Drebrin Regulates Acetylcholine Receptor Clustering and Organization of Microtubules at the Postsynaptic Machinery
Abstract
1. Introduction
2. Results
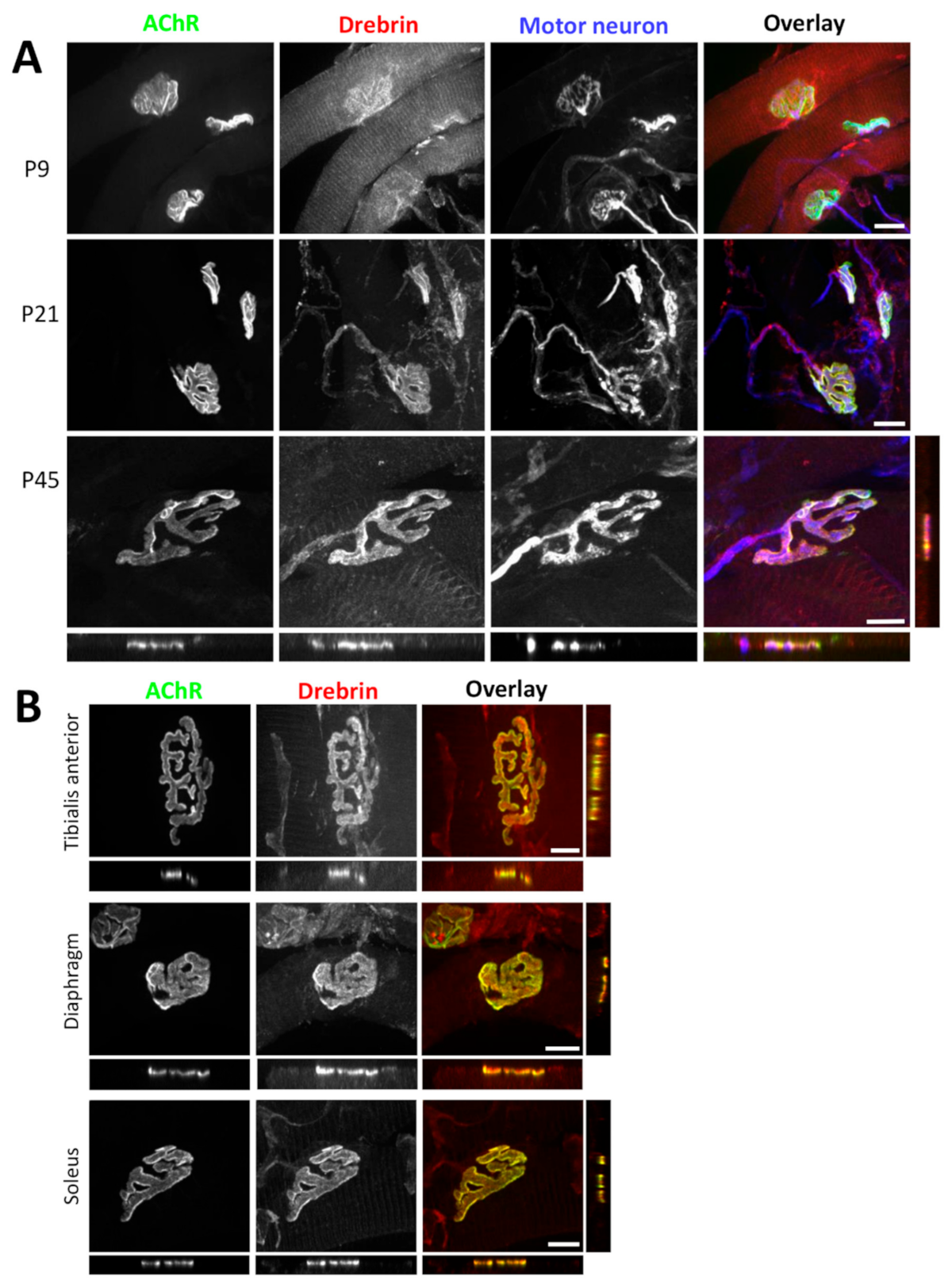
2.1. Drebrin Localizes to the AChR Compartment of Neuromuscular Junctions and to the Contractile Machinery of Muscle Sarcomeres
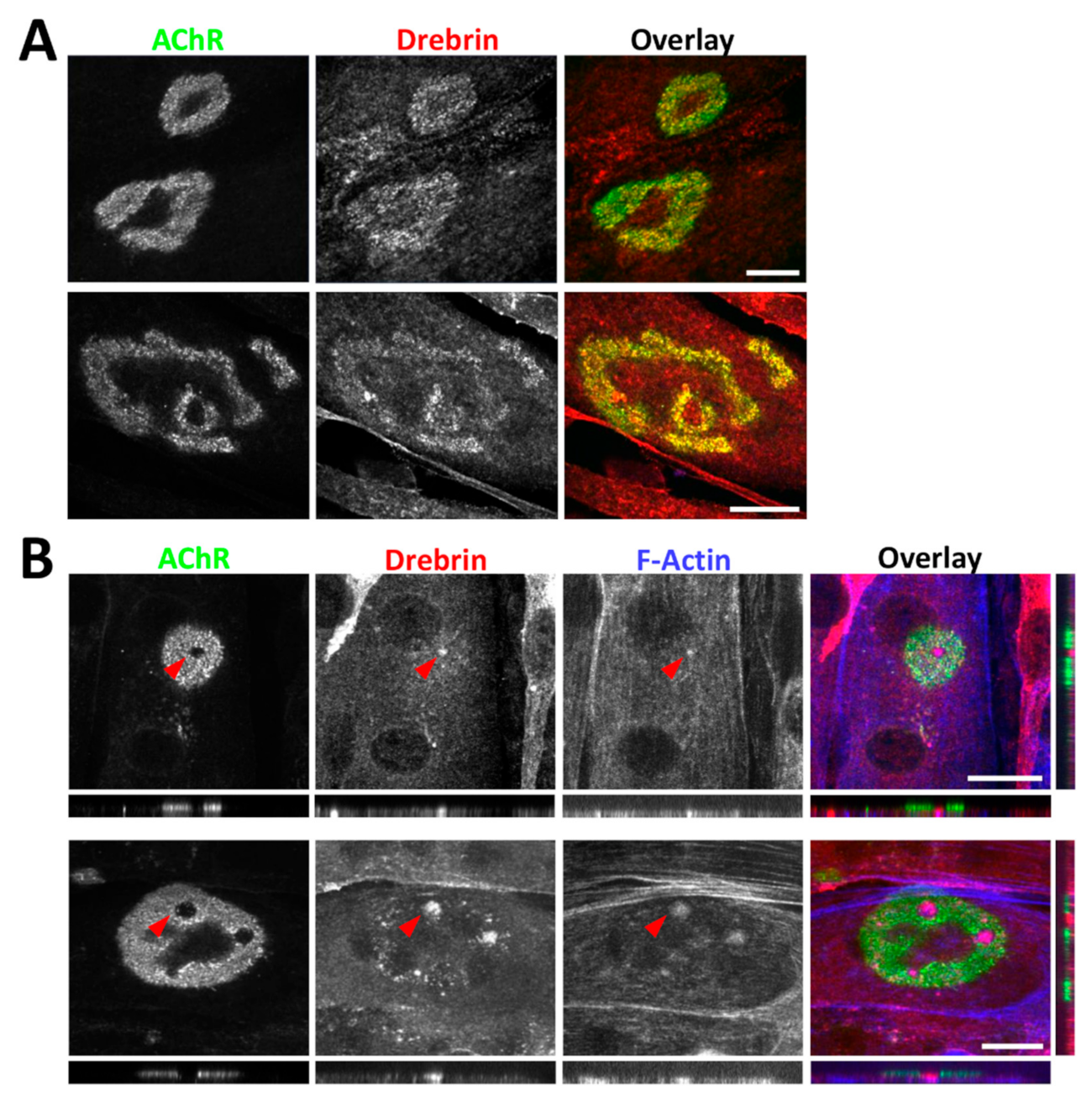
2.2. Drebrin Localizes to the Postsynaptic Machinery In Vitro
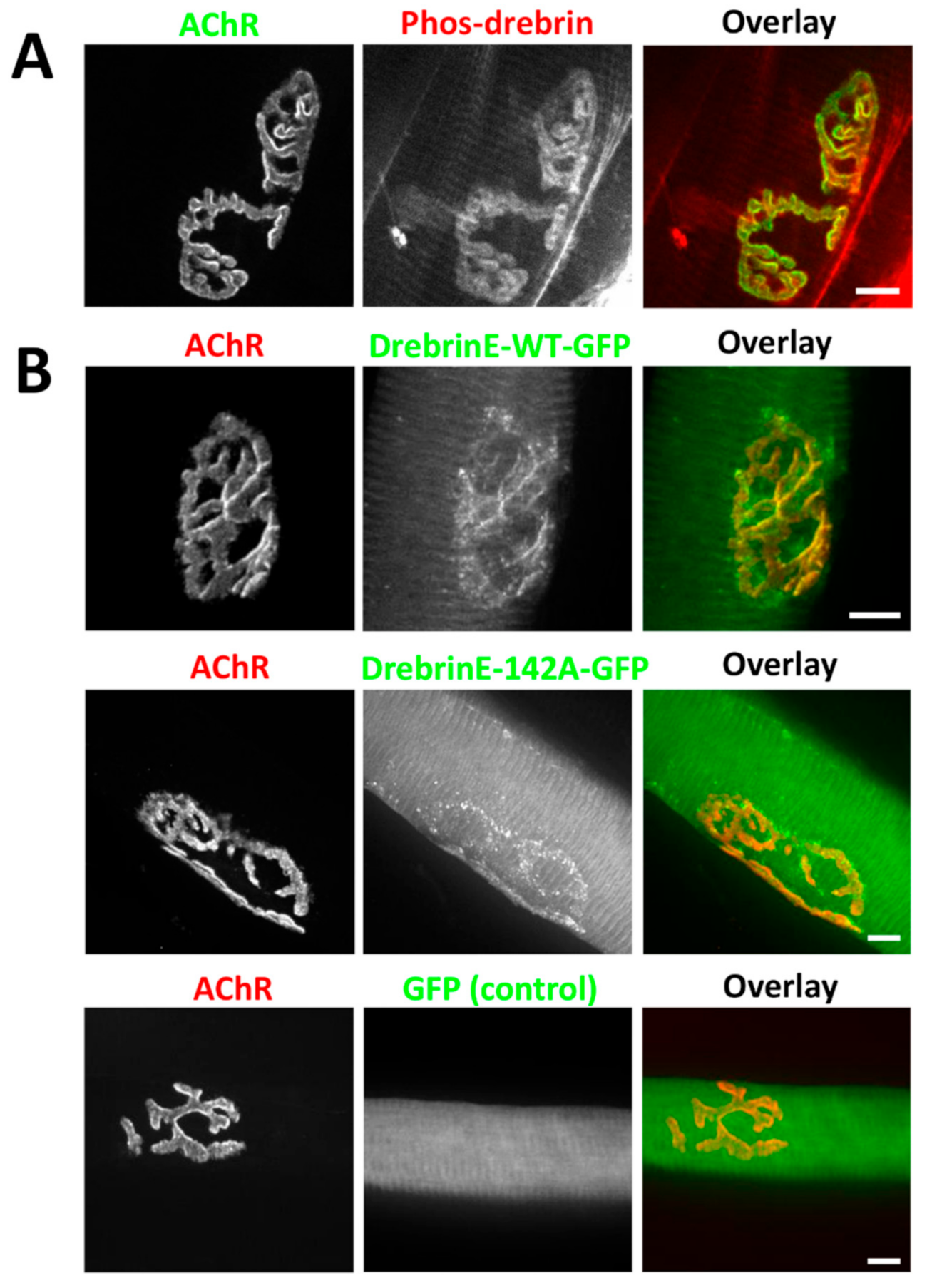
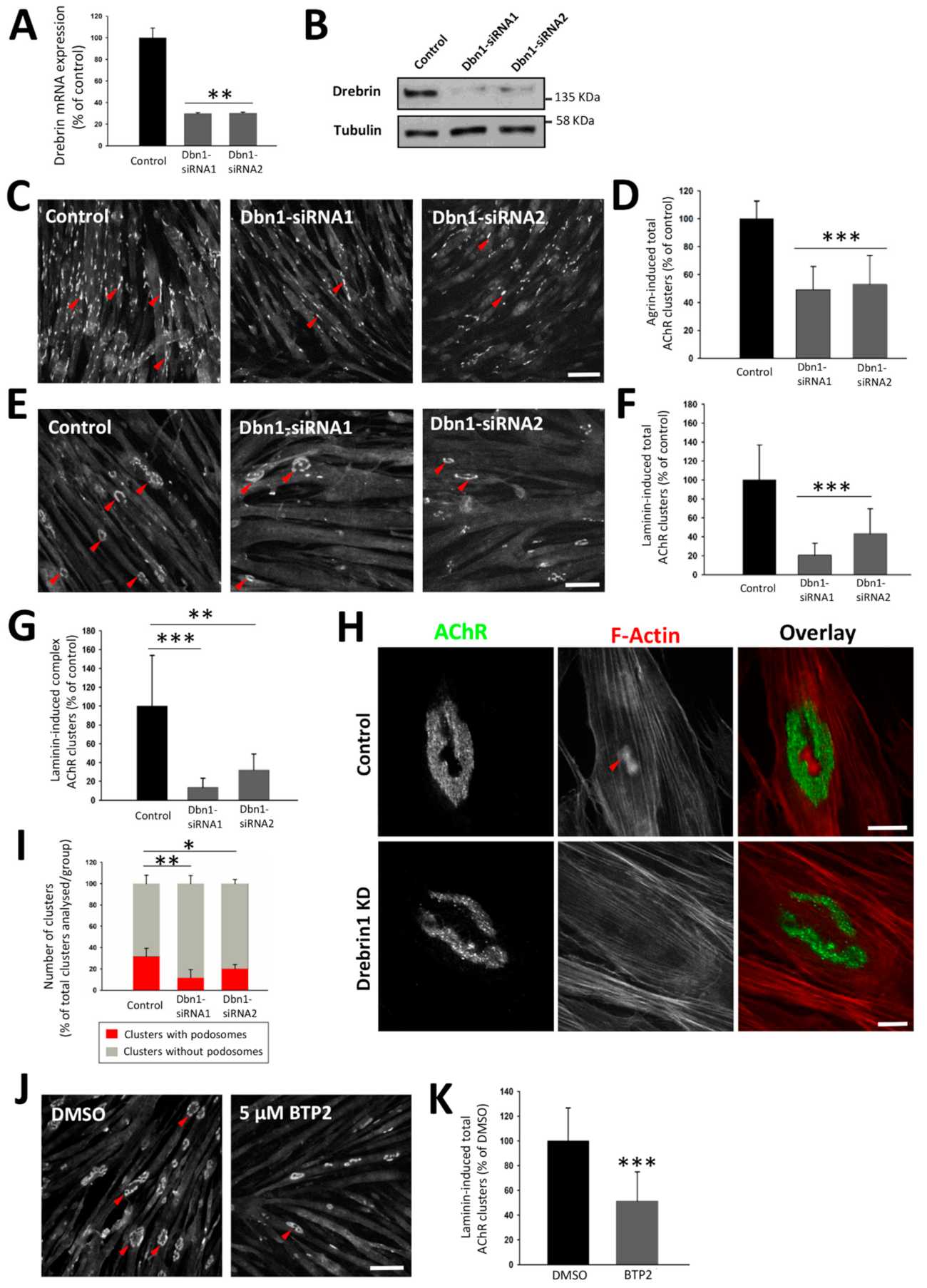
2.3. Drebrin Downregulation or Impairment of Drebrin Binding to F-Actin Compromises AChR Clustering In Vitro
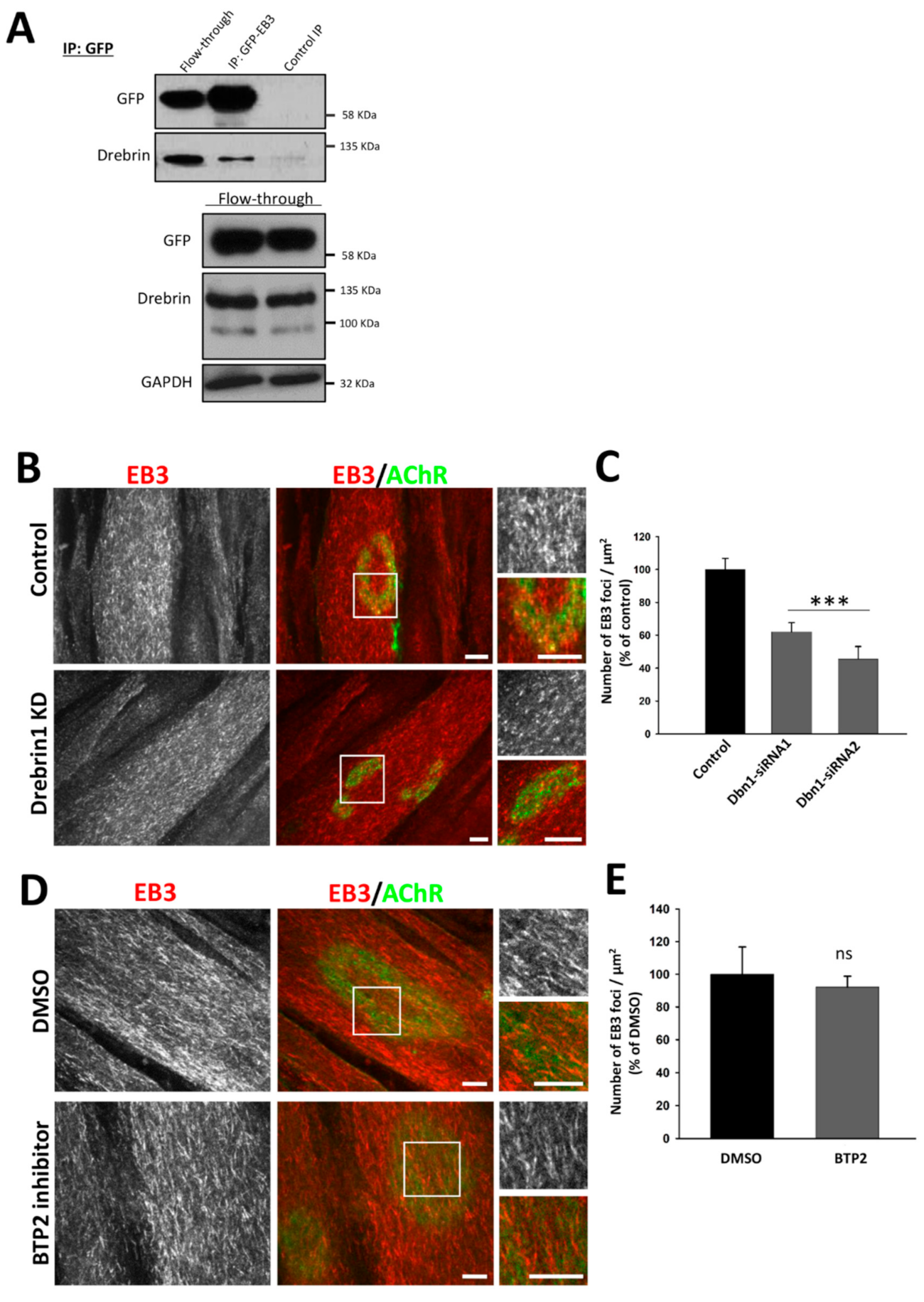
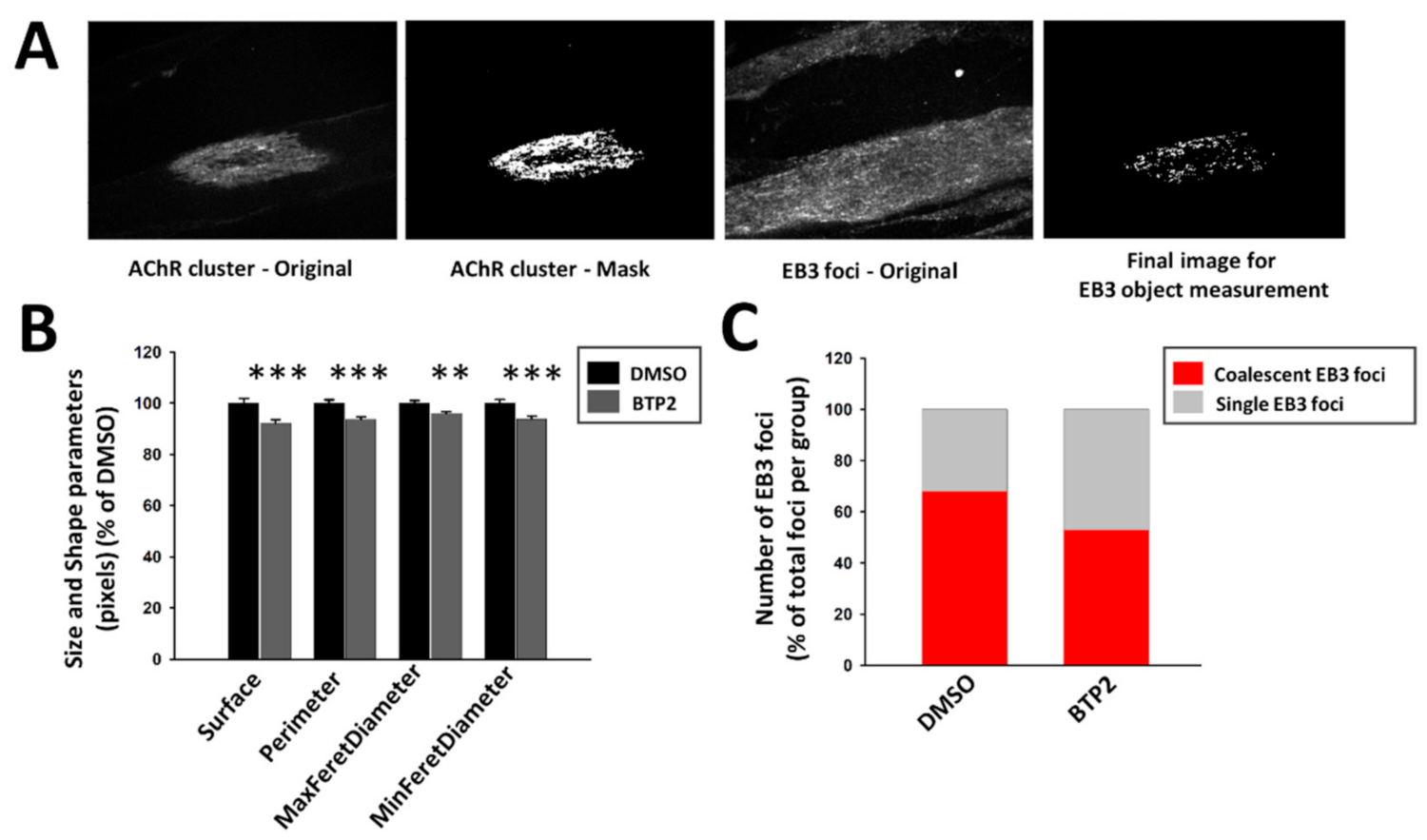
2.4. Drebrin Regulates Microtubule Capture at AChR Clusters
2.5. Cell Surface Delivery of AChRs Is Not Affected by Drebrin Knockdown
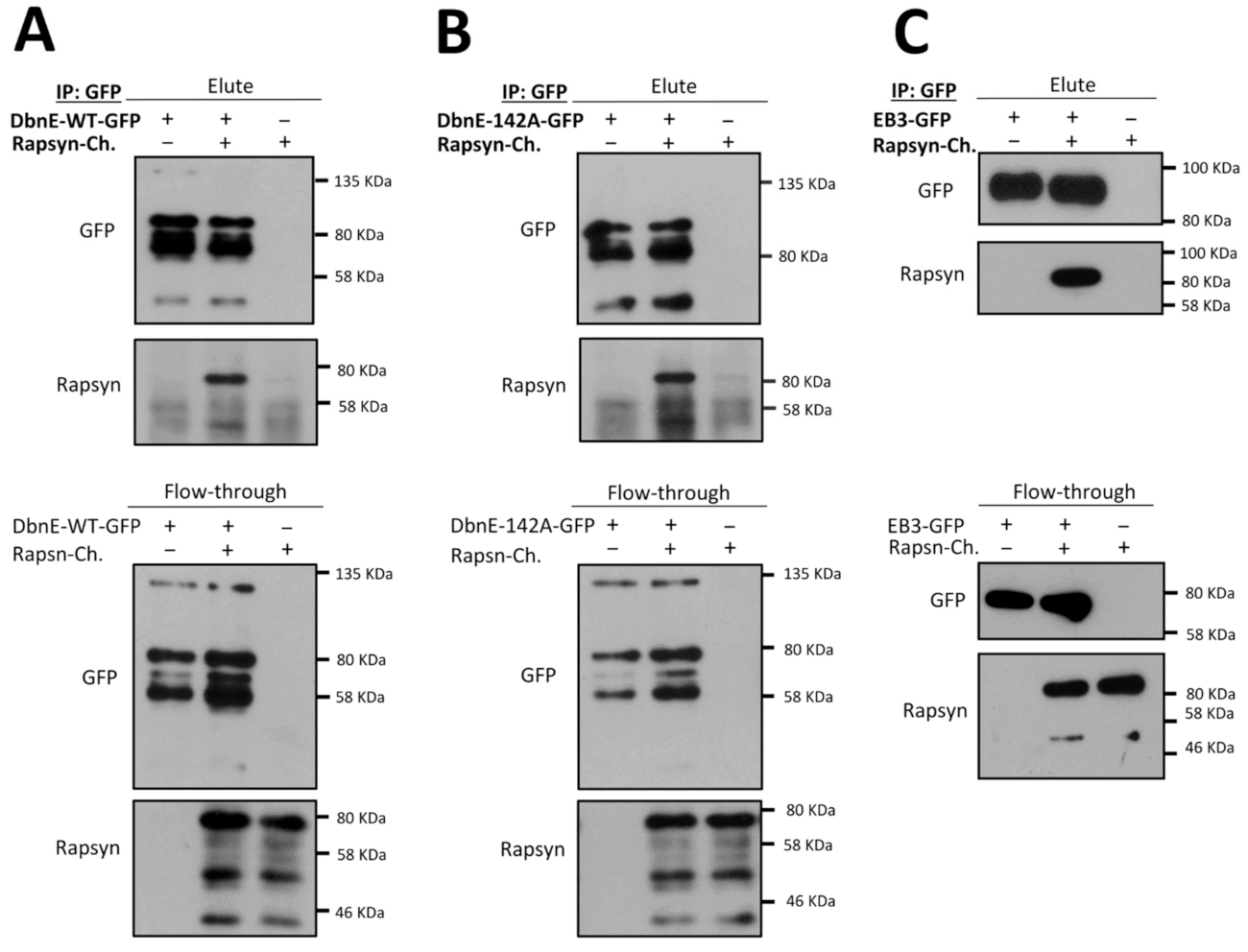
2.6. Drebrin Cooperates with AChR Clustering Regulator Rapsyn
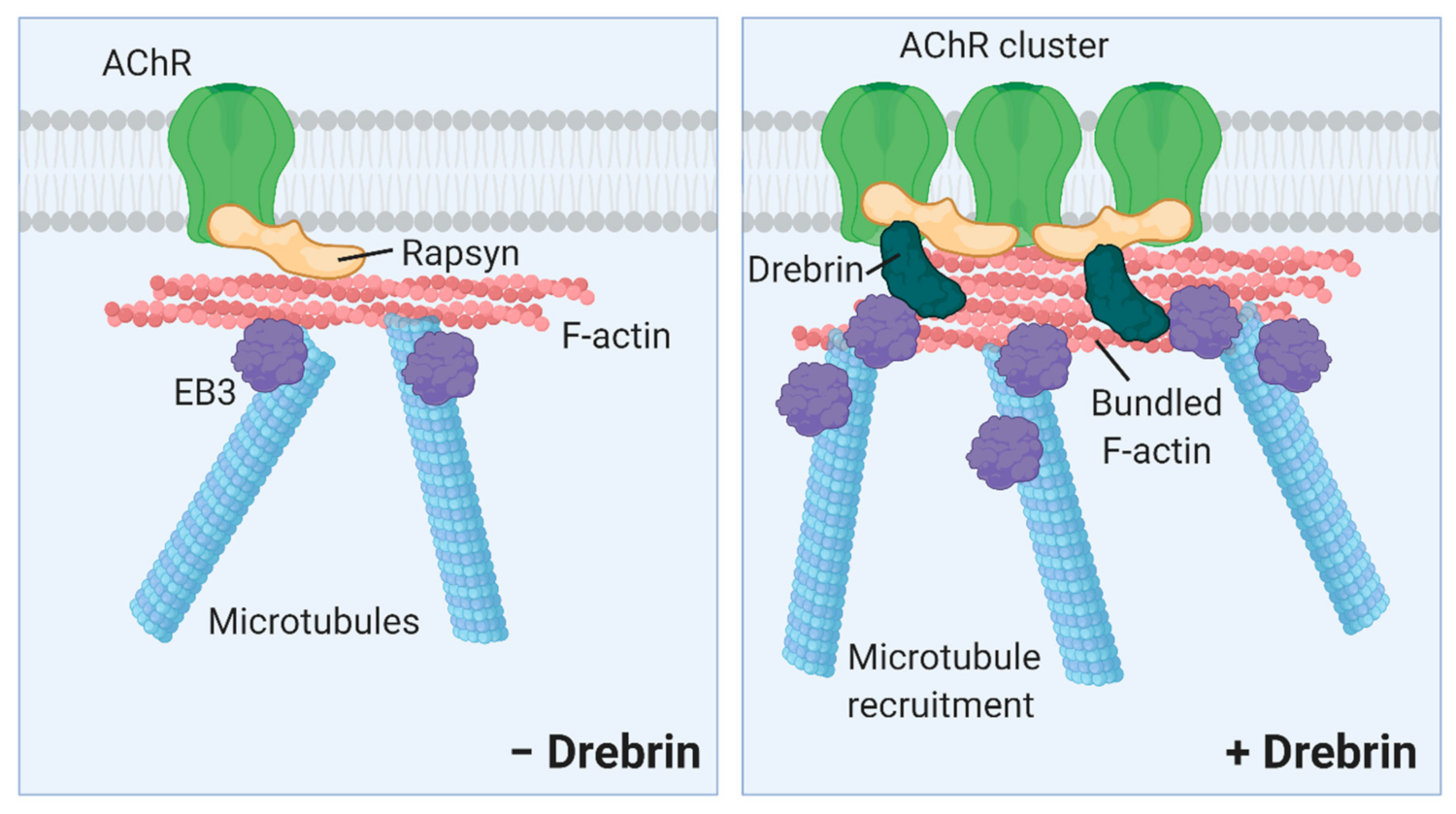
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell and Tissue Labeling
4.3. Microscopy and Image Analysis
4.4. Transfection with siRNA and Plasmids
4.5. Generation of Stable C2C12 Line Overexpressing EB3-GFP
4.6. AChR Pull-Down and Co-Immunoprecipitation
4.7. Western Blot
4.8. Muscle Electroporation
4.9. RNA Isolation and Quantitative PCR
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AChR | acetylcholine receptors |
| BSA | bovine serum albumin |
| BTP2 | 3,5-bis-trifluoromethyl pyrazole |
| BTX | bungarotoxin |
| CLASP2 | CLIP-associating protein 2 |
| CNS | central nervous system |
| DGC | dystrophin-glycoprotein complex |
| DMEM | in Dulbecco’s modified Eagle’s medium |
| DMSO | dimethyl sulfoxide |
| EB1/EB3 | end-binding protein 1 or 3 |
| ECM | extracellular matrix |
| EGFP | Enhanced green fluorescent protein |
| ELK | ETS-like protein 1, ETS domain transcription factor 1 |
| FBS | foetal bovine serum |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GEF | guanine nucleotide-exchange factor |
| HEK293 | human embryonic kidney cells 293 |
| LL5β | pleckstrin homology-like domain family B member 2 |
| MACF1 | microtubule-actin cross-linking factor 1 |
| MT | microtubule |
| NGS | normal goat serum |
| NMJ | neuromuscular junctions |
| PBS | phosphate buffered saline |
| PFA | paraformaldehyde |
| PIP3 | phosphatidylinositol-3,4,5-triphosphate |
| TBST | Tris buffered saline with Tween |
| TIP | plus-end-tracking proteins |
| WAVE1 | WASP-family verprolin-homologous protein 1 |
References
- Li, L.; Xiong, W.-C.; Mei, L. Neuromuscular Junction Formation, Aging, and Disorders. Annu. Rev. Physiol. 2018, 8014, 1–30. [Google Scholar] [CrossRef]
- Rodríguez Cruz, P.M.; Cossins, J.; Beeson, D.; Vincent, A. The Neuromuscular Junction in Health and Disease: Molecular Mechanisms Governing Synaptic Formation and Homeostasis. Front. Mol. Neurosci. 2020, 13, 1–22. [Google Scholar] [CrossRef]
- Araque, A.; Parpura, V.; Sanzgiri, R.P.; Haydon, P.G. Tripartite synapses: Glia, the unacknowledged partner. Trends Neurosci. 1999, 22, 208–215. [Google Scholar] [CrossRef]
- Alvarez-Suarez, P.; Gawor, M.; Prószyński, T.J. Perisynaptic schwann cells - The multitasking cells at the developing neuromuscular junctions. Semin. Cell Dev. Biol. 2020, 104, 31–38. [Google Scholar] [CrossRef]
- Balice-Gordon, R.J.; Lichtman, J.W. In vivo observations of pre- and postsynaptic changes during the transition from multiple to single innervation at developing neuromuscular junctions. J. Neurosci. 1993, 13, 834–855. [Google Scholar] [CrossRef]
- Kummer, T.T.; Misgeld, T.; Lichtman, J.W.; Sanes, J.R. Nerve-independent formation of a topologically complex postsynaptic apparatus. J. Cell Biol. 2004, 164, 1077–1087. [Google Scholar] [CrossRef]
- Marques, M.J.; Conchello, J.A.; Lichtman, J.W. From plaque to pretzel: Fold formation and acetylcholine receptor loss at the developing neuromuscular junction. J. Neurosci. 2000, 20, 3663–3675. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Fu, A.K.Y.; Ip, N.Y. Molecular mechanisms underlying maturation and maintenance of the vertebrate neuromuscular junction. Trends Neurosci. 2012, 35, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, G.C.; Zhang, B.; Xiong, W.C.; Mei, L. The Role of the Cytoskeleton in Neuromuscular Junction Formation. J. Mol. Neurosci. 2006, 30, 115–117. [Google Scholar] [CrossRef]
- Slater, C.R. Structural Factors Influencing the Efficacy of Neuromuscular Transmission. Ann. N. Y. Acad. Sci. 2008, 1132, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chevessier, F.; Girard, E.; Molgó, J.; Bartling, S.; Koenig, J.; Hantaï, D.; Witzemann, V. A mouse model for congenital myasthenic syndrome due to MuSK mutations reveals defects in structure and function of neuromuscular junctions. Hum. Mol. Genet. 2008, 17, 3577–3595. [Google Scholar] [CrossRef]
- Kong, L.; Wang, X.; Choe, D.W.; Polley, M.; Burnett, B.G.; Bosch-Marcé, M.; Griffin, J.W.; Rich, M.M.; Sumner, C.J. Impaired Synaptic Vesicle Release and Immaturity of Neuromuscular Junctions in Spinal Muscular Atrophy Mice. J. Neurosci. 2009, 29, 842–851. [Google Scholar] [CrossRef]
- Pratt, S.J.P.; Shah, S.B.; Ward, C.W.; Inacio, M.P.; Stains, J.P.; Lovering, R.M. Effects of in vivo injury on the neuromuscular junction in healthy and dystrophic muscles. J. Physiol. 2013, 591, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Selcen, D.; Milone, M.; Shen, X.-M.; Harper, C.M.; Stans, A.A.; Wieben, E.D.; Engel, A.G. Dok-7 Myasthenia: Phenotypic and Molecular Genetic Studies in 16 Patients. Ann. Neurol. 2008, 64, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Theroux, M.C.; Olivant, A.; Akins, R.E. C histomorphology of neuromuscular junction in Duchenne muscular dystrophy. Paediatr. Anaesth. 2008, 18, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Slater, C.R.; Fawcett, P.R.W.; Walls, T.J.; Lyons, P.R.; Bailey, S.J.; Beeson, D.; Young, C.; Gardner-Medwin, D. Pre- and post-synaptic abnormalities associated with impaired neuromuscular transmission in a group of patients with “limb-girdle myasthenia”. Brain 2006, 129, 2061–2076. [Google Scholar] [CrossRef]
- Proszynski, T.J.; Gingras, J.; Valdez, G.; Krzewski, K.; Sanes, J.R. Podosomes are present in a postsynaptic apparatus and participate in its maturation. Proc. Natl. Acad. Sci. USA 2009, 106, 18373–18378. [Google Scholar] [CrossRef]
- Proszynski, T.J.; Sanes, J.R. Amotl2 interacts with LL5 b, localizes to podosomes and regulates postsynaptic differentiation in muscle. J. Cell Sci. 2013, 126, 2225–2235. [Google Scholar] [CrossRef]
- Chan, Z.C.K.; Kwan, H.L.R.; Wong, Y.S.; Jiang, Z.; Zhou, Z.; Tam, K.W.; Chan, Y.S.; Chan, C.B.; Lee, C.W. Site-directed MT1-MMP trafficking and surface insertion regulate achr clustering and remodeling at developing NMJS. Elife 2020, 9, 1–33. [Google Scholar] [CrossRef]
- Wu, H.; Xiong, W.C.; Mei, L. To build a synapse: Signaling pathways in neuromuscular junction assembly. Development 2010, 137, 1017–1033. [Google Scholar] [CrossRef]
- Gawor, M.; Prószyński, T.J. The molecular cross talk of the dystrophin–glycoprotein complex. Ann. N. Y. Acad. Sci. 2018, 1412, 62–72. [Google Scholar] [CrossRef]
- Mohamed, A.S.; Swope, S.L. Phosphorylation and Cytoskeletal Anchoring of the Acetylcholine Receptor by Src Class Protein-tyrosine Kinases. Activation by Rapsyn. J. Biol. Chem. 1999, 274, 20529–20539. [Google Scholar] [CrossRef]
- Moransard, M.; Borges, L.S.; Willmann, R.; Marangi, P.A.; Brenner, H.R.; Ferns, M.J.; Fuhrer, C. Agrin Regulates Rapsyn Interaction with Surface Acetylcholine Receptors, and This Underlies Cytoskeletal Anchoring and Clustering. J. Biol. Chem. 2003, 278, 7350–7359. [Google Scholar] [CrossRef] [PubMed]
- Ohno, K.; Engel, A.G.; Shen, X.M.; Selcen, D.; Brengman, J.; Harper, C.M.; Tsujino, A.; Milone, M. Rapsyn mutations in humans cause endplate acetylcholine-receptor deficiency and myasthenic syndrome. Am. J. Hum. Genet. 2002, 70, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Oury, J.; Liu, Y.; Töpf, A.; Todorovic, S.; Hoedt, E.; Preethish-Kumar, V.; Neubert, T.A.; Lin, W.; Lochmüller, H.; Burden, S.J. MACF1 links Rapsyn to microtubule- and actin-binding proteins to maintain neuromuscular synapses. J. Cell Biol. 2019, 218, 1686–1705. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Han, J.; Bamburg, J.R.; Han, L.; Lynn, R.; Zheng, J.Q. Regulation of Acetylcholine Receptor Clustering by ADF/Cofilin- Directed Vesicular Trafficking. Nat. Neurosci. 2009, 12, 848–856. [Google Scholar] [CrossRef]
- Dai, Z.; Luo, X.; Xie, H.; Peng, H.B. The actin-driven movement and formation of acetylcholine receptor clusters. J. Cell Biol. 2000, 150, 1321–1334. [Google Scholar] [CrossRef]
- Luo, Z.G.; Wang, Q.; Zhou, J.Z.; Wang, J.; Luo, Z.; Liu, M.; He, X.; Wynshaw-Boris, A.; Xiong, W.C.; Lu, B.; et al. Regulation of AChR Clustering by Dishevelled Interacting with MuSK and PAK1. Neuron 2002, 35, 489–505. [Google Scholar] [CrossRef]
- Shi, L.; Butt, B.; Ip, F.C.F.; Dai, Y.; Jiang, L.; Yung, W.H.; Greenberg, M.E.; Fu, A.K.Y.; Ip, N.Y. Ephexin1 Is Required for Structural Maturation and Neurotransmission at the Neuromuscular Junction. Neuron 2010, 65, 204–216. [Google Scholar] [CrossRef]
- Sit, S.T.; Manser, E. Rho GTPases and their role in organizing the actin cytoskeleton. J. Cell Sci. 2011, 124, 679–683. [Google Scholar] [CrossRef]
- Weston, C.; Gordon, C.; Teressa, G.; Hod, E.; Ren, X.D.; Prives, J. Cooperative regulation by Rac and Rho of agrin-induced acetylcholine receptor clustering in muscle cells. J. Biol. Chem. 2003, 278, 6450–6455. [Google Scholar] [CrossRef]
- Weston, C.; Yee, B.; Hod, E.; Prives, J. Agrin-induced acetylcholine receptor clustering is mediated by the small guanosine triphosphatases Rac and Cdc42. J. Cell Biol. 2000, 150, 205–212. [Google Scholar] [CrossRef]
- Bernadzki, K.M.; Daszczuk, P.; Rojek, K.O.; Pęziński, M.; Gawor, M.; Pradhan, B.S.; de Cicco, T.; Bijata, M.; Bijata, K.; Włodarczyk, J.; et al. Arhgef5 Binds α-Dystrobrevin 1 and Regulates Neuromuscular Junction Integrity. Front. Mol. Neurosci. 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bernadzki, K.M.; Rojek, K.O.; Prószyński, T.J. Podosomes in muscle cells and their role in the remodeling of neuromuscular postsynaptic machinery. Eur. J. Cell Biol. 2014, 93, 478–485. [Google Scholar] [CrossRef]
- Lin, S.S.; Hsieh, T.L.; Liou, G.G.; Li, T.N.; Lin, H.C.; Chang, C.W.; Wu, H.Y.; Yao, C.K.; Liu, Y.W. Dynamin-2 Regulates Postsynaptic Cytoskeleton Organization and Neuromuscular Junction Development. Cell Rep. 2020, 33, 108310. [Google Scholar] [CrossRef] [PubMed]
- Jasmin, B.J.; Changeux, J.-P.; Cartaud, J. Compartmentalization of cold-stable and acetylated microtubules in the subsynaptic domain of chick skeletal muscle fibre. Nature 1990, 344, 673–675. [Google Scholar] [CrossRef]
- Schmidt, N.; Basu, S.; Sladecek, S.; Gatti, S.; van Haren, J.; Treves, S.; Pielage, J.; Galjart, N.; Brenner, H.R. Agrin regulates CLASP2-mediated capture of microtubules at the neuromuscular junction synaptic membrane. J. Cell Biol. 2012, 198, 421–437. [Google Scholar] [CrossRef]
- Basu, S.; Sladecek, S.; Martinez de la Peña y Valenzuela, I.; Akaaboune, M.; Smal, I.; Martin, K.; Galjart, N.; Brenner, H.R. CLASP2-dependent microtubule capture at the neuromuscular junction membrane requires LL5β and actin for focal delivery of acetylcholine receptor vesicles. Mol. Biol. Cell 2015, 26, 938–951. [Google Scholar] [CrossRef] [PubMed]
- Bernadzki, K.M.; Gawor, M.; Pęziński, M.; Mazurek, P.; Rędowicz, M.J.; Prószyński, T.J. Liprin-α-1 is a novel component of the murine neuromuscular junction and is involved in the organization of the postsynaptic machinery. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Osseni, A.; Ravel-Chapuis, A.; Thomas, J.L.; Gache, V.; Schaeffer, L.; Jasmin, B.J. HDAC6 regulates microtubule stability and clustering of AChRs at neuromuscular junctions. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Basu, S.; Sladecek, S.; Pemble, H.; Wittmann, T.; Slotman, J.A.; Van Cappellen, W.; Brenner, H.R.; Galjart, N. Acetylcholine receptor (AChR) clustering is regulated both by glycogen synthase kinase 3β (GSK3β)-dependent Phosphorylation and the level of CLIP-associated protein 2 (CLASP2) mediating the capture of microtubule plus-ends. J. Biol. Chem. 2014, 289, 30857–30867. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, G.C.; Luo, S.; Yang, Z.; Xiong, W.C.; Mei, L. Alpha-Actinin interacts with rapsyn in agrin-stimulated AChR clustering. Mol. Brain 2008, 1, 18. [Google Scholar] [CrossRef]
- Ghasemizadeh, A.; Christin, E.; Guiraud, A.; Couturier, N.; Risson, V.; Girard, E.; Jagla, C.; Soler, C.; Laddada, L.; Sanchez, C.; et al. Skeletal muscle MACF1 maintains myonuclei and mitochondria localization through microtubules to control muscle functionalities. bioRxiv 2019. [Google Scholar] [CrossRef]
- Ivanov, A.; Esclapez, M.; Pellegrino, C.; Shirao, T.; Ferhat, L. Drebrin A regulates dendritic spine plasticity and synaptic function in mature cultured hippocampal neurons. J. Cell Sci. 2009, 122, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, C.; Aoki, C.; Kojima, N.; Yamazaki, H.; Shirao, T. Drebrin A content correlates with spine head size in the adult mouse cerebral cortex. J. Comp. Neurol. 2007, 503, 618–626. [Google Scholar] [CrossRef]
- Shan, Y.; Farmer, S.M.; Wray, S. Drebrin regulates cytoskeleton dynamics in migrating neurons through interaction with CXCR4. Proc. Natl. Acad. Sci. USA 2021, 118, 1–12. [Google Scholar] [CrossRef]
- Trivedi, N.; Stabley, D.R.; Cain, B.; Howell, D.; Laumonnerie, C.; Ramahi, J.S.; Temirov, J.; Kerekes, R.A.; Gordon-Weeks, P.R.; Solecki, D.J. Drebrin-mediated microtubule-actomyosin coupling steers cerebellar granule neuron nucleokinesis and migration pathway selection. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Zhao, B.; Meka, D.P.; Scharrenberg, R.; König, T.; Schwanke, B.; Kobler, O.; Windhorst, S.; Kreutz, M.R.; Mikhaylova, M.; Calderon De Anda, F. Microtubules Modulate F-actin Dynamics during Neuronal Polarization. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Sekino, Y.; Tanaka, S.; Mizui, T.; Kishi, S.; Shirao, T. Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J. Neurosci. 2003, 23, 6586–6595. [Google Scholar] [CrossRef]
- Hayashi, K.; Ishikawa, R.; Ye, L.H.; He, X.L.; Takata, K.; Kohama, K.; Shirao, T. Modulatory role of drebrin on the cytoskeleton within dendritic spines in the rat cerebral cortex. J. Neurosci. 1996, 16, 7161–7170. [Google Scholar] [CrossRef]
- Asada, H.; Uyemura, K.; Shirao, T. Actin-Binding protein, drebrin, accumulates in submembranous regions in parallel with neuronal differentiation. J. Neurosci. Res. 1994, 38, 149–159. [Google Scholar] [CrossRef]
- Majoul, I.; Shirao, T.; Sekino, Y.; Duden, R. Many faces of drebrin: From building dendritic spines and stabilizing gap junctions to shaping neurite-like cell processes. Histochem. Cell Biol. 2007, 127, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Majoul, I.V.; Ernesti, J.S.; Butkevich, E.V.; Duden, R.; Drebrins and connexins: A biomedical perspective. Drebrins and connexins: A biomedical perspective. In Drebrin—From Structure and Function to Physiological and Pathological Roles; Springer Japan KK: Tokyo, Japan, 2017; pp. 225–247. ISBN 9784431565505. [Google Scholar]
- Peitsch, W.K.; Grund, C.; Kuhn, C.; Schnölzer, M.; Spring, H.; Schmelz, M.; Franke, W.W. Drebrin is a widespread actin-associating protein enriched at junctional plaques, defining a specific microfilament anchorage system in polar epithelial cells. Eur. J. Cell Biol. 1999, 78, 767–778. [Google Scholar] [CrossRef]
- Kobayashi, S.; Shirao, T.; Sasaki, T. Drebrin expression is increased in spinal motoneurons of rats after axotomy. Neurosci. Lett. 2001, 311, 165–168. [Google Scholar] [CrossRef]
- Geraldo, S.; Khanzada, U.K.; Parsons, M.; Chilton, J.K.; Gordon-Weeks, P.R. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell Biol. 2008, 10, 1181–1189. [Google Scholar] [CrossRef]
- Mancini, A.; Sirabella, D.; Zhang, W.; Yamazaki, H.; Shirao, T.; Krauss, R.S. Regulation of myotube formation by the actin-binding factor drebrin. Skelet. Muscle 2011, 1, 10–13. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber types in Mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Chakkalakal, J.V.; Nishimune, H.; Ruas, J.L.; Spiegelman, B.M.; Sanes, J.R. Retrograde influence of muscle fibers on their innervation revealed by a novel marker for slow motoneurons. Development 2010, 137, 3489–3499. [Google Scholar] [CrossRef]
- Jones, R.A.; Reich, C.D.; Dissanayake, K.N.; Kristmundsdottir, F.; Findlater, G.S.; Ribchester, R.R.; Simmen, M.W.; Gillingwater, T.H. NMJ-morph reveals principal components of synaptic morphology influencing structure-function relationships at the neuromuscular junction. Open Biol. 2016, 6. [Google Scholar] [CrossRef]
- Lee, Y. il Differences in the constituent fiber types contribute to the intermuscular variation in the timing of the developmental synapse elimination. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Blank, S.; Chen, V.; Hamilton, N.; Salerno, T.A.; Ianuzzo, C.D. Biochemical characteristics of mammalian myocardia. Respir. Physiol. 1988, 74, 115–126. [Google Scholar] [CrossRef]
- Brust, M. Fatigue and caffeine effects in fast-twitch and slow-twitch muscles of the mouse. Pflügers Arch. Eur. J. Physiol. 1976, 367, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Hodge, K.; Powers, S.K.; Coombes, J.; Fletcher, L.; Demirel, H.A.; Dodd, S.L.; Martin, D. Bioenergetic characteristics of the costal and crural diaphragm in mammals. Respir. Physiol. 1997, 109, 149–154. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Zhang, W.J.; Medler, S. The continuum of hybrid IIX/IIB fibers in normal mouse muscles: MHC isoform proportions and spatial distribution within single fibers. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, 1582–1591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanabe, K.; Yamazaki, H.; Inaguma, Y.; Asada, A.; Kimura, T.; Takahashi, J.; Taoka, M.; Ohshima, T.; Furuichi, T.; Isobe, T.; et al. Phosphorylation of Drebrin by Cyclin-dependent Kinase 5 and Its role in neuronal migration. PLoS One 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Worth, D.C.; Daly, C.N.; Geraldo, S.; Oozeer, F.; Gordon-Weeks, P.R. Drebrin contains a cryptic F-actin-bundling activity regulated by Cdk5 phosphorylation. J. Cell Biol. 2013, 202, 793–806. [Google Scholar] [CrossRef]
- Capes, E.M.; Loaiza, R.; Valdivia, H.H. Ryanodine Receptors. Skelet. Muscle 2011, 1, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Luther, P.K. The vertebrate muscle Z-disc: Sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 2009, 30, 171–185. [Google Scholar] [CrossRef]
- Wang, L.; Geist, J.; Grogan, A.; Hu, L.Y.R.; Kontrogianni-Konstantopoulos, A. Thick filament protein network, functions, and disease association. Compr. Physiol. 2018, 8, 631–709. [Google Scholar] [CrossRef]
- Pęziński, M.; Daszczuk, P.; Pradhan, B.S.; Lochmüller, H.; Prószyński, T.J. An improved method for culturing myotubes on laminins for the robust clustering of postsynaptic machinery. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Madhavan, R.; Gong, Z.L.; Ma, J.J.; Chan, A.W.S.; Peng, H.B. The function of cortactin in the clustering of acetylcholine receptors at the vertebrate neuromuscular junction. PLoS One 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Mercer, J.C.; Qi, Q.; Mottram, L.F.; Law, M.; Bruce, D.; Iyer, A.; Morales, J.L.; Yamazaki, H.; Shirao, T.; R, B.; et al. Chemico-Genetic Identification of Drebrin as a Regulator of Calcium Responses. Int. J. Biochem. 2010, 42, 337–345. [Google Scholar] [CrossRef]
- Kowarz, E.; Löscher, D.; Marschalek, R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015, 10, 647–653. [Google Scholar] [CrossRef]
- Gautam, M.; Noakes, P.G.; Mudd, J.; Nichol, M.; Chu, G.C.; Sanes, J.R.; Merlie, J.P. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature 1995, 377, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cao, Y.; Wu, H.; Ye, X.; Zhu, Z.; Xing, G.; Shen, C.; Barik, A.; Zhang, B.; Xie, X.; et al. Enzymatic Activity of the Scaffold Protein Rapsyn for Synapse Formation. Neuron 2016, 92, 1007–1019. [Google Scholar] [CrossRef]
- Benarroch, L.; Bonne, G.; Rivier, F.; Hamroun, D. The 2020 version of the gene table of neuromuscular disorders (nuclear genome). Neuromuscul. Disord. 2019, 29, 980–1018. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ip, F.C.F.; Shi, L.; Zhang, Z.; Tang, H.; Ng, Y.P.; Ye, W.C.; Fu, A.K.Y.; Ip, N.Y. Coronin 6 regulates acetylcholine receptor clustering through modulating receptor anchorage to actin cytoskeleton. J. Neurosci. 2014, 34, 2413–2421. [Google Scholar] [CrossRef]
- Hanamura, K. Drebrin in neuronal migration and axonal growth. In Drebrin—From Structure and Function to Physiological and Pathological Roles; Shirao, T., Sekino, Y., Eds.; Springer Japan KK: Tokyo, Japan, 2017; ISBN 9784431565505. [Google Scholar]
- Kojima, N.; Shirao, T. Synaptic dysfunction and disruption of postsynaptic drebrin-actin complex: A study of neurological disorders accompanied by cognitive deficits. Neurosci. Res. 2007, 58, 1–5. [Google Scholar] [CrossRef]
- Gordon-Weeks, P.R. The role of the drebrin/EB3/Cdk5 pathway in dendritic spine plasticity, implications for Alzheimer’s disease. Brain Res. Bull. 2016, 126, 293–299. [Google Scholar] [CrossRef]
- Fu, A.K.Y.; Ip, F.C.F.; Fu, W.Y.; Cheung, J.; Wang, J.H.; Yung, W.H.; Ip, N.Y. Aberrant motor axon projection, acetylcholine receptor clustering, and neurotransmission in cyclin-dependent kinase 5 null mice. Proc. Natl. Acad. Sci. USA 2005, 102, 15224–15229. [Google Scholar] [CrossRef]
- Lin, W.; Dominguez, B.; Yang, J.; Aryal, P.; Brandon, E.P.; Gage, F.H.; Lee, K.F. Neurotransmitter acetylcholine negatively regulates neuromuscular synapse formation by a Cdk5-dependent mechanism. Neuron 2005, 46, 569–579. [Google Scholar] [CrossRef]
- Ehrengruber, M.U.; Kato, A.; Inokuchi, K.; Hennou, S. Homer/Vesl Proteins and Their Roles in CNS Neurons. Mol. Neurobiol. 2004, 29, 213–227. [Google Scholar] [CrossRef]
- Foa, L.; Rajan, I.; Haas, K.; Wu, G.Y.; Brakeman, P.; Worley, P.; Cline, H. The scaffold protein, Homer1b/c, regulates axon pathfinding in the central nervous system in vivo. Nat. Neurosci. 2001, 4, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y. Drebrin-Homer Interaction at An Atomic Scale. Structure 2019, 27, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.; Li, J.; Yang, Q.; Feng, Z.; Li, Y.; Yang, H.; Yu, C.; Wan, J.; Liu, W.; et al. Homer Tetramer Promotes Actin Bundling Activity of Drebrin. Structure 2019, 27, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi-Yamaguchi, Y.; Sato, Y.; Sakai, R.; Mizutani, A.; Knöpfel, T.; Mori, N.; Mikoshiba, K.; Furuichi, T. Interaction of Cupidin/Homer2 with two actin cytoskeletal regulators, Cdc42 small GTPase and Drebrin, in dendritic spines. BMC Neurosci. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Salanova, M.; Bortoloso, E.; Schiffl, G.; Gutsmann, M.; Belavy, D.L.; Felsenberg, D.; Furlan, S.; Volpe, P.; Blottner, D. Expression and regulation of Homer in human skeletal muscle during neuromuscular junction adaptation to disuse and exercise. FASEB J. 2011, 25, 4312–4325. [Google Scholar] [CrossRef] [PubMed]
- Salanova, M.; Volpe, P.; Blottner, D. Homer protein family regulation in skeletal muscle and neuromuscular adaptation. IUBMB Life 2013, 65, 769–776. [Google Scholar] [CrossRef]
- Nahm, M.; Long, A.A.; Paik, S.K.; Kim, S.; Bae, Y.C.; Broadie, K.; Lee, S. The Cdc42-selective GAP Rich regulates postsynaptic development and retrograde BMP transsynaptic signaling. J. Cell Biol. 2010, 191, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Weston, C.A.; Teressa, G.; Weeks, B.S.; Prives, J. Agrin and laminin induce acetylcholine receptor clustering by convergent, Rho GTPase-dependent signaling pathways. J. Cell Sci. 2007, 120, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Lansbergen, G.; Grigoriev, I.; Mimori-Kiyosue, Y.; Ohtsuka, T.; Higa, S.; Kitajima, I.; Demmers, J.; Galjart, N.; Houtsmuller, A.B.; Grosveld, F.; et al. CLASPs Attach Microtubule Plus Ends to the Cell Cortex through a Complex with LL5β. Dev. Cell 2006, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Xing, G.; Xiong, W.C.; Mei, L. Rapsyn as a signaling and scaffolding molecule in neuromuscular junction formation and maintenance. Neurosci. Lett. 2020, 731, 135013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Luo, S.; Dong, X.P.; Zhang, X.; Liu, C.; Luo, Z.; Xiong, W.C.; Mei, L. Β-Catenin Regulates Acetylcholine Receptor Clustering in Muscle Cells Through Interaction With Rapsyn. J. Neurosci. 2007, 27, 3968–3973. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ruan, N.J.; Qian, L.; Lei, W.L.; Chen, F.; Luo, Z.G. Wnt/β-catenin signaling suppresses Rapsyn expression and inhibits acetylcholine receptor clustering at the neuromuscular junction. J. Biol. Chem. 2008, 283, 21668–21675. [Google Scholar] [CrossRef]
- Grintsevich, E.E.; Reisler, E. Drebrin Inhibits Cofilin-Induced Severing of F-Actin. Cytoskeleton 2014, 71, 472–483. [Google Scholar] [CrossRef]
- Ishikawa, R. Biochemistry of drebrin and its binding to actin filaments. In Drebrin - From Structure and Function to Physiological and Pathological Roles; Shirao, T., Sekino, Y., Eds.; Springer Japan KK: Tokyo, Japan, 2017; pp. 37–47. ISBN 9784431565505. [Google Scholar]
- Ishikawa, R.; Hayashi, K.; Shirao, T.; Xue, Y.; Takagi, T.; Sasaki, Y.; Kohama, K. Drebrin, a Development-associated Brain Protein from Rat Embryo, Causes the Dissociation of Tropomyosin from Actin Filaments. J. Biol. Chem. 1994, 269, 29928–29933. [Google Scholar] [CrossRef]
- Mikati, M.A.; Grintsevich, E.E.; Reisler, E. Drebrin-induced Stabilization of Actin Filaments. J. Biol. Chem. 2013, 288, 19926–19938. [Google Scholar] [CrossRef]
- Harris, A.R.; Jreij, P.; Belardi, B.; Joffe, A.M.; Bausch, A.R.; Fletcher, D.A. Biased localization of actin binding proteins by actin filament conformation. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Dart, A.E.; Worth, D.C.; Muir, G.; Chandra, A.; Morris, J.D.; McKee, C.; Verrill, C.; Bryant, R.J.; Gordon-Weeks, P.R. The drebrin/EB3 pathway drives invasive activity in prostate cancer. Oncogene 2017, 36, 4111–4123. [Google Scholar] [CrossRef]
- Roth, D.; Fitton, B.P.; Chmel, N.P.; Wasiluk, N.; Straube, A. Spatial positioning of EB family proteins at microtubule tips involves distinct nucleotide-dependent binding properties. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.R.; Kang, I.H.; Wheeler, D.B.; Lindquist, R.A.; Papallo, A.; Sabatini, D.M.; Golland, P.; Carpenter, A.E. CellProfiler Analyst: Data exploration and analysis software for complex image-based screens. BMC Bioinformatics 2008, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, T.; Slemmer, J.; Hoogenraad, C.C.; Lansbergen, G.; Dortland, B.; De Zeeuw, C.I.; Grosveld, F.; Van Cappellen, G.; Akhmanova, A.; Galjart, N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 2003, 23, 2655–2664. [Google Scholar] [CrossRef]
- Mátés, L.; Chuah, M.K.L.; Belay, E.; Jerchow, B.; Manoj, N.; Acosta-Sanchez, A.; Grzela, D.P.; Schmitt, A.; Becker, K.; Matrai, J.; et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009, 41, 753–761. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Suarez, P.; Nowak, N.; Protasiuk-Filipunas, A.; Yamazaki, H.; Prószyński, T.J.; Gawor, M. Drebrin Regulates Acetylcholine Receptor Clustering and Organization of Microtubules at the Postsynaptic Machinery. Int. J. Mol. Sci. 2021, 22, 9387. https://doi.org/10.3390/ijms22179387
Alvarez-Suarez P, Nowak N, Protasiuk-Filipunas A, Yamazaki H, Prószyński TJ, Gawor M. Drebrin Regulates Acetylcholine Receptor Clustering and Organization of Microtubules at the Postsynaptic Machinery. International Journal of Molecular Sciences. 2021; 22(17):9387. https://doi.org/10.3390/ijms22179387
Chicago/Turabian StyleAlvarez-Suarez, Paloma, Natalia Nowak, Anna Protasiuk-Filipunas, Hiroyuki Yamazaki, Tomasz J. Prószyński, and Marta Gawor. 2021. "Drebrin Regulates Acetylcholine Receptor Clustering and Organization of Microtubules at the Postsynaptic Machinery" International Journal of Molecular Sciences 22, no. 17: 9387. https://doi.org/10.3390/ijms22179387
APA StyleAlvarez-Suarez, P., Nowak, N., Protasiuk-Filipunas, A., Yamazaki, H., Prószyński, T. J., & Gawor, M. (2021). Drebrin Regulates Acetylcholine Receptor Clustering and Organization of Microtubules at the Postsynaptic Machinery. International Journal of Molecular Sciences, 22(17), 9387. https://doi.org/10.3390/ijms22179387





