, A.; Baral, V.; Pingault, V.; Huylebroeck, D.; Bondurand, N. Differentiation of Mouse Enteric Nervous System Progenitor Cells Is Controlled by Endothelin 3 and Requires Regulation of Ednrb by SOX10 and ZEB2. Gastroenterology 2017, 152, 1139–1150.e4. [Google Scholar] [CrossRef] [PubMed]
- Nagy, N.; Goldstein, A.M. Endothelin-3 regulates neural crest cell proliferation and differentiation in the hindgut enteric nervous system. Dev. Biol. 2006, 293, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Barlow, A.; de Graaff, E.; Pachnis, V. Enteric nervous system progenitors are coordinately controlled by the G protein-coupled receptor EDNRB and the receptor tyrosine kinase RET. Neuron 2003, 40, 905–916. [Google Scholar] [CrossRef]
- Hearn, C.J.; Murphy, M.; Newgreen, D. GDNF and ET-3 differentially modulate the numbers of avian enteric neural crest cells and enteric neurons in vitro. Dev. Biol. 1998, 197, 93–105. [Google Scholar] [CrossRef]
- Bondurand, N.; Natarajan, D.; Barlow, A.; Thapar, N.; Pachnis, V. Maintenance of mammalian enteric nervous system progenitors by SOX10 and endothelin 3 signalling. Development 2006, 133, 2075–2086. [Google Scholar] [CrossRef]
- Gazquez, E.; Watanabe, Y.; Broders-Bondon, F.; Paul-Gilloteaux, P.; Heysch, J.; Baral, V.; Bondurand, N.; Dufour, S. Endothelin-3 stimulates cell adhesion and cooperates with β1-integrins during enteric nervous system ontogenesis. Sci. Rep. 2016, 6, 37877. [Google Scholar] [CrossRef] [PubMed]
- Rothman, T.P.; Chen, J.; Howard, M.J.; Costantini, F.; Schuchardt, A.; Pachnis, V.; Gershon, M.D. Increased expression of laminin-1 and collagen (IV) subunits in the aganglionic bowel of ls/ls, but not c-ret −/− mice. Dev. Biol. 1996, 178, 498–513. [Google Scholar] [CrossRef][Green Version]
- Tang, C.S.; Li, P.; Lai, F.P.; Fu, A.X.; Lau, S.T.; So, M.T.; Lui, K.N.; Li, Z.; Zhuang, X.; Yu, M.; et al. Identification of Genes Associated With Hirschsprung Disease, Based on Whole-Genome Sequence Analysis, and Potential Effects on Enteric Nervous System Development. Gastroenterology 2018, 155, 1908–1922.e5. [Google Scholar] [CrossRef]
- Salomon, R.; Attié, T.; Pelet, A.; Bidaud, C.; Eng, C.; Amiel, J.; Sarnacki, S.; Goulet, O.; Ricour, C.; Nihoul-Fékété, C.; et al. Germline mutations of the RET ligand GDNF are not sufficient to cause Hirschsprung disease. Nat. Genet. 1996, 14, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, J.M.; Ling, A.Y.; Turner, T.N.; Sosa, M.X.; Krumm, N.; Chatterjee, S.; Kapoor, A.; Coe, B.P.; Nguyen, K.H.; Gupta, N.; et al. Molecular Genetic Anatomy and Risk Profile of Hirschsprung’s Disease. N. Engl. J. Med. 2019, 380, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Sribudiani, Y.; Chauhan, R.K.; Alves, M.M.; Petrova, L.; Brosens, E.; Harrison, C.; Wabbersen, T.; de Graaf, B.M.; Rügenbrink, T.; Burzynski, G.; et al. Identification of Variants in RET and IHH Pathway Members in a Large Family With History of Hirschsprung Disease. Gastroenterology 2018, 155, 118–129.e6. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Barcelo, M.M.; Tang, C.S.; Ngan, E.S.; Lui, V.C.; Chen, Y.; So, M.T.; Leon, T.Y.; Miao, X.P.; Shum, C.K.; Liu, F.Q.; et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proc. Natl. Acad. Sci. USA 2009, 106, 2694–2699. [Google Scholar] [CrossRef]
- Jiang, Q.; Arnold, S.; Heanue, T.; Kilambi, K.P.; Doan, B.; Kapoor, A.; Ling, A.Y.; Sosa, M.X.; Guy, M.; Jiang, Q.; et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am. J. Hum. Genet. 2015, 96, 581–596. [Google Scholar] [CrossRef]
- Kapoor, A.; Jiang, Q.; Chatterjee, S.; Chakraborty, P.; Sosa, M.X.; Berrios, C.; Chakravarti, A. Population variation in total genetic risk of Hirschsprung disease from common RET, SEMA3 and NRG1 susceptibility polymorphisms. Hum. Mol. Genet. 2015, 24, 2997–3003. [Google Scholar] [CrossRef]
- Phusantisampan, T.; Sangkhathat, S.; Phongdara, A.; Chiengkriwate, P.; Patrapinyokul, S.; Mahasirimongkol, S. Association of genetic polymorphisms in the RET-protooncogene and NRG1 with Hirschsprung disease in Thai patients. J. Hum. Genet. 2012, 57, 286–293. [Google Scholar] [CrossRef]
- Tang, C.S.-M.; Tang, W.-K.; So, M.-T.; Miao, X.-P.; Leung, B.M.-C.; Yip, B.H.-K.; Leon, T.Y.-Y.; Ngan, E.S.-W.; Lui, V.C.-H.; Chen, Y.; et al. Fine Mapping of the NRG1 Hirschsprung’s Disease Locus. PLoS ONE 2011, 6, e16181. [Google Scholar] [CrossRef][Green Version]
- Tang, C.S.; Gui, H.; Kapoor, A.; Kim, J.H.; Luzon-Toro, B.; Pelet, A.; Burzynski, G.; Lantieri, F.; So, M.T.; Berrios, C.; et al. Trans-ethnic meta-analysis of genome-wide association studies for Hirschsprung disease. Hum. Mol. Genet. 2016, 25, 5265–5275. [Google Scholar] [CrossRef]
- Fadista, J.; Lund, M.; Skotte, L.; Geller, F.; Nandakumar, P.; Chatterjee, S.; Matsson, H.; Granström, A.L.; Wester, T.; Salo, P.; et al. Genome-wide association study of Hirschsprung disease detects a novel low-frequency variant at the RET locus. Eur. J. Hum. Genet. 2018, 26, 561–569. [Google Scholar] [CrossRef]
- Le, T.-L.; Galmiche, L.; Levy, J.; Suwannarat, P.; Hellebrekers, D.M.E.I.; Morarach, K.; Boismoreau, F.; Theunissen, T.E.J.; Lefebvre, M.; Pelet, A.; et al. Dysregulation of the NRG1/ERBB pathway causes a developmental disorder with gastrointestinal dysmotility in humans. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Tang, C.S.; Ngan, E.S.; Tang, W.K.; So, M.T.; Cheng, G.; Miao, X.P.; Leon, T.Y.; Leung, B.M.; Hui, K.J.; Lui, V.H.; et al. Mutations in the NRG1 gene are associated with Hirschsprung disease. Hum. Genet. 2012, 131, 67–76. [Google Scholar] [CrossRef]
- Yang, J.; Duan, S.; Zhong, R.; Yin, J.; Pu, J.; Ke, J.; Lu, X.; Zou, L.; Zhang, H.; Zhu, Z.; et al. Exome sequencing identified NRG3 as a novel susceptible gene of Hirschsprung’s disease in a Chinese population. Mol. Neurobiol. 2013, 47, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Gui, H.; Tang, W.K.; So, M.T.; Proitsi, P.; Sham, P.C.; Tam, P.K.; Ngan, E.S.; Sau-Wai Ngan, E.; Cherny, S.S.; Garcia-Barceló, M.M. RET and NRG1 interplay in Hirschsprung disease. Hum. Genet. 2013, 132, 591–600. [Google Scholar] [CrossRef]
- Birchmeier, C.; Nave, K.A. Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia 2008, 56, 1491–1497. [Google Scholar] [CrossRef] [PubMed]
- Newbern, J.; Birchmeier, C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Semin. Cell Dev. Biol. 2010, 21, 922–928. [Google Scholar] [CrossRef]
- Uesaka, T.; Nagashimada, M.; Enomoto, H. Neuronal Differentiation in Schwann Cell Lineage Underlies Postnatal Neurogenesis in the Enteric Nervous System. J. Neurosci. 2015, 35, 9879–9888. [Google Scholar] [CrossRef]
- Uesaka, T.; Okamoto, M.; Nagashimada, M.; Tsuda, Y.; Kihara, M.; Kiyonari, H.; Enomoto, H. Enhanced enteric neurogenesis by Schwann cell precursors in mouse models of Hirschsprung disease. Glia 2021, 69, 2575–2590. [Google Scholar] [CrossRef] [PubMed]
- Holloway, E.M.; Czerwinski, M.; Tsai, Y.-H.; Wu, J.H.; Wu, A.; Childs, C.J.; Walton, K.D.; Sweet, C.W.; Yu, Q.; Glass, I.; et al. Mapping Development of the Human Intestinal Niche at Single-Cell Resolution. Cell Stem Cell 2021, 28, 568–580.e4. [Google Scholar] [CrossRef] [PubMed]
- Jardé, T.; Chan, W.H.; Rossello, F.J.; Kaur Kahlon, T.; Theocharous, M.; Kurian Arackal, T.; Flores, T.; Giraud, M.; Richards, E.; Chan, E.; et al. Mesenchymal Niche-Derived Neuregulin-1 Drives Intestinal Stem Cell Proliferation and Regeneration of Damaged Epithelium. Cell Stem Cell 2020, 27, 646–662.e7. [Google Scholar] [CrossRef] [PubMed]
- Luzón-Toro, B.; Fernández, R.M.; Torroglosa, A.; de Agustín, J.C.; Méndez-Vidal, C.; Segura, D.I.; Antiñolo, G.; Borrego, S. Mutational spectrum of semaphorin 3A and semaphorin 3D genes in Spanish Hirschsprung patients. PLoS ONE 2013, 8, e54800. [Google Scholar] [CrossRef][Green Version]
- Heanue, T.A.; Pachnis, V. Expression profiling the developing mammalian enteric nervous system identifies marker and candidate Hirschsprung disease genes. Proc. Natl. Acad. Sci. USA 2006, 103, 6919–6924. [Google Scholar] [CrossRef]
- Fu, A.X.; Lui, K.N.; Tang, C.S.; Ng, R.K.; Lai, F.P.; Lau, S.T.; Li, Z.; Gracia-Barcelo, M.M.; Sham, P.; Tam, P.K.; et al. Whole-genome analysis of noncoding genetic variations identifies multi-scale regulatory element perturbations associated with Hirschsprung disease. Genome Res. 2020, 30, 1618–1632. [Google Scholar] [CrossRef]
- Gao, T.; Wright-Jin, E.C.; Sengupta, R.; Anderson, J.B.; Heuckeroth, R.O. Cell-autonomous retinoic acid receptor signaling has stage-specific effects on mouse enteric nervous system. JCI Insight 2021, 6, e145854. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Sato, Y.; Lyons-Warren, A.; Zhang, B.; Kane, M.A.; Napoli, J.L.; Heuckeroth, R.O. Vitamin A facilitates enteric nervous system precursor migration by reducing Pten accumulation. Development 2010, 137, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.K.H.; Chung, P.H.Y.; St Peter, S.D.; Gayer, C.P.; Ford, H.R.; Tam, G.C.H.; Wong, K.K.Y.; Pakarinen, M.P.; Davenport, M. Advances in paediatric gastroenterology. Lancet 2017, 390, 1072–1082. [Google Scholar] [CrossRef]
- Parikh, D.H.; Tam, P.K.; Van Velzen, D.; Edgar, D. Abnormalities in the distribution of laminin and collagen type IV in Hirschsprung’s disease. Gastroenterology 1992, 102 (Pt 1), 1236–1241. [Google Scholar] [CrossRef]
- Parikh, D.H.; Tam, P.K.; Van Velzen, D.; Edgar, D. The extracellular matrix components, tenascin and fibronectin, in Hirschsprung’s disease: An immunohistochemical study. J. Pediatr. Surg. 1994, 29, 1302–1306. [Google Scholar] [CrossRef]
- Parikh, D.H.; Tam, P.K.; Lloyd, D.A.; Van Velzen, D.; Edgar, D.H. Quantitative and qualitative analysis of the extracellular matrix protein, laminin, in Hirschsprung’s disease. J. Pediatr. Surg. 1992, 27, 991–995, discussion 5–6. [Google Scholar] [CrossRef]
- Zheng, Y.; Lv, X.; Wang, D.; Gao, N.; Zhang, Q.; Li, A. Down-regulation of fibronectin and the correlated expression of neuroligin in hirschsprung disease. Neurogastroenterol. Motil. 2017, 29, e13134. [Google Scholar] [CrossRef]
- Gao, N.; Hou, P.; Wang, J.; Zhou, T.; Wang, D.; Zhang, Q.; Mu, W.; Lv, X.; Li, A. Increased Fibronectin Impairs the Function of Excitatory/Inhibitory Synapses in Hirschsprung Disease. Cell Mol. Neurobiol. 2020, 40, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Wang, J.; Zhang, Q.; Zhou, T.; Mu, W.; Hou, P.; Wang, D.; Lv, X.; Li, A. Aberrant Distributions of Collagen I, III, and IV in Hirschsprung Disease. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 450–456. [Google Scholar] [CrossRef]
- Payette, R.F.; Tennyson, V.M.; Pomeranz, H.D.; Pham, T.D.; Rothman, T.P.; Gershon, M.D. Accumulation of components of basal laminae: Association with the failure of neural crest cells to colonize the presumptive aganglionic bowel of ls/ls mutant mice. Dev. Biol. 1988, 125, 341–360. [Google Scholar] [CrossRef]
- Druckenbrod, N.R.; Epstein, M.L. Age-dependent changes in the gut environment restrict the invasion of the hindgut by enteric neural progenitors. Development 2009, 136, 3195–3203. [Google Scholar] [CrossRef] [PubMed]
- Breau, M.A.; Pietri, T.; Eder, O.; Blanche, M.; Brakebusch, C.; Fässler, R.; Thiery, J.P.; Dufour, S. Lack of beta1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development 2006, 133, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Chernousov, M.A.; Kaufman, S.J.; Stahl, R.C.; Rothblum, K.; Carey, D.J. Alpha7beta1 integrin is a receptor for laminin-2 on Schwann cells. Glia 2007, 55, 1134–1144. [Google Scholar] [CrossRef]
- Nagy, N.; Mwizerwa, O.; Yaniv, K.; Carmel, L.; Pieretti-Vanmarcke, R.; Weinstein, B.M.; Goldstein, A.M. Endothelial cells promote migration and proliferation of enteric neural crest cells via beta1 integrin signaling. Dev. Biol. 2009, 330, 263–272. [Google Scholar] [CrossRef]
- Akbareian, S.E.; Nagy, N.; Steiger, C.E.; Mably, J.D.; Miller, S.A.; Hotta, R.; Molnar, D.; Goldstein, A.M. Enteric neural crest-derived cells promote their migration by modifying their microenvironment through tenascin-C production. Dev. Biol. 2013, 382, 446–456. [Google Scholar] [CrossRef]
- Nagy, N.; Barad, C.; Hotta, R.; Bhave, S.; Arciero, E.; Dora, D.; Goldstein, A.M. Collagen 18 and agrin are secreted by neural crest cells to remodel their microenvironment and regulate their migration during enteric nervous system development. Development 2018, 145, dev160317. [Google Scholar] [CrossRef]
- Soret, R.; Mennetrey, M.; Bergeron, K.F.; Dariel, A.; Neunlist, M.; Grunder, F.; Faure, C.; Silversides, D.W.; Pilon, N. A collagen VI–dependent pathogenic mechanism for Hirschsprung’s disease. J. Clin. Investig. 2015, 125, 4483–4496. [Google Scholar] [CrossRef] [PubMed]
- Breau, M.A.; Dahmani, A.; Broders-Bondon, F.; Thiery, J.P.; Dufour, S. Beta1 integrins are required for the invasion of the caecum and proximal hindgut by enteric neural crest cells. Development 2009, 136, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Nishida, S.; Yoshizaki, H.; Yasui, Y.; Kuwahara, T.; Kiyokawa, E.; Kohno, M. Collagen VI suppresses fibronectin-induced enteric neural crest cell migration by downregulation of focal adhesion proteins. Biochem. Biophys. Res. Commun. 2018, 495, 1461–1467. [Google Scholar] [CrossRef]
- Tang, C.S.; Zhuang, X.; Lam, W.Y.; Ngan, E.S.; Hsu, J.S.; Yu, M.; So, M.T.; Cherny, S.S.; Ngo, N.D.; Sham, P.C.; et al. Uncovering the genetic lesions underlying the most severe form of Hirschsprung disease by whole-genome sequencing. Eur. J. Hum. Genet. 2018, 26, 818–826. [Google Scholar] [CrossRef]
- Grove, M.; Brophy, P.J. FAK is required for Schwann cell spreading on immature basal lamina to coordinate the radial sorting of peripheral axons with myelination. J. Neurosci. 2014, 34, 13422–13434. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Menacho, I.; Morandi, A.; Mologni, L.; Boender, P.; Gambacorti-Passerini, C.; Magee, A.I.; Hofstra, R.M.; Knowles, P.; McDonald, N.Q.; Isacke, C.M. Focal adhesion kinase (FAK) binds RET kinase via its FERM domain, priming a direct and reciprocal RET-FAK transactivation mechanism. J. Biol. Chem. 2011, 286, 17292–17302. [Google Scholar] [CrossRef]
| Gene | Phenotype | Mode of Inheritance a | Variant Spectrum |
|---|---|---|---|
| Cellular niche factors | |||
| GDNF | Isolated HSCR | AD | Rare |
| EDN3 | Isolated HSCR | AR/AD | Rare |
| Waardenburg syndrome | AD | Rare | |
| ECE1 | HSCR with cardiac, craniofacial, and autonomic defects | AD | Rare |
| SOX10 | Isolated HSCR | AD | Rare |
| Waardenburg syndrome | AD | Rare | |
| PHOX2B | Congenital Central Hypoventilation Syndrome with HSCR | AD | Rare |
| Isolated HSCR | Additive | Common | |
| NRG1 | Isolated HSCR | AD and additive | Rare and common |
| SEMA3C/D | Isolated HSCR | AD and additive | Rare and common |
| ECM niche factors | |||
| COL6A2 | Isolated HSCR | AD | Rare |
| COL6A3 | Isolated HSCR | AD | Rare |
| COL1A2 | Isolated HSCR | AD | Rare |
| LAMB2 | Isolated HSCR | AD | Rare |
| LAMA5 | Isolated HSCR | AD | Rare |
| ITGA2 | Isolated HSCR | AD | Rare |
| ITGB4 | Isolated HSCR | AD | Rare and low frequency |
| ITGB5 | Isolated HSCR | AD | Rare |
| PTK2/FAK | Isolated HSCR | AD | Rare |
| VCL | Isolated HSCR | AD | Rare |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Y.; Tam, P.K.-H.; Tang, C.S.-M. Roles of Enteric Neural Stem Cell Niche and Enteric Nervous System Development in Hirschsprung Disease. Int. J. Mol. Sci. 2021, 22, 9659. https://doi.org/10.3390/ijms22189659
Ji Y, Tam PK-H, Tang CS-M. Roles of Enteric Neural Stem Cell Niche and Enteric Nervous System Development in Hirschsprung Disease. International Journal of Molecular Sciences. 2021; 22(18):9659. https://doi.org/10.3390/ijms22189659
Chicago/Turabian StyleJi, Yue, Paul Kwong-Hang Tam, and Clara Sze-Man Tang. 2021. "Roles of Enteric Neural Stem Cell Niche and Enteric Nervous System Development in Hirschsprung Disease" International Journal of Molecular Sciences 22, no. 18: 9659. https://doi.org/10.3390/ijms22189659
APA StyleJi, Y., Tam, P. K.-H., & Tang, C. S.-M. (2021). Roles of Enteric Neural Stem Cell Niche and Enteric Nervous System Development in Hirschsprung Disease. International Journal of Molecular Sciences, 22(18), 9659. https://doi.org/10.3390/ijms22189659






