Human IPSC-Derived Model to Study Myelin Disruption
Abstract
:1. Introduction
2. Results
2.1. Selection of Chemicals for Assay Development
2.1.1. Literature Review Results
2.1.2. Selection of Test Chemicals from Literature Review
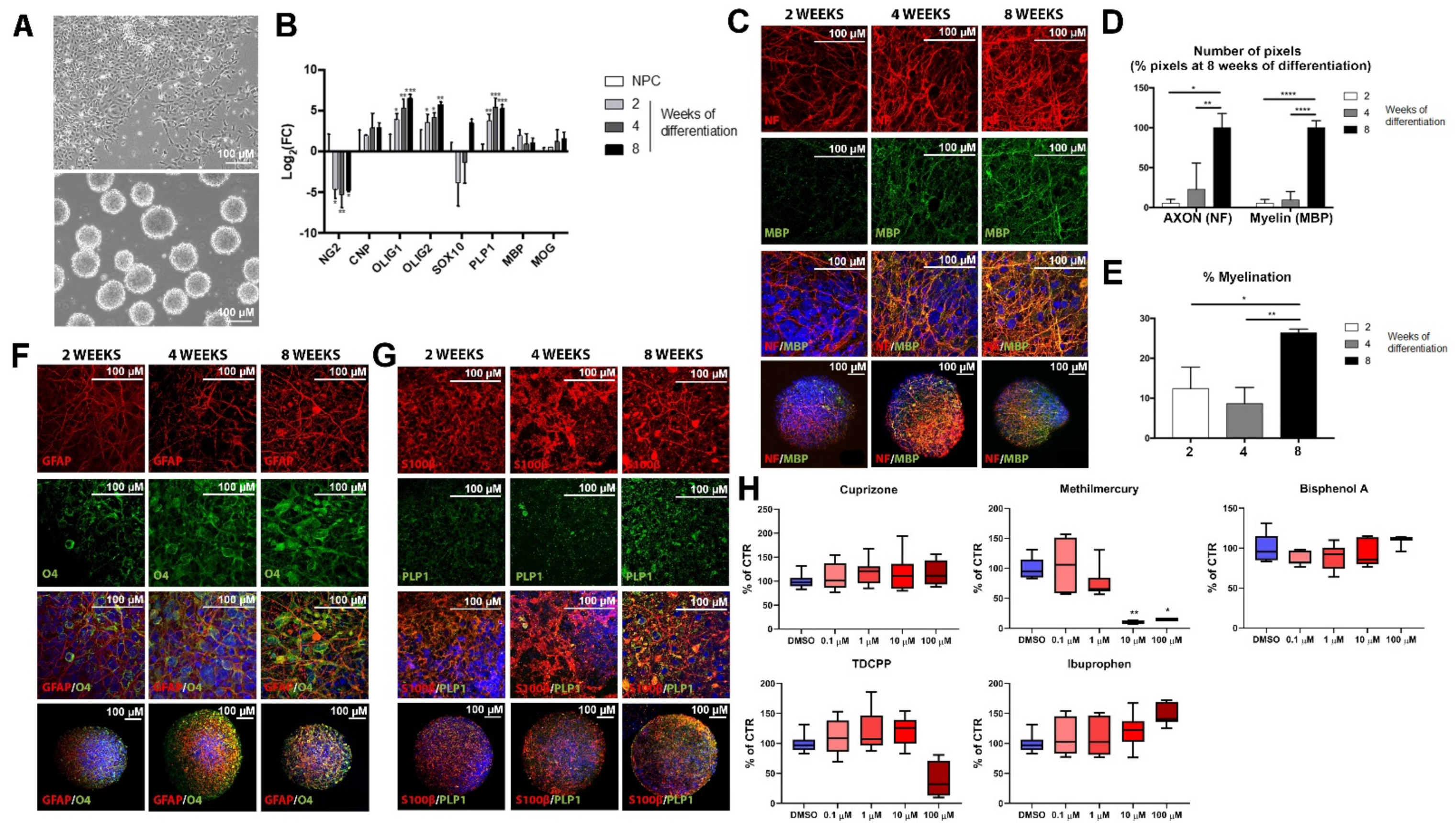
2.2. Maturation of Oligodendrocyte and Expression of Myelin-Related Markers during BrainSphere Differentiation
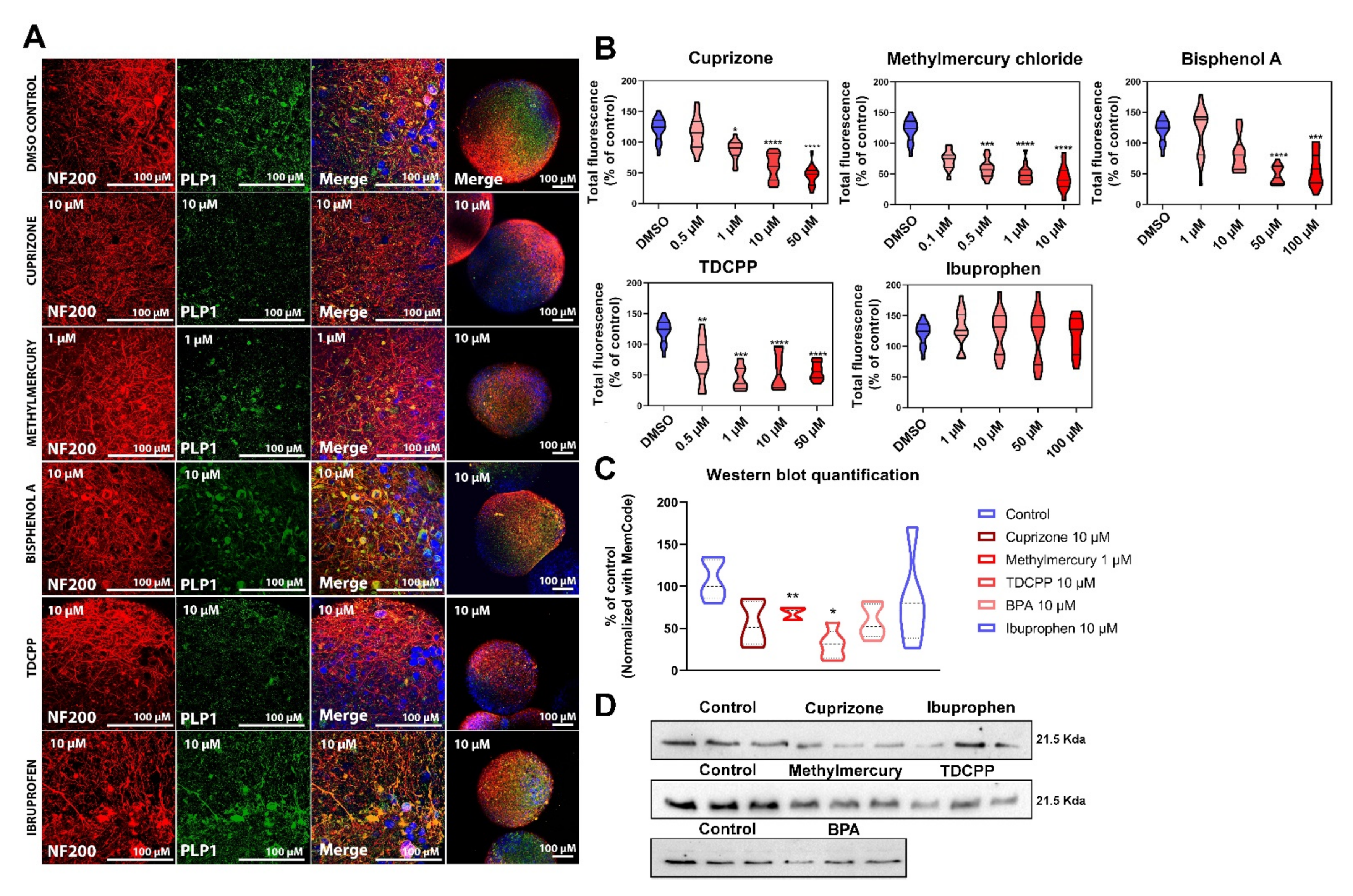
2.3. Exposure to DNT Selected Compounds Alter Myelin in BrainSpheres
2.3.1. Cytotoxicity Assessment of Test Chemicals
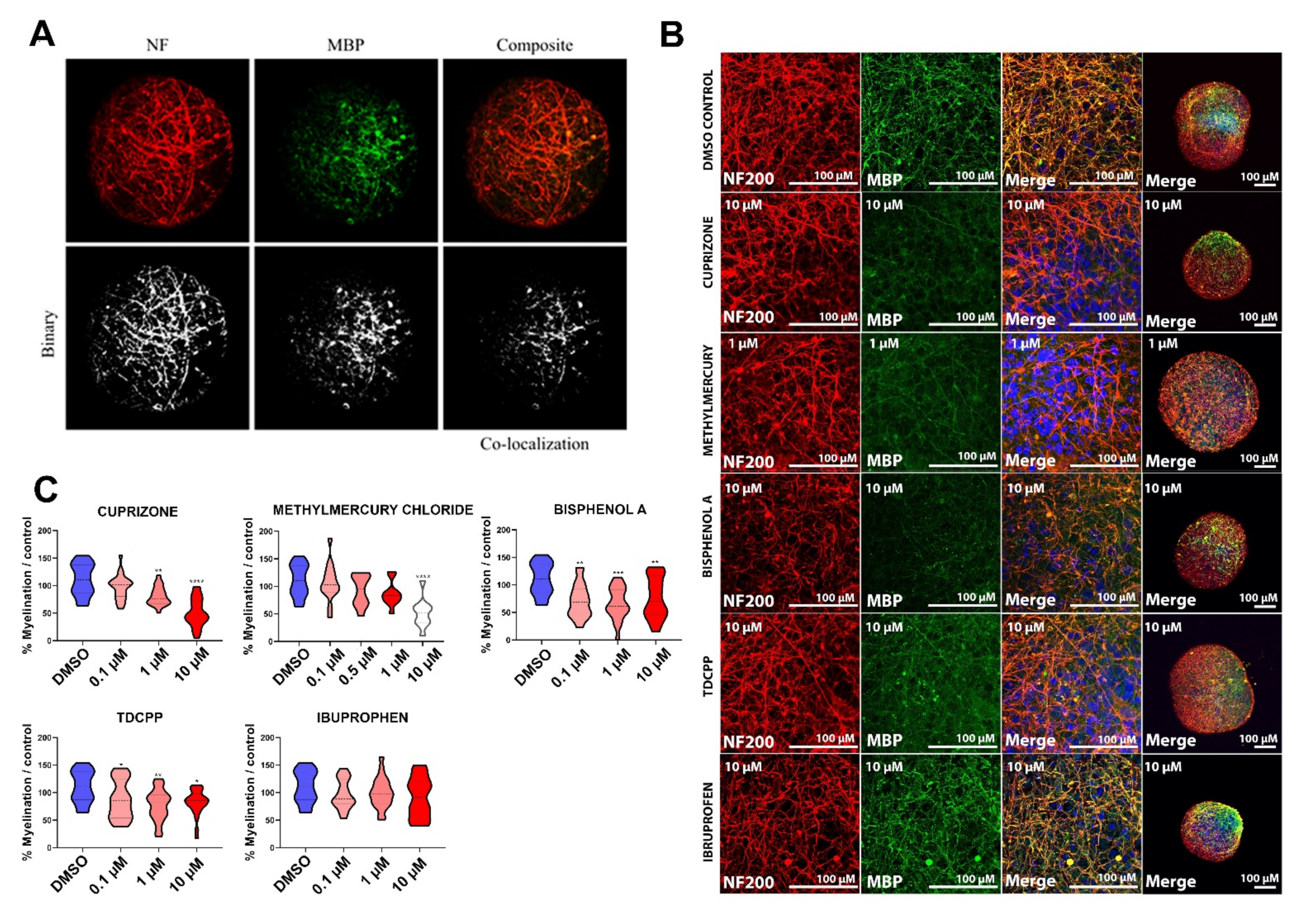
2.3.2. Myelin Is Affected after Exposure of BrainSpheres to Chemicals Inducing DNT
3. Discussion
4. Materials and Methods
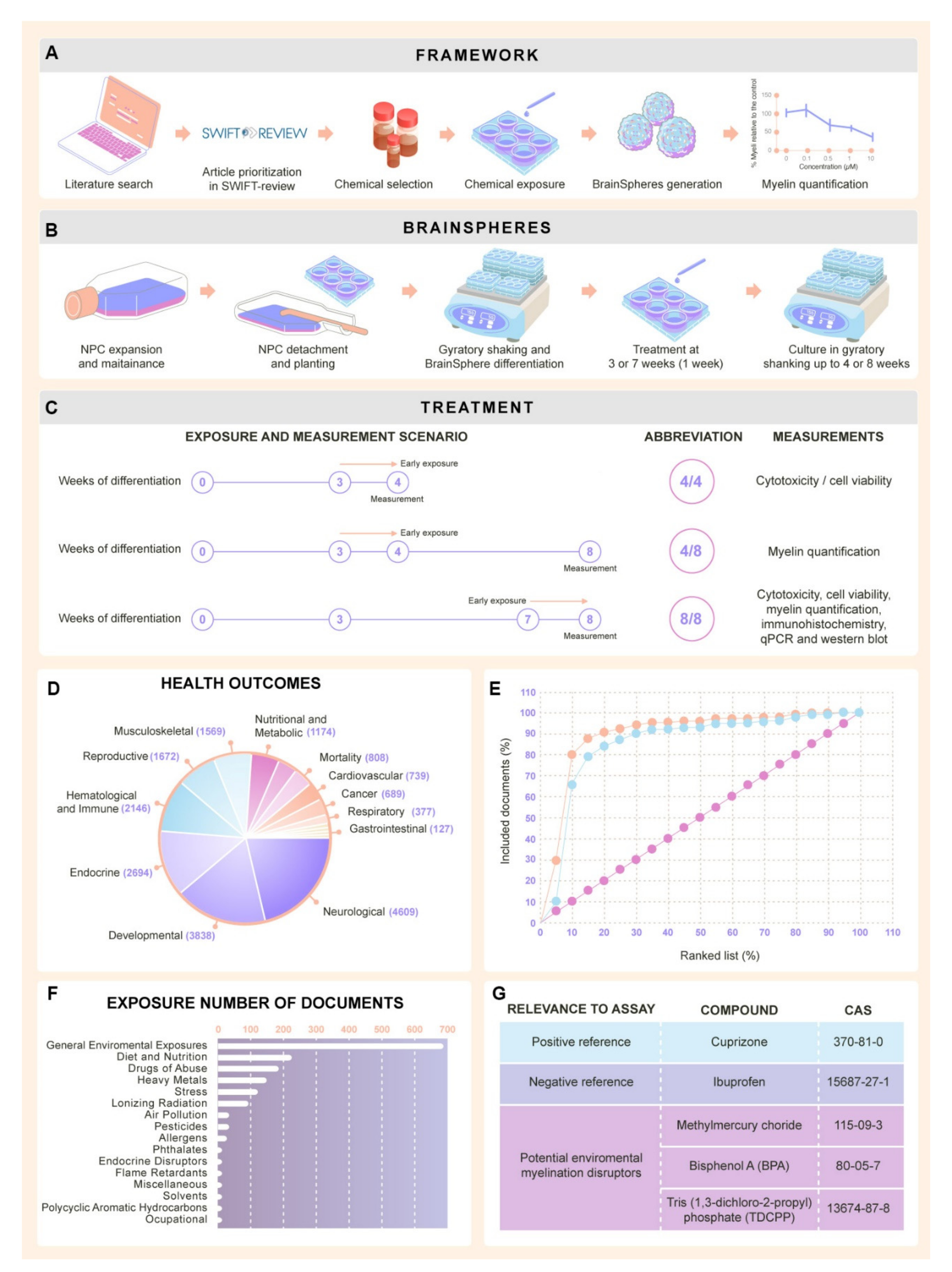
4.1. Literature Review to Identify Test Chemicals
Study Prioritization and Selection
4.2. Generation of Neural Progenitor Cells from Human Induced Pluripotent Stem Cells
4.3. Generation of the 3D Human Brain Model (BrainSpheres)
4.4. Chemical Exposure
4.5. Cell Viability Assessment
4.6. Immunocytochemical Staining and Confocal Imaging
4.7. Myelin Quantification
4.8. Gene Expression Analysis
4.9. Western Blot
4.10. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baron, W.; Hoekstra, D. On the biogenesis of myelin membranes: Sorting, trafficking and cell polarity. FEBS Lett. 2010, 584, 1760–1770. [Google Scholar] [CrossRef] [Green Version]
- Nave, K.A. Myelination and support of axonal integrity by glia. Nature 2010, 468, 244–252. [Google Scholar] [CrossRef]
- Fields, R.D. A new mechanism of nervous system plasticity: Activity-dependent myelination. Nat. Rev. Neurosci. 2015, 16, 756–767. [Google Scholar] [CrossRef]
- Bourbon-Teles, J.; Bells, S.; Jones, D.K.; Coulthard, E.; Rosser, A.; Metzler-Baddeley, C. Myelin Breakdown in Human Huntington’s Disease: Multi-Modal Evidence from Diffusion MRI and Quantitative Magnetization Transfer. Neuroscience 2019, 403, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Dean, D.C.; Sojkova, J.; Hurley, S.; Kecskemeti, S.; Okonkwo, O.; Bendlin, B.B.; Theisen, F.; Johnson, S.C.; Alexander, A.L.; Gallagher, C.L. Alterations of Myelin Content in Parkinson’s Disease: A Cross-Sectional Neuroimaging Study. PLoS ONE 2016, 11, e0163774. [Google Scholar] [CrossRef] [PubMed]
- Nasrabady, S.E.; Rizvi, B.; Goldman, J.E.; Brickman, A.M. White matter changes in Alzheimer’s disease: A focus on myelin and oligodendrocytes. Acta Neuropathol. Commun. 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Duncan, I.D.; Radcliff, A.B. Inherited and acquired disorders of myelin: The underlying myelin pathology. Exp. Neurol. 2016, 283, 452–475. [Google Scholar] [CrossRef]
- Barak, B.; Zhang, Z.C.; Liu, Y.Y.; Nir, A.; Trangle, S.S.; Ennis, M.; Levandowski, K.M.; Wang, D.Q.; Quast, K.; Boulting, G.L.; et al. Neuronal deletion of Gtf2i, associated with Williams syndrome, causes behavioral and myelin alterations rescuable by a remyelinating drug (vol 22, pg 700, 2019). Nat. Neurosci. 2019, 22, 1197. [Google Scholar] [CrossRef]
- Nir, A.; Barak, B. White matter alterations in Williams syndrome related to behavioral and motor impairments. Glia 2021, 69, 5–19. [Google Scholar] [CrossRef]
- Barateiro, A.; Fernandes, A. Temporal oligodendrocyte lineage progression: In vitro models of proliferation, differentiation and myelination. BBA-Mol. Cell Res. 2014, 1843, 1917–1929. [Google Scholar] [CrossRef] [Green Version]
- Chanoumidou, K.; Mozafari, S.; Baron-Van Evercooren, A.; Kuhlmann, T. Stem cell derived oligodendrocytes to study myelin diseases. Glia 2020, 68, 705–720. [Google Scholar] [CrossRef]
- Sim, F.J.; Windrem, M.S.; Goldman, S.A. Fate determination of adult human glial progenitor cells. Neuron Glia Biol. 2009, 5, 45–55. [Google Scholar] [CrossRef] [Green Version]
- de Monasterio-Schrader, P.; Jahn, O.; Tenzer, S.; Wichert, S.P.; Patzig, J.; Werner, H.B. Systematic approaches to central nervous system myelin. Cell Mol. Life Sci. 2012, 69, 2879–2894. [Google Scholar] [CrossRef] [PubMed]
- Meredith, R.M. Sensitive and critical periods during neurotypical and aberrant neurodevelopment: A framework for neurodevelopmental disorders. Neurosci. Biobehav. Rev. 2015, 50, 180–188. [Google Scholar] [CrossRef]
- Landrigan, P.J.; Goldman, L.R. Children’s Vulnerability To Toxic Chemicals: A Challenge and Opportunity to Strengthen Health and Environmental Policy. Health Aff. 2011, 30, 842–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- USEPA. America’s Children and the Environment (ACE), 3rd ed.; USEPA: Washington, DC, USA, 2013. [Google Scholar]
- Ferguson, A.; Penney, R.; Solo-Gabriele, H. A Review of the Field on Children’s Exposure to Environmental Contaminants: A Risk Assessment Approach. Int. J. Environ. Res. Public Health 2017, 14, 65. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Hogberg, H.T.; Leist, M.; Hartung, T. Developmental Neurotoxicity—Challenges in the 21st Century and In Vitro Opportunities. ALTEX Altern. Anim. Exp. 2014, 31, 129–156. [Google Scholar]
- Andersen, S.L. Trajectories of brain development: Point of vulnerability or window of opportunity? Neurosci. Biobehav. Rev. 2003, 27, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Grandjean, P.; Landrigan, P.J. Neurobehavioural effects of developmental toxicity. Lancet Neurol. 2014, 13, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Boyle, C.A.; Boulet, S.; Schieve, L.A.; Cohen, R.A.; Blumberg, S.J.; Yeargin-Allsopp, M.; Visser, S.; Kogan, M.D. Trends in the Prevalence of Developmental Disabilities in US Children, 1997–2008. Pediatrics 2011, 127, 1034–1042. [Google Scholar] [CrossRef] [Green Version]
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G.; et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef]
- Danielson, M.L.; Bitsko, R.H.; Ghandour, R.M.; Holbrook, J.R.; Kogan, M.D.; Blumberg, S.J. Prevalence of Parent-Reported ADHD Diagnosis and Associated Treatment Among U.S. Children and Adolescents, 2016. J. Clin. Child. Adolesc. Psychol. 2018, 47, 199–212. [Google Scholar] [CrossRef]
- Hansen, S.N.; Schendel, D.E.; Parner, E.T. Explaining the increase in the prevalence of autism spectrum disorders: The proportion attributable to changes in reporting practices. JAMA Pediatr. 2015, 169, 56–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visser, S.N.; Danielson, M.L.; Bitsko, R.H.; Holbrook, J.R.; Kogan, M.D.; Ghandour, R.M.; Perou, R.; Blumberg, S.J. Trends in the Parent-Report of Health Care Provider-Diagnosed and Medicated Attention-Deficit/Hyperactivity Disorder: United States, 2003–2011. J. Am. Acad. Child. Psychiatry 2014, 53, 34–46. [Google Scholar] [CrossRef] [Green Version]
- Ardhanareeswaran, K.; Mariani, J.; Coppola, G.; Abyzov, A.; Vaccarino, F.M. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat. Rev. Neurol. 2017, 13, 265–278. [Google Scholar] [CrossRef]
- Grandjean, P.; Landrigan, P.J. Developmental neurotoxicity of industrial chemicals. Lancet 2006, 368, 2167–2178. [Google Scholar] [CrossRef]
- Heyer, D.B.; Meredith, R.M. Environmental toxicology: Sensitive periods of development and neurodevelopmental disorders. Neurotoxicology 2017, 58, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Schettler, T. Toxic threats to neurologic development of children. Environ. Health Perspect. 2001, 109, 813–816. [Google Scholar] [CrossRef]
- Bondy, S.C.; Campbell, A. Developmental neurotoxicology. J. Neurosci. Res. 2005, 81, 605–612. [Google Scholar] [CrossRef] [Green Version]
- Lanphear, B.P.; Hornung, R.; Khoury, J.; Yolton, K.; Baghurstl, P.; Bellinger, D.C.; Canfield, R.L.; Dietrich, K.N.; Bornschein, R.; Greene, T.; et al. Low-level environmental lead exposure and children’s intellectual function: An international pooled analysis. Environ. Health Perspect. 2005, 113, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Kadereit, S.; Zimmer, B.; van Thriel, C.; Hengstler, J.G.; Leist, M. Compound selection for in vitro modeling of developmental neurotoxicity. Front. Biosci-Landmark 2012, 17, 2442–2460. [Google Scholar] [CrossRef] [Green Version]
- Makris, S.L.; Raffaele, K.; Allen, S.; Bowers, W.J.; Hass, U.; Alleva, E.; Calamandrei, G.; Sheets, L.; Amcoff, P.; Delrue, N.; et al. A Retrospective Performance Assessment of the Developmental Neurotoxicity Study in Support of OECD Test Guideline 426. Environ. Health Perspect 2009, 117, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bal-Price, A.; Crofton, K.M.; Leist, M.; Allen, S.; Arand, M.; Buetler, T.; Delrue, N.; FitzGerald, R.E.; Hartung, T.; Heinonen, T.; et al. International STakeholder NETwork (ISTNET): Creating a developmental neurotoxicity (DNT) testing road map for regulatory purposes. Arch. Toxicol. 2015, 89, 269–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crofton, K.M.; Mundy, W.R.; Shafer, T.J. Developmental neurotoxicity testing: A path forward. Congenit. Anom. 2012, 52, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Zhang, Q.; Carmichael, P.L.; Boekelheide, K.; Andersen, M.E. Toxicity Testing in the 21(st) Century: Defining New Risk Assessment Approaches Based on Perturbation of Intracellular Toxicity Pathways. PLoS ONE 2011, 6, e20887. [Google Scholar] [CrossRef]
- Leist, M.; Hartung, T.; Nicotera, P. The dawning of a new age of toxicology. ALTEX-Altern. Tierexp. 2008, 25, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Hartung, T.; Leist, M. Food for thought ... on the evolution of toxicology and the phasing out of animal testing. ALTEX 2008, 25, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Hartung, T.; McBride, M. Food for Thought ... on mapping the human toxome. ALTEX 2011, 28, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Leist, M.; Hasiwa, N.; Rovida, C.; Daneshian, M.; Basketter, D.; Kimber, I.; Clewell, H.; Gocht, T.; Goldberg, A.; Busquet, F.; et al. Consensus Report on the Future of Animal-Free Systemic Toxicity Testing. ALTEX-Altern. Anim. Exp. 2014, 31, 341–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, S.A.; Altman, J.; Russo, R.J.; Zhang, X. Timetables of Neurogenesis in the Human Brain Based on Experimentally Determined Patterns in the Rat. Neurotoxicology 1993, 14, 83–144. [Google Scholar]
- Rice, D.; Barone, S., Jr. Critical periods of vulnerability for the developing nervous system: Evidence from humans and animal models. Environ. Health Perspect. 2000, 108 (Suppl. 3), 511–533. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, E.; Alm, H.; Baumann, J.; Geerts, L.; Håkansson, H.; Masjosthusmann, S.; Witters, H. Literature review on in vitro and alternative Developmental Neurotoxicity (DNT) testing methods. EFSA Support. Publ. 2015, 12, 778E. [Google Scholar] [CrossRef]
- Chan, J.R.; Watkins, T.A.; Cosgaya, J.M.; Zhang, C.Z.; Chen, L.; Reichardt, L.F.; Shooter, E.M.; Barres, B.A. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron 2004, 43, 183–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lariosa-Willingham, K.D.; Rosler, E.S.; Tung, J.S.; Dugas, J.C.; Collins, T.L.; Leonoudakis, D. Development of a central nervous system axonal myelination assay for high throughput screening. BMC Neurosci. 2016, 17, 16. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Colognato, H.; Ffrench-Constant, C. Contrasting effects of mitogenic growth factors on myelination in neuron-oligodendrocyte co-cultures. Glia 2007, 55, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Zheng, B.; Kimberly, S.L.; Cai, Z.; Rhodes, P.G.; Lin, R.C. Neuron-oligodendrocyte myelination co-culture derived from embryonic rat spinal cord and cerebral cortex. Brain Behav. 2012, 2, 53–67. [Google Scholar] [CrossRef]
- Ristola, M.; Sukki, L.; Azevedo, M.M.; Seixas, A.I.; Relvas, J.B.; Narkilahti, S.; Kallio, P. A compartmentalized neuron-oligodendrocyte co-culture device for myelin research: Design, fabrication and functionality testing. J. Micromech. Microeng. 2019, 29, 065009. [Google Scholar] [CrossRef]
- Clark, A.J.; Kaller, M.S.; Galino, J.; Willison, H.J.; Rinaldi, S.; Bennett, D.L.H. Co-cultures with stem cell-derived human sensory neurons reveal regulators of peripheral myelination. Brain 2017, 140, 898–913. [Google Scholar] [CrossRef] [Green Version]
- Barateiro, A.; Domingues, H.S.; Fernandes, A.; Relvas, J.B.; Brites, D. Rat Cerebellar Slice Cultures Exposed to Bilirubin Evidence Reactive Gliosis, Excitotoxicity and Impaired Myelinogenesis that Is Prevented by AMPA and TNF-alpha Inhibitors. Mol. Neurobiol. 2014, 49, 424–439. [Google Scholar] [CrossRef]
- Hill, R.A.; Medved, J.; Patel, K.D.; Nishiyama, A. Organotypic Slice Cultures to Study Oligodendrocyte Dynamics and Myelination. JoVE-J. Vis. Exp. 2014, 90, e51835. [Google Scholar] [CrossRef] [Green Version]
- Miron, V.E.; Ludwin, S.K.; Darlington, P.J.; Jarjour, A.A.; Soliven, B.; Kennedy, T.E.; Antel, J.P. Fingolimod (FTY720) Enhances Remyelination Following Demyelination of Organotypic Cerebellar Slices. Am. J. Pathol. 2010, 176, 2682–2694. [Google Scholar] [CrossRef]
- Zurich, M.G.; Stanzel, S.; Kopp-Schneider, A.; Prieto, P.; Honegger, P. Evaluation of aggregating brain cell cultures for the detection of acute organ-specific toxicity. Toxicol. In Vitro 2013, 27, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Zurich, M.G.; Honegger, P.; Schilter, B.; Costa, L.G.; Monnet-Tschudi, F. Use of aggregating brain cell cultures to study developmental effects of organophosphorus insecticides. Neurotoxicology 2000, 21, 599–605. [Google Scholar] [PubMed]
- Honegger, P.; Zurich, M.G. Preparation and Use of Serum-Free Aggregating Brain Cell Cultures for Routine Neurotoxicity Screening. Neuromethods 2011, 56, 105–128. [Google Scholar] [CrossRef]
- Guentert-Lauber, B.; Monnet-Tschudi, F.; Omlin, F.X.; Favrod, P.; Honegger, P. Serum-free aggregate cultures of rat CNS glial cells: Biochemical, immunocytochemical and morphological characterization. Dev. Neurosci. 1985, 7, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Kerman, B.E.; Kim, H.J.; Padmanabhan, K.; Mei, A.; Georges, S.; Joens, M.S.; Fitzpatrick, J.A.J.; Jappelli, R.; Chandross, K.J.; August, P.; et al. In vitro myelin formation using embryonic stem cells. Development 2015, 142, 2213–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chesnut, M.; Hartung, T.; Hogberg, H.; Pamies, D. Human Oligodendrocytes and Myelin In Vitro to Evaluate Developmental Neurotoxicity. Int. J. Mol. Sci. 2021, 22, 7929. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, M.; Nevin, Z.S.; Shick, H.E.; Garrison, E.; Clarkson-Paredes, C.; Karl, M.; Clayton, B.L.L.; Factor, D.C.; Allan, K.C.; Barbar, L.; et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods 2018, 15, 700–706. [Google Scholar] [CrossRef]
- Kim, H.; Xu, R.; Padmashri, R.; Dunaevsky, A.; Liu, Y.; Dreyfus, C.F.; Jiang, P. Pluripotent Stem Cell-Derived Cerebral Organoids Reveal Human Oligodendrogenesis with Dorsal and Ventral Origins. Stem Cell Rep. 2019, 12, 890–905. [Google Scholar] [CrossRef] [Green Version]
- Pamies, D.; Barreras, P.; Block, K.; Makri, G.; Kumar, A.; Wiersma, D.; Smirnova, L.; Zhang, C.; Bressler, J.; Christian, K.M.; et al. A Human Brain Microphysiological System Derived from Induced Pluripotent Stem Cells to Study Neurological Diseases and Toxicity. ALTEX-Altern. Anim. Exp. 2017, 34, 362–376. [Google Scholar] [CrossRef] [Green Version]
- Marton, R.M.; Miura, Y.; Sloan, S.A.; Li, Q.Y.; Revah, O.; Levy, R.J.; Huguenard, J.R.; Pasca, S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019, 22, 484–491. [Google Scholar] [CrossRef] [PubMed]
- James, O.G.; Selvaraj, B.T.; Magnani, D.; Burr, K.; Connick, P.; Barton, S.K.; Vasistha, N.A.; Hampton, D.W.; Story, D.; Smigiel, R.; et al. iPSC-derived myelinoids to study myelin biology of humans. Dev. Cell 2021, 56, 1346–1358.e1346. [Google Scholar] [CrossRef] [PubMed]
- Hogberg, H.T.; Bressler, J.; Christian, K.M.; Harris, G.; Makri, G.; O’Driscoll, C.; Pamies, D.; Smirnova, L.; Wen, Z.X.; Hartung, T. Toward a 3D model of human brain development for studying gene/environment interactions. Stem Cell Res. Ther. 2013, 4, S4. [Google Scholar] [CrossRef] [Green Version]
- Plummer, S.; Wallace, S.; Ball, G.; Lloyd, R.; Schiapparelli, P.; Quinones-Hinojosa, A.; Hartung, T.; Pamies, D. A Human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine. Sci. Rep. UK 2019, 9, 1407. [Google Scholar] [CrossRef]
- Zhou, Q.J.; Nino, D.F.; Yamaguchi, Y.; Wang, S.X.; Fulton, W.B.; Jia, H.P.; Lu, P.; Prindle, T.; Pamies, D.; Morris, M.; et al. Necrotizing enterocolitis induces T lymphocyte-mediated injury in the developing mammalian brain. Sci. Transl. Med. 2021, 13, eaay6621. [Google Scholar] [CrossRef]
- Leite, P.E.C.; Pereira, M.R.; Harris, G.; Pamies, D.; dos Santos, L.M.G.; Granjeiro, J.M.; Hogberg, H.T.; Hartung, T.; Smirnova, L. Suitability of 3D human brain spheroid models to distinguish toxic effects of gold and poly-lactic acid nanoparticles to assess biocompatibility for brain drug delivery. Part. Fibre Toxicol. 2019, 16, 22. [Google Scholar] [CrossRef]
- Pamies, D.; Block, K.; Lau, P.; Gribaldo, L.; Pardo, C.A.; Barreras, P.; Smirnova, L.; Wiersma, D.; Zhao, L.; Harris, G.; et al. Rotenone exerts developmental neurotoxicity in a human brain spheroid model. Toxicol. Appl. Pharm. 2018, 354, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Harris, G.; Smirnova, L.; Zufferey, V.; Sa, R.; Baldino Russo, F.; Baleeiro Beltrao Braga, P.C.; Chesnut, M.; Zurich, M.G.; Hogberg, H.T.; et al. Antidepressant Paroxetine Exerts Developmental Neurotoxicity in an iPSC-Derived 3D Human Brain Model. Front. Cell Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Cammer, W. The neurotoxicant, cuprizone, retards the differentiation of oligodendrocytes in vitro. J. Neurol. Sci. 1999, 168, 116–120. [Google Scholar] [CrossRef]
- Gospe, S.M.; Zhou, S.S. Toluene abuse embryopathy: Longitudinal neurodevelopmental effects of prenatal exposure to toluene in rats. Reprod. Toxicol. 1998, 12, 119–126. [Google Scholar] [CrossRef]
- Tiwari, S.K.; Agarwal, S.; Chauhan, L.K.; Mishra, V.N.; Chaturvedi, R.K. Bisphenol-A impairs myelination potential during development in the hippocampus of the rat brain. Mol. Neurobiol. 2015, 51, 1395–1416. [Google Scholar] [CrossRef]
- Xu, X.B.; Fan, S.J.; He, Y.; Ke, X.; Song, C.; Xiao, Y.; Zhang, W.H.; Zhang, J.Y.; Yin, X.P.; Kato, N.; et al. Loss of Hippocampal Oligodendrocytes Contributes to the Deficit of Contextual Fear Learning in Adult Rats Experiencing Early Bisphenol A Exposure. Mol. Neurobiol. 2017, 54, 4524–4536. [Google Scholar] [CrossRef]
- Dach, K.; Bendt, F.; Huebenthal, U.; Giersiefer, S.; Lein, P.J.; Heuer, H.; Fritsche, E. BDE-99 impairs differentiation of human and mouse NPCs into the oligodendroglial lineage by speciesspecific modes of action. Sci Rep. UK 2017, 7, 4861. [Google Scholar] [CrossRef]
- Schreiber, T.; Gassmann, K.; Gotz, C.; Hubenthal, U.; Moors, M.; Krause, G.; Merk, H.F.; Nguyen, N.H.; Scanlan, T.S.; Abel, J.; et al. Polybrominated Diphenyl Ethers Induce Developmental Neurotoxicity in a Human in Vitro Model: Evidence for Endocrine Disruption. Environ. Health Perspect. 2010, 118, 572–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bichenkov, E.; Ellingson, J.S. Ethanol exerts different effects on myelin basic protein and 2′,3′-cyclic nucleotide 3′-phosphodiesterase expression in differentiating CG-4 oligodendrocytes. Brain Res. Dev. Brain Res. 2001, 128, 9–16. [Google Scholar] [CrossRef]
- Miller, M.W.; al-Rabiai, S. Effects of prenatal exposure to ethanol on the number of axons in the pyramidal tract of the rat. Alcohol. Clin. Exp. Res. 1994, 18, 346–354. [Google Scholar] [CrossRef]
- Roskam, S.; Koch, M. Effects of neonatal and peripubertal ethanol treatment on various aspects of adult rat behavior and brain anatomy. Int. J. Dev. Neurosci. 2009, 27, 249–256. [Google Scholar] [CrossRef]
- Zoeller, R.T.; Butnariu, O.V.; Fletcher, D.L.; Riley, E.P. Limited postnatal ethanol exposure permanently alters the expression of mRNAS encoding myelin basic protein and myelin-associated glycoprotein in cerebellum. Alcohol. Clin. Exp. Res. 1994, 18, 909–916. [Google Scholar] [CrossRef]
- Padhi, B.K.; Pelletier, G. Perturbation of myelin basic protein (Mbp) splice variant expression in developing rat cerebellum following perinatal exposure to methylmercury. Toxicol. Lett. 2012, 213, 374–380. [Google Scholar] [CrossRef]
- Wang, Q.; Lai, N.L.; Wang, X.; Guo, Y.; Lam, P.K.; Lam, J.C.; Zhou, B. Bioconcentration and transfer of the organophorous flame retardant 1,3-dichloro-2-propyl phosphate causes thyroid endocrine disruption and developmental neurotoxicity in zebrafish larvae. Environ. Sci. Technol. 2015, 49, 5123–5132. [Google Scholar] [CrossRef]
- Azeez, I.A.; Olopade, F.; Laperchia, C.; Andrioli, A.; Scambi, I.; Onwuka, S.K.; Bentivoglio, M.; Olopade, J.O. Regional Myelin and Axon Damage and Neuroinflammation in the Adult Mouse Brain After Long-Term Postnatal Vanadium Exposure. J. Neuropathol. Exp. Neurol. 2016, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Soazo, M.; Garcia, G.B. Vanadium exposure through lactation produces behavioral alterations and CNS myelin deficit in neonatal rats. Neurotoxicol. Teratol. 2007, 29, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Todorich, B.; Olopade, J.O.; Surguladze, N.; Zhang, X.; Neely, E.; Connor, J.R. The mechanism of vanadium-mediated developmental hypomyelination is related to destruction of oligodendrocyte progenitors through a relationship with ferritin and iron. Neurotox. Res. 2011, 19, 361–373. [Google Scholar] [CrossRef]
- Deng, W.; McKinnon, R.D.; Poretz, R.D. Lead exposure delays the differentiation of oligodendroglial progenitors in vitro. Toxicol. Appl. Pharmacol. 2001, 174, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Schneider, J.S. Effects of lead exposure on proliferation and differentiation of neural stem cells derived from different regions of embryonic rat brain. Neurotoxicology 2004, 25, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Torkildsen, O.; Brunborg, L.A.; Myhr, K.M.; Bo, L. The cuprizone model for demyelination. Acta Neurol. Scand. 2008, 117, 72–76. [Google Scholar] [CrossRef]
- Matsushima, G.K.; Morell, P. The neurotoxicant, cuprizone, as a model to study demyelination and remyelination in the central nervous system. Brain Pathol. 2001, 11, 107–116. [Google Scholar] [CrossRef]
- Venturini, G. Enzymic activities and sodium, potassium and copper concentrations in mouse brain and liver after cuprizone treatment in vivo. J. Neurochem. 1973, 21, 1147–1151. [Google Scholar] [CrossRef]
- Stapleton, H.M.; Klosterhaus, S.; Eagle, S.; Fuh, J.; Meeker, J.D.; Blum, A.; Webster, T.F. Detection of organophosphate flame retardants in furniture foam and U.S. house dust. Environ. Sci. Technol. 2009, 43, 7490–7495. [Google Scholar] [CrossRef] [Green Version]
- Dishaw, L.V.; Hunter, D.L.; Padnos, B.; Padilla, S.; Stapleton, H.M. Developmental exposure to organophosphate flame retardants elicits overt toxicity and alters behavior in early life stage zebrafish (Danio rerio). Toxicol. Sci. Off. J. Soc. Toxicol. 2014, 142, 445–454. [Google Scholar] [CrossRef] [Green Version]
- Aschner, M.; Ceccatelli, S.; Daneshian, M.; Fritsche, E.; Hasiwa, N.; Hartung, T.; Hogberg, H.T.; Leist, M.; Li, A.; Mundi, W.R.; et al. Reference compounds for alternative test methods to indicate developmental neurotoxicity (DNT) potential of chemicals: Example lists and criteria for their selection and use. ALTEX 2017, 34, 49–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parepally, J.M.; Mandula, H.; Smith, Q.R. Brain uptake of nonsteroidal anti-inflammatory drugs: Ibuprofen, flurbiprofen, and indomethacin. Pharm. Res. 2006, 23, 873–881. [Google Scholar] [CrossRef]
- Sandstrom, J.; Eggermann, E.; Charvet, I.; Roux, A.; Toni, N.; Greggio, C.; Broyer, A.; Monnet-Tschudi, F.; Stoppini, L. Development and characterization of a human embryonic stem cell-derived 3D neural tissue model for neurotoxicity testing. Toxicol In Vitro 2017, 38, 124–135. [Google Scholar] [CrossRef]
- Fancy, S.P.; Chan, J.R.; Baranzini, S.E.; Franklin, R.J.; Rowitch, D.H. Myelin regeneration: A recapitulation of development? Annu. Rev. Neurosci. 2011, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Holzhütter, H.; Archer, G.; Dami, N.; Lovell, D.P.; Saltelli, A.; Sjostrom, M. Recommendations for the Application of Biostatistical Methods During the Development and Validation of Alternative Toxicological Methods: ECVAM Biostatistics Task Force Report 1. ATLA 1996, 24, 511–530. [Google Scholar] [CrossRef]
- Chiang, C.H.; Su, Y.; Wen, Z.; Yoritomo, N.; Ross, C.A.; Margolis, R.L.; Song, H.; Ming, G.I. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol. Psychiatr. 2011, 16, 358–360. [Google Scholar] [CrossRef]
- Wen, Z.X.; Nguyen, H.N.; Guo, Z.Y.; Lalli, M.A.; Wang, X.Y.; Su, Y.J.; Kim, N.S.; Yoon, K.J.; Shin, J.; Zhang, C.; et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014, 515, 414. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| String | Combined with OR | Combined with OR | Combined with OR | ||
|---|---|---|---|---|---|
| 1 | Embryonic and fetal development, embryonic structures, embryonic, embryo, embryos, embryology, fetal, fetus, pregnancy, gestation, gestational, in utero, prenatal neonatal, neonate, perinatal, postnatal, infant, adolescent, child, fetal brain, human development, developing brain, neurodevelopmental, neurodevelopment | AND | Pharmacological and toxicological phenomena and processes, toxicology, toxicity, toxicity tests, toxicant, toxicants, toxin, toxins, toxic actions, neurotoxicant, neurotoxicants, neurotoxin, neurotoxins, neurotoxins, pharmacology, pharmacologic actions, specialty uses of chemicals, organic chemicals, inorganic chemicals, environment and public health, exposure, exposures, environmental chemical, environmental chemicals, environmental health, hazard, hazards, hazardous, xenobiotics | AND | Myelin, oligodendroglia, myelin sheath, myelinogenesis, myelination, myelin proteins, oligodendrocyte, oligodendrocytes, oligodendrogenesis, white matter, dysmyelination, dysmyelinating, demyelination, demyelinating |
| 2 | Developmental neurotoxicant, developmental neurotoxicants, developmental neurotoxin, developmental neurotoxin, developmental neurotoxicity, neurodevelopmental toxicity, neurodevelopmental disorder, prenatal injuries, maternal exposure, teratogen, teratogens, teratogenic | ||||
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| DNT is investigated with appropriate exposure scenarios (e.g., prenatal maternal, infant, or childhood exposure (in vivo), exposure during cell proliferation, differentiation, migration, myelination, or synaptogenesis (in vitro)) | DNT is not investigated |
| A single chemical exposure is reported with a clearly identified chemical name or CAS number, or a chemical mixture is reported with human relevance | A chemical exposure is not reported, a mixture of chemicals is reported without human relevance, chemicals are not clearly identified, or exposures are psychosocial (e.g., stress or socioeconomic status), physical (e.g., radiation, particulate matter, or nanomaterials), or intrinsic biological traits (e.g., genetic mutations) |
| At least two dose or concentration levels are tested or a single dose is tested but was chosen based on previous experience with multiple doses or on human exposure levels | One dose or concentration is tested but was not chosen based on previous experience with multiple doses or on human exposure levels |
| DNT was evident at doses or concentrations lower than those which cause maternal toxicity (in vivo), general toxicity (in vivo), or cytotoxicity (in vitro) | The relationship between DNT and other forms of toxicity were not described or DNT was only evident at doses that also caused maternal toxicity (in vivo), general toxicity (in vivo), or cytotoxicity (in vitro) |
| A chemical was characterized as a developmental neurotoxicant using endpoints associated with myelination in the central nervous system (e.g., markers or levels of oligodendrocyte differentiation or MBP gene expression), or a chemical was tested using endpoints associated with myelination during neurodevelopment but found to have no effect | A chemical was not characterized as a developmental neurotoxicant or was characterized using endpoints not associated with myelination, or only with peripheral nervous system myelination |
| Studies with appropriate negative and solvent controls or control groups | Studies without appropriate negative and solvent controls or control groups |
| Name | Abbreviation | Brand | Reference |
|---|---|---|---|
| Neurofilament 200 | NF | Sigma | N4142 |
| Myelin basic protein | MBP | BioLegend | 808402 |
| Proteolipid protein 1 | PLP1 | Biorad | MCA839G |
| Oligodendrocyte marker 4 | O4 | R&D systems | MAB1326 |
| Glial fibrillary acidic protein | GFAP * | Sigma | G9269 |
| S100 calcium-binding protein B | S100B * | Abcam | ab52642 |
| Gene Name | Abbreviation | Taqman® Assay ID |
|---|---|---|
| Neural-glial antigen 2 (chondroitin sulfate proteoglycan 4) | NG2 (CSPG4) | Hs00361541_g1 |
| Oligodendrocyte transcription factor 1 | OLIG1 | Hs00744293_s1 |
| Oligodendrocyte transcription factor 2 | OLIG2 | Hs00300164_s1 |
| SOX-10 transcription factor | SOX10 | Hs00366918_m1 |
| Adenomatous polyposis coli | APC | Hs01568269_m1 |
| Proteolipid protein 1 | PLP1 | Hs00166914_m1 |
| 2′,3′-Cyclic-nucleotide 3′-phosphodiesterase | CNP | Hs00263981_m1 |
| Myelin oligodendrocyte glycoprotein | MOG | Hs01555268_m1 |
| Myelin basic protein | MBP | Hs00921945_m1 |
| Housekeeping Genes | Abbreviation | Taqman® Assay ID |
| β-actin | ACTB | Hs01060665_g1 |
| 18S | 18S | Hs999999_01 |
| Glyceraldehyde 3-phosphate dehydrogenase | GAPDH | Hs02786624_g1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chesnut, M.; Paschoud, H.; Repond, C.; Smirnova, L.; Hartung, T.; Zurich, M.-G.; Hogberg, H.T.; Pamies, D. Human IPSC-Derived Model to Study Myelin Disruption. Int. J. Mol. Sci. 2021, 22, 9473. https://doi.org/10.3390/ijms22179473
Chesnut M, Paschoud H, Repond C, Smirnova L, Hartung T, Zurich M-G, Hogberg HT, Pamies D. Human IPSC-Derived Model to Study Myelin Disruption. International Journal of Molecular Sciences. 2021; 22(17):9473. https://doi.org/10.3390/ijms22179473
Chicago/Turabian StyleChesnut, Megan, Hélène Paschoud, Cendrine Repond, Lena Smirnova, Thomas Hartung, Marie-Gabrielle Zurich, Helena T. Hogberg, and David Pamies. 2021. "Human IPSC-Derived Model to Study Myelin Disruption" International Journal of Molecular Sciences 22, no. 17: 9473. https://doi.org/10.3390/ijms22179473
APA StyleChesnut, M., Paschoud, H., Repond, C., Smirnova, L., Hartung, T., Zurich, M.-G., Hogberg, H. T., & Pamies, D. (2021). Human IPSC-Derived Model to Study Myelin Disruption. International Journal of Molecular Sciences, 22(17), 9473. https://doi.org/10.3390/ijms22179473