Abstract
Short-chain dehydrogenase/reductase (SDR) belongs to the NAD(P)(H)-dependent oxidoreductase superfamily. Limited investigations reveal that SDRs participate in diverse metabolisms. A genome-wide identification of the SDR gene family in M. truncatula was conducted. A total of 213 MtSDR genes were identified, and they were distributed on all chromosomes unevenly. MtSDR proteins were categorized into seven subgroups based on phylogenetic analysis and three types including ‘classic’, ‘extended’, and ‘atypical’, depending on the cofactor-binding site and active site. Analysis of the data from M. truncatula Gene Expression Atlas (MtGEA) showed that above half of MtSDRs were expressed in at least one organ, and lots of MtSDRs had a preference in a tissue-specific expression. The cis-acting element responsive to plant hormones (salicylic acid, ABA, auxin, MeJA, and gibberellin) and stresses were found in the promoter of some MtSDRs. Many genes of MtSDR7C, MtSDR65C, MtSDR110C, MtSDR114C, and MtSDR108E families were responsive to drought, salt, and cold. The study provides useful information for further investigation on biological functions of MtSDRs, especially in abiotic stress adaptation, in the future.
1. Introduction
Short-chain dehydrogenases/reductases (SDRs) belong to the NAD(P)(H)-dependent oxidoreductase protein superfamily [1,2]. This old family of metabolic enzymes is present in all organisms [3,4]. Although SDR proteins show diversity in structure and function, they have three common properties: (1) a cofactor binding site (TGxxxGxG), (2) a catalytical residue motif YxxxK, and (3) a conserved three-dimensional structure made up of “Rossmann fold” a-sheet with a-helices on both sides [5]. The SDR superfamily is usually divided into five types: ‘classical’, ‘extended’, ‘atypical’, ‘unknown’, and ‘divergent’, respectively, based on the structural characteristics [1,6].
A limited investigation on SDR in plants revealed that the SDR superfamily is involved in either primary or secondary metabolisms, such as fatty acid synthesis and elongation [7,8], chlorophyll biosynthesis or retrogradation [9,10], and terpenoids [11], steroids [12], phenolics [13], and alkaloids synthesis and metabolism [14]. The secondary metabolites of plants can be accumulated in response to stress in the form of chemical defense to provide a protective barrier. AtSDR1, being named as ABSCISIC ACID DEFICIENT2 (ABA2), catalyzes the multistep reaction from xanthoxin to abscisic aldehyde that is the key step of ABA biosynthesis [15,16,17]. AtSDR3 is homologous to AtSDR1, which confers resistance to Pst DC3000 by regulating AtPR-1 gene expression in a salicylic-acid-dependent pathway [18]. CaMNR1 (menthone: (1)-(3S)-neomenthol reductase) catalyzes menthone reduction to generate neomenthol using NADP as a cofactor. Overexpressing CaMNR1 improves the immunity to both bacterial and fungal pathogens in Capsicum annuum [19]. However, the role of SDR family members in abiotic stress adaptation has not been reported.
Legumes are important for both humans as food and livestock as feed [20,21]. Medicago truncatula is a model plant of legumes due to its modest genome size, diploid genome, and annual and autogamous nature, and it is easy to be transformed [22,23,24]. Given the potential importance of the SDR family, the genome-wide identification of SDR genes was conducted in M. truncatula in the present study to understand their gene structure, phylogenetic relationship, and common protein features as well as tissue-specific expression and responses to abiotic stress. The investigation will provide basic information for further comprehensive approaches on diverse functions of SDR in M. truncatula in the future.
2. Results
2.1. Identification and Classification of MtSDR Genes
With 178 Arabidopsis SDR protein sequences as baits, BLAST (Basic Local Alignment Search Tool) was applied to search for the target sequence of M. truncatula in order to comprehensively identify the SDR coding genes of M. truncatula. We used the hidden Markov model (HMM) profile of SDR (Pfam: PF00106, PF01370, and PF01073) [25] from the Pfam database to search the protein sequences in M. truncatula. The candidate SDR-encoding genes were manually chosen and functionally annotated based on the closest A. thaliana homology. Finally, a total of 213 MtSDRs were identified and were categorized into 42 families.
The gene locus ID, protein sequence length (SL), molecular weight (MW), and isoelectric point (pI) were summarized in Table S1. Most of MtSDRs (88%) were divided into three main types, ‘classical’, ‘extended’, and ‘divergent’, while a minority (10%) was classified into unknown or atypical types. Five MtSDRs (Medtr3g101500.1, Medtr4g086400.1, Medtr4g086410.1, Medtr6g088500.1, and Medtr8g044240.1) showing up as homologous to At4G333601.1, At1G49671.1, and At4G132501.1 had low HMM scores and no structural data. MtSDR proteins had the average length of 340 amino acids with arrangement from 185 amino acids for MtSDR114C6 to 799 amino acids for MtSDR2E1. The MWs varied from 20.41 kDa (MtSDR114C6) to 89.92 kDa (MtSDR2E1), with an average of 37.40 kDa. The pI of the MtSDRs varied from 4.69 (MtSDR114C11) to 9.96 (MtSDR50E3), with an average of 7.14 (Table S1).
2.2. Phylogenetic Relationships of MtSDR Proteins
To explore the evolutionary relationship among MtSDR proteins, the unrooted phylogenetic tree of 213 MtSDRs was aligned using MEGA-X software based on the sequences of MtSDR proteins (Table S2). The chromosomal location and gene function annotation of MtSDRs was analyzed using TBtools software. Based on the phylogenetic tree, MtSDRs were classified into three primary groups (classical SDR fold, atypical members, and extended SDR fold) and seven subgroups, with 115 MtSDRs in classical SDR fold, 17 MtSDRs in atypical, and 81 MtSDRs in extended SDR fold (Figure 1, Table S2). Moreover, the MtSDRs in the classical SDR fold were further classified into three subgroups (cluster C1, 2, and 3) consisting of 50, 45, and 20 members, respectively. The MtSDRs in extended SDR fold were further classified into three subgroups (cluster E1, 2, and 3) consisting of 18, 28, and 35 members, respectively (Figure 1 and Figure S1).
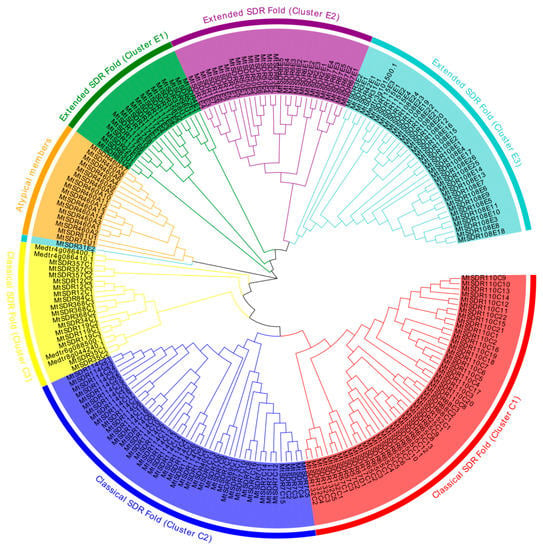
Figure 1.
Phylogenetic tree of the domain relationship of MtSDRs. The different colors perform the different groups (or subgroups) of MtSDR.
2.3. Ten Conserved Motifs Were Examined in MtSDR Proteins
We searched for the conserved motifs in 213 MtSDR proteins aiming to evaluate the functional regions of MtSDR proteins. Ten conserved motifs, namely motifs 1 to 10, were identified and indicated in the colored boxes based on their scale with sizes varying from 14 to 39 aa residues in width (Figure 2). Motif 1 encodes the conserved cofactor-binding site of ‘classic’, ‘extended’, and ‘divergent’ domains, motif 2 encodes the conserved active site of ‘classical’, ‘extended’, and ‘atypical’ domains. Motif 1 and motif 2 were observed in all MtSDRs (Figure S1). The other conserved motifs (motifs 3 to 10) also existed in MtSDR proteins with varying numbers from one to eight.
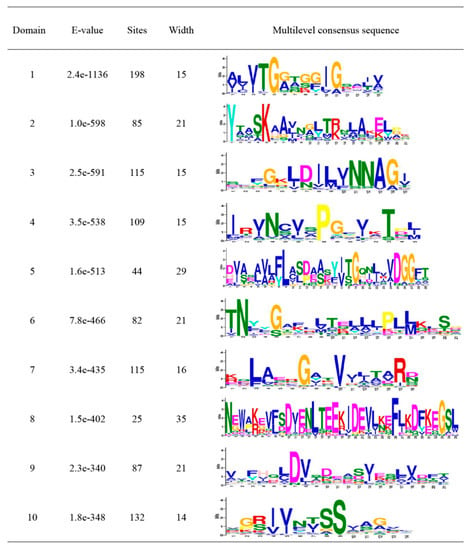
Figure 2.
Details of Ten Motifs of MtSDRs.
2.4. Chromosomal Location (Distribution) and Expansion (Duplication) Analysis of MtSDRs
In order to understand the distribution of MtSDRs in the genome, BLAST was used to locate 213 MtSDR genes on the corresponding chromosomes. MtSDRs were present on all chromosomes, while the arrangement and density of MtSDRs on every chromosome were uneven (Figure 3). The maximal number of MtSDRs (21.6%) was located on chromosome 4, whereas the minimum number of MtSDRs (6.1%) was located on chromosome 6. There was a high density of MtSDRs located on chromosomes 3, 4, 7, and 8.
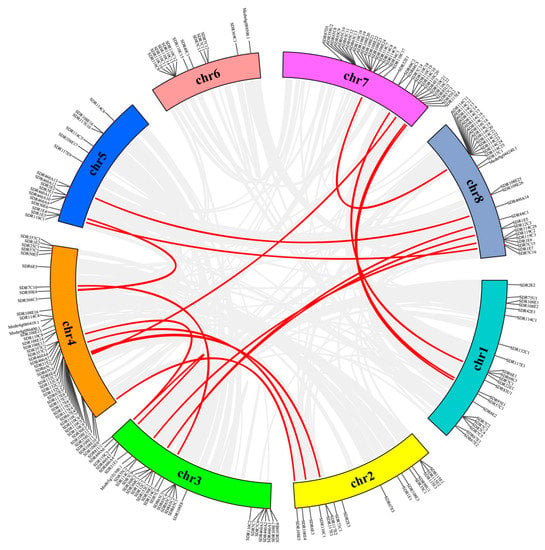
Figure 3.
Synteny analysis of interchromosomal relationships of MtSDR genes. Red lines indicate duplicated gene pairs.
The duplication patterns (tandem and segmental repeats) were analyzed using BLASTp and MCScanX (Multiple Collinear Scan toolkit). Twenty gene pairs were found to be paralogous in MtSDRs, and they belonged to segmental duplications (Table S3). In addition, no tandem duplication was observed. The results showed that segmental duplication was the crucial driving strength for the evolution of MtSDRs.
Collinearity diagrams among MtSDRs were analyzed using gene duplication analysis. A total of 20 pairs of gene duplications were identified (Figure 4). The orthologous relationship between 213 MtSDR genes and 178 AtSDR genes from Arabidopsis or 315 GmSDR genes from Glycin max was analyzed. The results showed that there were 83 pairs of orthologous SDRs between M. truncatula and A. thaliana and 275 pairs between M. truncatula and G. max (Figure 4). The synonymous substitution rate (Ks) and non-synonymous substitution rate (Ka) of the MtSDR gene pairs were calculated using TBtools software. The Ka/Ks value can demonstrate whether the select pressure functions on protein coding MtSDRs. The Ka/Ks value of all the paralogous MtSDR gene pairs was below 1 (Table S3), implying that purifying selection existed in the gene pairs.
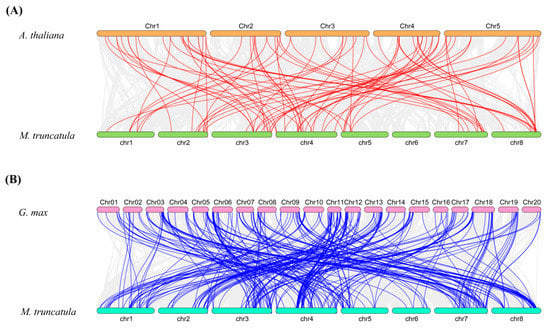
Figure 4.
Synteny analysis of MtSDR genes in the genomes between M. truncatula and A. thaliana or G.max. (A) M. truncatula and A. thaliana; (B) M. truncatula and G.max. The gray lines show collinear blocks. The red and blue lines indicate the syntenic gene pairs between M. truncatula and A. thaliana or G. max, respectively.
2.5. Analysis of the Expression Pattern of MtSDRs in Different Organs
The spatial expression profile was analyzed based on microarray data from M. truncatula Gene Expression Atlas (MtGEA, https://mtgea.noble.org/v3/, accessed on 5 April 2021) of 156 MtSDRs in flowers, leaves, petioles, pods, roots, stems, and seeds (Table S4), while the information of 57 MtSDR genes was not available. Above half of MtSDRs were expressed in at least one organ. Transcript of most of the genes in the SDR108E family was observed in roots (Figure 5). Some MtSDRs genes exhibited specific tissue expression. MtSDR1E6, 65C7, 65C11, 114C3, and 114C7 were preferentially expressed in flowers; MtSDR460A14, 108E19, 114C15, and 110C2 were specifically expressed in roots; while MtSDR57C3 and MtSDR119C3 were only expressed in seeds. The transcript levels of MtSDR57C3 and MtSDR119C3 were increased from 12 to 24 d, but decreased at 36 d after pollination, indicating that they might be associated with seed development.
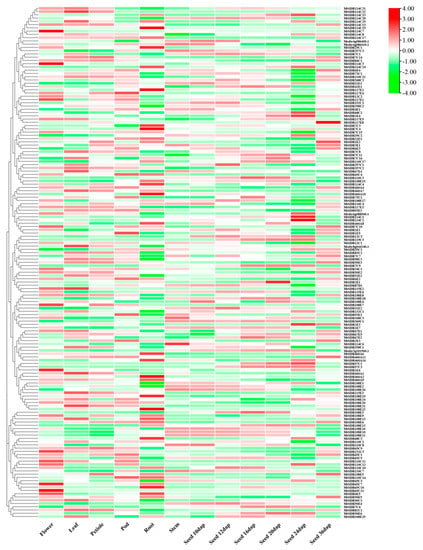
Figure 5.
Heat map of MtSDRs in different organs. The expression data of MtSDRs in flowers, leaves, petioles, flowers, roots, stems, and seeds were collected from M. truncatula Gene Expression Atlas (MtGEA, https://mtgea.noble.org/v3/, accessed on 5 April 2021). The relative expression of heat map was characterized by log2 transformed.
2.6. Analysis of Cis-Acting Elements in the Promoter Region of MtSDRs
The cis-acting elements in the 2000bp sequence of the promoter region were analyzed using PlantCARE software. Thirteen types cis-acting elements associated with responses to stresses and phytohormones including wound responsive element (WUN-motif and WRE3), circadian responsive element (Circadia), drought responsive element (MBS), low-temperature responsive element (LTR), auxin responsive element (TGA-element and AuxRR-core), gibberellin responsive element (TATC-box, GARE-motif, and P-box), salicylic acid responsive element (TCA-element), MeJA responsive element (CGTCA-motif and TGACG-motif), and ABA response element (ABRE) were found in the promoters (Figure 6, Table S5). A total of 280 wound responsive elements, 43 circadian responsive elements, 137 drought responsive elements, 89 low-temperature responsive elements, 128 auxin responsive elements, 151 gibberellin responsive elements, 131 salicylic acid responsive elements, 500 MeJA responsive elements, and 435 ABA responsive elements were identified in all MtSDR genes, indicating that MtSDRs participated in plant growth, development, and stress responses.
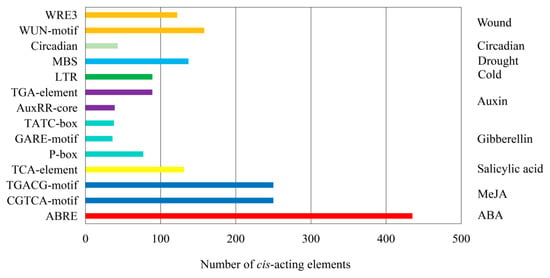
Figure 6.
The number of cis-acting elements was tested in the promoter region of MtSDRs. The name of cis-acting elements was listed on the left side of the image, and the corresponding function annotation was listed on the right side.
2.7. Expression Pattern of MtSDRs in Response to Abiotic Stresses
MtSDRs genes in five subfamilies including SDR108E (dihydroflavonol 4-reductase), SDR110C (ABA2 xanthoxin dehydrogenase family), MtSDR7C (chloroplast protein import translocon), MtSDR65C (tropinone reductase), and MtSDR114C (menthone/salutaridine reductase) in response to salt and drought were analyzed. The expression data were obtained from M. truncatula Gene Expression Atlas (MtGEA, https://mtgea.noble.org/v3/, accessed on 5 April 2021) (Table S6). Of the 16 genes in the MtSDR114C family, 12 were upregulated after 6 h of salt treatment, and three were upregulated after 48 h (Figure 7). Of the 11 genes in MtSDR110C families, six were upregulated after 6 h of salt treatment, three were upregulated after 48 h, and two were downregulated. Most of the genes in SDR108E and SDR65C families were upregulated after 6 h or 48 h of salt stress, while transcript levels of nine genes (75%) in the MtSDR7C family were decreased after salt stress. In addition, most genes in the MtSDR114C family were upregulated by drought in roots, followed by downregulation after rewatering, while most genes in MtSDR460A family were downregulated after drought treatment, followed by upregulation after rewatering.
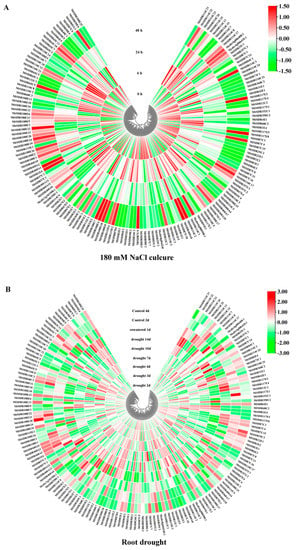
Figure 7.
Heat map of MtSDRs expression in response to salt and drought stress. The expression data were collected from M. truncatula Gene Expression Atlas (MtGEA, https://mtgea.noble.org/v3/, accessed on 5 April 2021). (A) Two-week-old seedlings were treated with 180 mM NaCl for 0, 6, 24, and 48 h, respectively. (B) Twenty-four-day-old seedlings were withholding irrigation for soil drying for 14 d, followed by 1 d of rewatering.
In order to understand the response of MtSDR genes to the cold, six MtSDR genes containing LTR cis-acting elements in the promoter region were analyzed. The results showed that MtSDR65C8 and Mt132C6 transcripts were increased after 6 h of cold treatment and reached a peak at 12 h (Figure 8). MtSDR7C6 and Mt110C15 transcripts were induced after 12 h of cold treatment, MtSDR108E18 transcript was induced after 24 h, while MtSDR108E3 transcript was greatly reduced after 2 h of cold treatment. The results implied that the MtSDR genes were cold-responsive.
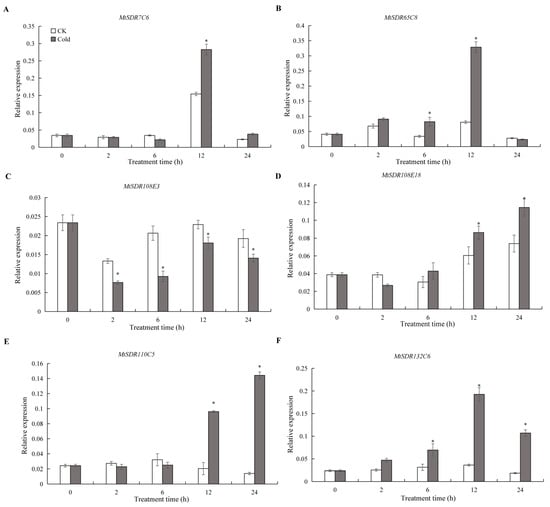
Figure 8.
Relative expression of MtSDR7C6 (A), SDR65C8 (B), SDR108E3 (C), SDR108E18 (D), SDR110C15 (E), and SDR132C6 (F) in response to cold treatment. Four-week-old seedlings were placed in a growth chamber at 4 °C for 0, 2, 6, 12, and 24 h. Mean values and standard errors were calculated from three biological replicates. * indicates significant difference between cold treatment and control using one-way ANOVA at p ≤ 0.05.
2.8. Subcellular Localization of MtSDRs
The salt or drought responsive genes MtSDR7C16, 65C9, 108E24, 110C17, and 114C22 were selected for investigation for protein subcellular localization. GFP fluorescence signal was disseminated throughout the cell, while the signal of PM-marker protein was localized on the plasma membrane (Figure 9). The signal of MtSDR7C16, 65C9, 108E24, 110C17, and 114C22 fused with GFP was not co-localized with plasma membrane localization protein AtAKT1, indicating that MtSDR7C16, 65C9, 108E24, 110C17, and 114C22 were localized in the cytoplasm.
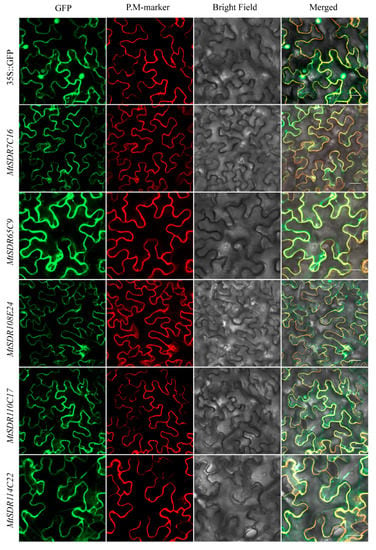
Figure 9.
Subcellular localization of MtSDRs. The injected tobacco was used for observation. PM-marker: plasma membrane localization protein AtAKT1. Bars = 20 µm.
3. Discussion
The SDR superfamily is found in all organisms. The SDR superfamily has a conservative “Rossmann fold” structure and shows low sequence similarity among members, which makes it difficult to identify [26,27,28]. 213 MtSDR genes were identified in M. truncatula using the HMM models and similarity searches, which is similar to that in rice (227 OsSDRs) and maize (230 ZmSDRs) but different from that in G. max (315 GmSDRs, A. thaliana (178 AtSDRs)) [5]. The analysis revealed that MtSDR proteins had diverse variations in sequence length, molecular weight, and isoelectric points.
Ten conserved motifs were identified in MtSDR proteins. Among them, motif 1 and motif 2 that encode the conserved cofactor-binding site and the conserved active site existed in almost all MtSDR proteins. MtSDR genes were irregularly distributed on chromosomes. Homologous gene pairs were found in a few MtSDRs, indicating that the members of the MtSDR superfamily had low pairwise sequence identity. Compared to Arabidopsis, strikingly, based on the syntenic map of M. truncatula and G. max, 116 MtSDRs were shown to have 212 corresponding G. max orthologs. Therefore, the expansion of the MtSDR gene family may have appeared before the separation of G. max. The process of genetic evolution was probably reestablished by analyzing collinearity of the genomic sequences of SDR family in the same genome or between different genomes [29].
Gene functions are closely related to tissue-specific expression. Most of the genes in the SDR108E family were highly expressed in root, suggesting that SDR108E may be important for the function of roots. MtSDR67E5 was highly expressed in all test tissues. MtSDR67E5 is homologous to UDP-D-apiose/UDP-D-xylose synthases (AXS2) confers synthesis of Rhamnogalacturonan-II (RG-II), which is one of major pectin types in M. truncatula [30]. SDR proteins were involved in many processes of primary and secondary metabolism [31]. DVR (3,8-divinyl protochlorophyllide an 8-vinyl reductase) was an enzyme belonging to the extended SDR fold, participating in synthesis of monovinyl derivatives of chlorophyll in Arabidopsis [32]. Two NAD-dependent 3β-HSD/D (At1g47290 and At2g26260), belonging to the SDR31E family, are involved in sterol biosynthesis in Arabidopsis [12,33]. The members in SDR31E family in M. truncatula are homologous to those in SDR31E family in Arabidopsis, implying that they might have similar functions.
The potential role of MtSDR genes in abiotic stress responses was analyzed in the study. Regulatory elements in the promoter region are associated in the regulation of gene expression, being temporal, spatial, and cell-specific [34]. The cis-elements responsive to abiotic stresses and hormones were identified in the promoter regions of MtSDRs. Plants can alleviate the damage caused by stress by enhancing the accumulation of secondary metabolites [31]. Remarkably, the four families responsive to abiotic stress were involved in secondary metabolism: MtSDR65C (tropinone reductase), SDR108E (Dihydroflavonol 4-reductase) family, SDR110C (ABA2 xanthoxin dehydrogenase family), and MtSDR114C (menthone/salutaridine reductase). The conversion of xanthoxin to abscisic aldehyde was catalyzed by ABA2 (SDR1) in Arabidopsis. MtSDR110C2, the homolog of AtSDR1, was highly expressed in roots, suggesting that it may also involve in the conversion of ABA [15]. Dihydroflavonol 4-reductase (SDR108E) was one of the key regulatory enzymes of flavan-3-ols biosynthesis and protects plants against oxidative damage in transgenic tobacco [35]. Members in MtSDR7C, MtSDR65C, MtSDR110C, MtSDR114C, and MtSDR108E families were up- or down-regulated in response to salt and drought stresses, while the expression of MtSDR7C6, Mt65C8, Mt108E18, Mt110C15, and Mt132C6 genes was altered after cold treatment. The results suggest that MtSDRs may be associated with abiotic stress adaptation. The above genes should be selected for further studies on the role of abiotic stress tolerance in M. truncatula.
4. Materials and Methods
4.1. Identification of MtSDR Genes
Arabidopsis SDR protein sequences (178) were used as baits to search for the target sequence of M. truncatula. The sequences of SDR genes were gained from genome databases of Arabidopsis (TAIR, http://www.arabidopsis.org/, accessed on 1 April 2021) and M. truncatula (http://www.medicagohapmap.org/, accessed on 1 April 2021), respectively. The candidate SDR-encoding genes were chosen and annotated based on the closest A. thaliana homology. Three Pfam HMMs were used to select the genome set of predicted proteins: PF00106, PF01370, and PF01073 using HMMER3 (http://hmmer.org/, accessed on 5 April 2021) [25]. The decision rules for SDR inventory were employed as described by Moummou et al. [5]. The MtSDRs protein sequences were predicted using SMART (http://smart.embl-heidelberg.de/, accessed on 5 April 2021) [36] and InterPro (http://www.ebi.ac. uk/interpro/, accessed on 5 April 2021), while the redundant sequences were removed. The physicochemical properties of MtSDRs including protein sequence length (SL), molecular weight (MW), and isoelectric point (PI) were analyzed using the ExPasy online tool (http://web.expasy.org/protparam/, accessed on 5 April 2021).
4.2. Phylogenetic Analysis and Multiple Sequence Alignment
The multiple sequence alignment of the MtSDR domains was analyzed using ClustalW algorithm [37]. Phylogenetic analysis of SDRs in M. truncatula was conducted using MEGA-X with the maximum-likelihood (ML) method in 1000 bootstrap replicates [38].
4.3. Motif Analysis
MEME (http://meme.nbcr.net/meme/intro.html, accessed on 5 April 2021) was applied to analyze SDR conserved motifs [39]. The results were presented using TBtools software [40].
4.4. Chromosomal Distribution and Synteny Correlation Analysis
The circos diagram was illustrated using the genome annotation file for analysis of chromosomal locations of MtSDR genes. The physical position of the MtSDR genes on the chromosome was mapped using TBtools software. The synteny correlation analysis of SDR genes between the homologs in M. truncatula and A. thaliana or G. max were verified and visualized using TBtools software.
4.5. Analysis of Microarray Expression Profile
Microarray data on the expression profile of MtSDR genes in different tissues and responses to drought and salt were extracted from the MtGEA (https://mtgea.noble.org/v3/, accessed on 5 April 2021). The heatmap was drawn using the TBtools software.
4.6. Analysis of cis-Acting Regulatory Elements
The promoter sequences (2000 bp) upstream of the transcription start site of all MtSDRs were downloaded from the M. truncatula genome database and analyzed using PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 6 April 2021) [41].
4.7. Analysis of MtSDRs Expression in Response to Cold
M. truncatula (Jemalong) A17 seedlings were grown in a greenhouse (16 h light/8 h dark, 20 to 28 °C) for four weeks as previously described [42]. The plants were transferred to a growth chamber at 4 °C for 24 h under 12 h light for cold treatment. Leaves (0.1 g) were frozen in liquid nitrogen and used for extraction of total RNA and synthesis of cDNA using the TIANGEN total RNA Kit and TaKaRa PrimeScript RT kit with gDNA Eraser, respectively, as described by the manufacturer’s instructions. Primers were designed using the PrimerQuest tool (http://www.idtdna.com/Primerquest/Home/Index, accessed on 6 April 2021) and listed in Table S7. qRT-PCR was conducted using Thermal Cycler Dice Real Time System II (Takara, Japan). The MtActin7 (Medtr3g095530) gene was amplified as the internal control to normalize the amount of the template. Relative expression was calculated using the normalized (2−∆∆Ct) value.
4.8. Analysis of Subcellular Localization
The coding sequences of several MtSDRs without terminal codon were amplified and fused with GFP by cloning to vector pCAMBIA-1305, being driven by the CaMV 35S promoter. The constructed vector or empty vector as control was co-transformed with 35S::mCherry vector or 35S::AtAKT1-mCherry vector to the abaxial surface of 4-week-old N. benthamiana leaves using an Agrobacterium-mediated method. Fluorescence was observed 72 h after transformation using a confocal laser scanning microscope (Zeiss LSM800, Germany).
5. Conclusions
A total of 213 MtSDR proteins were identified from M. truncatula. They were classified into three primary groups (classical SDR fold, atypical members, and extended SDR fold). Segmental duplication was the crucial driving strength for MtSDRs evolution. In addition, MtSDR proteins have undergone strong purification pressures during their evolution. Above half of MtSDRs were expressed in at least one organ, and lots of MtSDRs had a preference for tissue expression. The cis-acting element responsive to plant hormones (salicylic acid, ABA, auxin, MeJA, and gibberellin) and stresses were found in the promoter of some MtSDRs. Many genes of MtSDR7C, MtSDR65C, MtSDR110C, MtSDR114C, and MtSDR108E families were responsive to drought, salt, and cold. The study provides useful information for further investigation on biological functions of MtSDRs, especially in abiotic stress adaptation, in the future.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22179498/s1.
Author Contributions
Z.G. and S.Y. designed and wrote the article; S.Y. and Q.S. performed the experiments; S.Y., P.Z., J.W. and Y.S. collected data and performed most of the data analysis. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number: 31971766).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kallberg, Y.; Oppermann, U.; Persson, B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models. FEBS J. 2010, 277, 2375–2386. [Google Scholar] [CrossRef]
- Jornvall, H.; Hedlund, J.; Bergman, T.; Oppermann, U.; Persson, B. Superfamilies SDR and MDR: From early ancestry to present forms. Emergence of three lines, a Zn-metalloenzyme, and distinct variabilities. Biochem. Biophys. Res. Commun. 2010, 396, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Filling, C.; Berndt, K.D.; Benach, J.; Knapp, S.; Prozorovski, T.; Nordling, E.; Ladenstein, R.; Jornvall, H.; Oppermann, U. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 2002, 277, 25677–25684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavanagh, K.; Jornvall, H.; Persson, B.; Oppermann, U. The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef] [Green Version]
- Moummou, H.; Kallberg, Y.; Tonfack, L.B.; Persson, B.; van der Rest, B. The Plant Short-Chain Dehydrogenase (SDR) superfamily: Genome-wide inventory and diversification patterns. BMC Plant Biol. 2012, 12, 219. [Google Scholar] [CrossRef] [Green Version]
- Kallberg, Y.; Oppermann, U.; Jornvall, H.; Persson, B. Short-chain dehydrogenase/reductase (SDR) relationships: A large family with eight clusters common to human, animal, and plant genomes. Protein Sci. 2002, 11, 636–641. [Google Scholar] [CrossRef] [Green Version]
- Beaudoin, F.; Wu, X.Z.; Li, F.L.; Haslam, R.P.; Markham, J.E.; Zheng, H.Q.; Napier, J.A.; Kunst, L. Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme a reductase candidates of the fatty acid elongase. Plant Physiol. 2009, 150, 1174–1191. [Google Scholar] [CrossRef] [Green Version]
- Ohlrogge, J.B.; Jaworski, J.G. Regulation of fatty acid synthesis. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 1997, 48, 109–136. [Google Scholar] [CrossRef] [Green Version]
- Paddock, T.N.; Mason, M.E.; Lima, D.F.; Armstrong, G.A. Arabidopsis protochlorophyllide oxidoreductase A (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol. Biol. 2010, 72, 445–457. [Google Scholar] [CrossRef]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Ringer, K.L.; Davis, E.M.; Croteau, R. Monoterpene metabolism. Cloning, expression and characterization of (−)-isopiperitenol/(−)-carveol dehydrogenase of Peppermint and Spearmint. Plant Physiol. 2005, 137, 863–872. [Google Scholar] [CrossRef] [Green Version]
- Rahier, A.; Bergdoll, M.; Genot, G.; Bouvier, F.; Camara, B. Homology modeling and site-directed mutagenesis reveal catalytic key amino acids of 3 beta-Hydroxysteroid-Dehydrogenase/C4-Decarboxylase from Arabidopsis. Plant Physiol. 2009, 149, 1872–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacombe, E.; Hawkins, S.; VanDoorsselaere, J.; Piquemal, J.; Goffner, D.; Poeydomenge, O.; Boudet, A.M.; GrimaPettenati, J. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships. Plant J. 1997, 11, 429–441. [Google Scholar] [CrossRef]
- Ziegler, J.; Voigtlander, S.; Schmidt, J.; Kramell, R.; Miersch, O.; Ammer, C.; Gesell, A.; Kutchan, T.M. Comparative transcript and alkaloid profiling in Papaver species identifies a short chain dehydrogenase/reductase involved in morphine biosynthesis. Plant J. 2006, 48, 177–192. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Apostolova, N.; Belles, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodriguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rook, F.; Corke, F.; Card, R.; Munz, G.; Smith, C.; Bevan, M.W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001, 26, 421–433. [Google Scholar] [CrossRef]
- Huijser, C.; Kortstee, A.; Pego, J.; Weisbeek, P.; Wisman, E.; Smeekens, S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 2000, 23, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.-G.; Lin, N.-C.; Hsiao, Y.-Y.; Kuo, C.-H.; Chang, P.-F.; Deng, W.-L.; Chiang, M.-H.; Shen, H.-L.; Chen, C.-Y.; Cheng, W.-H. The Arabidopsis short-chain dehydrogenase/reductase 3, an ABSCISIC ACID DEFICIENT 2 homolog, is involved in plant defense responses but not in ABA biosynthesis. Plant Physiol. Biochem. 2012, 51, 63–73. [Google Scholar] [CrossRef]
- Choi, H.W.; Lee, B.G.; Kim, N.H.; Park, Y.; Lim, C.W.; Song, H.K.; Hwang, B.K. A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol. 2008, 148, 383–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gepts, P.; Beavis, W.D.; Brummer, E.C.; Shoemaker, R.C.; Stalker, H.T.; Weeden, N.F.; Young, N.D. Legumes as a model plant family. Genomics for food and feed report of the cross-legume advances through genomics conference. Plant Physiol. 2005, 137, 1228–1235. [Google Scholar] [CrossRef] [Green Version]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valot, B.; Gianinazzi, S.; Eliane, D.G. Sub-cellular proteomic analysis of a Medicago truncatula root microsomal fraction. Phytochemistry 2004, 65, 1721–1732. [Google Scholar] [CrossRef]
- Young, N.D.; Udvardi, M. Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 2009, 12, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.D.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Min, T.P.; Kasahara, H.; Bedgar, D.L.; Youn, B.Y.; Lawrence, P.K.; Gang, D.R.; Halls, S.C.; Park, H.J.; Hilsenbeck, J.L.; Davin, L.B.; et al. Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases. J. Biol. Chem. 2003, 278, 50714–50723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, H.; Dixon, R.A.; Wang, X.Q. Crystal structure of vestitone reductase from alfalfa (Medicago sativa L.). J. Mol. Biol. 2007, 369, 265–276. [Google Scholar] [CrossRef]
- Wang, X.Q.; He, X.Z.; Lin, J.Q.; Shao, H.; Chang, Z.Z.; Dixon, R.A. Crystal structure of isoflavone reductase from alfalfa (Medicago sativa L.). J. Mol. Biol. 2006, 358, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.H.; Ebert, B.; Zhang, B.C.; Liu, H.B.; Zhang, Y.T.; Zeng, W.; Rautengarten, C.; Li, H.L.; Chen, X.Y.; Bacic, A.; et al. UDP-Api/UDP-Xyl synthases affect plant development by controlling the content of UDP-Api to regulate the RG-II-borate complex. Plant J. 2020, 104, 252–267. [Google Scholar] [CrossRef]
- Tonfack, L.; Moummou, H.; Latché, A.; Youmbi, E.; Benichou, M.; Pech, J.-C.; van der Rest, B. The plant SDR superfamily: Involvement in primary and secondary metabolism. Curr. Top. Plant Biol. 2011, 12, 41–53. [Google Scholar]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabiopsis thalinana. Plant Cell Physiol. 2005, 46, S89. [Google Scholar]
- Rahier, A.; Darnet, S.; Bouvier, F.; Camara, B.; Bard, M. Molecular and enzymatic characterizations of novel bifunctional 3 beta-hydroxysteroid dehydrogenases/C-4 decarboxylases from Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 27264–27277. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, W.-Z.; Song, L.-F.; Zou, J.-J.; Su, Z.; Wu, W.-H. Transcriptome analyses show Changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008, 148, 1201–1211. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Nadda, G.; Kumar, S.; Yadav, S.K. Transgenic Tobacco Overexpressing Tea cDNA Encoding Dihydroflavonol 4-Reductase and Anthocyanidin Reductase Induces Early Flowering and Provides Biotic Stress Tolerance. PLoS ONE 2013, 8, e65535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART 6: Recent updates and new developments. Nucleic Acids Res. 2009, 37, D229–D232. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yu, S.; Guo, Z. Calmodulin-Like (CML) Gene Family in Medicago truncatula: Genome-Wide Identification, Characterization and Expression Analysis. Int. J. Mol. Sci. 2020, 21, 7142. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).