circBTBD7 Promotes Immature Porcine Sertoli Cell Growth through Modulating miR-24-3p/MAPK7 Axis to Inactivate p38 MAPK Signaling Pathway
Abstract
:1. Introduction
2. Results
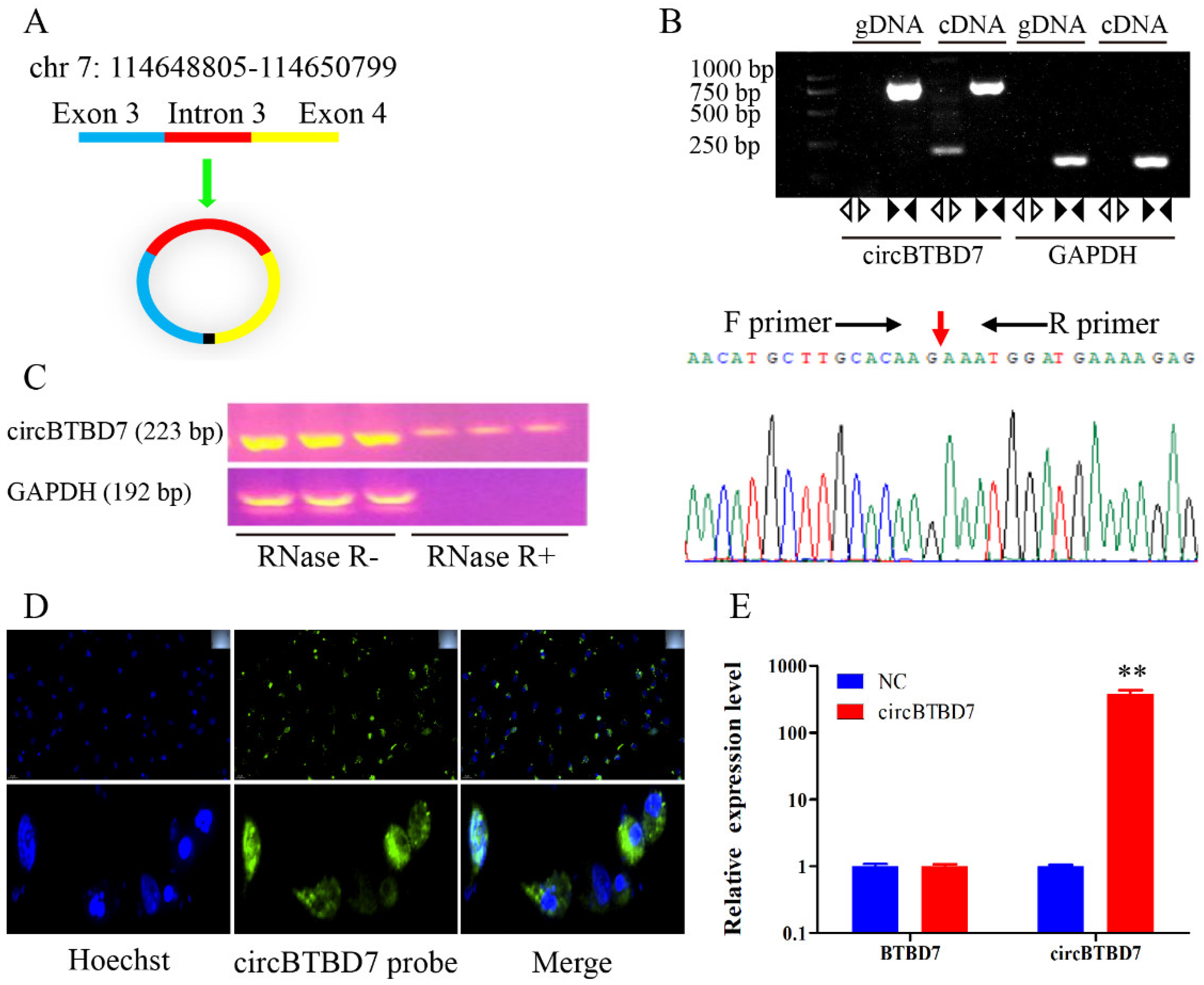
2.1. Characteristics of circBTBD7 in Immature Porcine Sertoli Cells
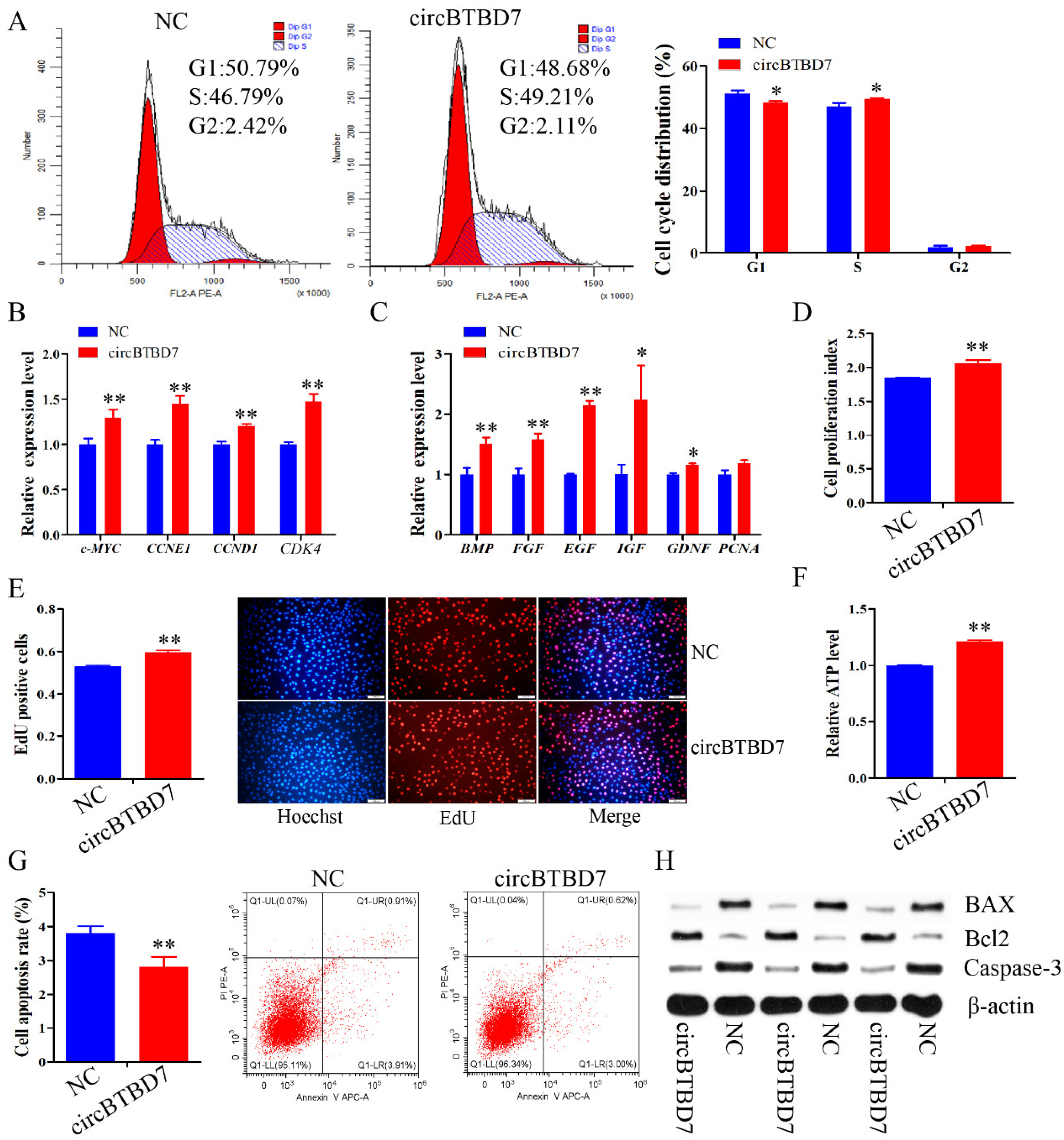
2.2. circBTBD7 Promotes Proliferation and Inhibits Apoptosis of Immature Porcine Sertoli Cells
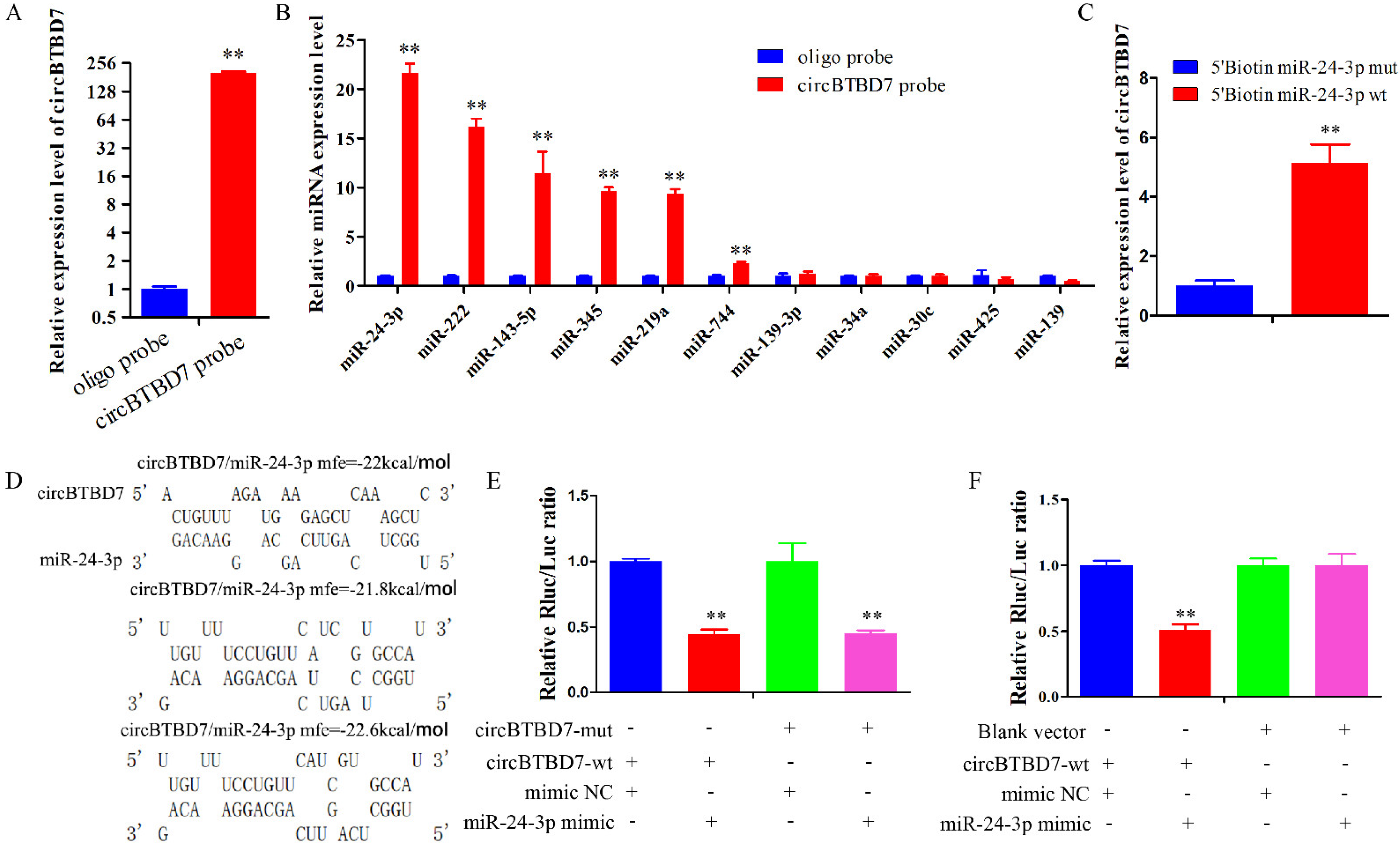
2.3. circBTBD7 Acts a Sponge for miR-24-3p
2.4. miR-24-3p Suppresses Proliferation, Elevates Apoptosis of Immature Porcine Sertoli Cells
2.5. miR-24-3p Antagonizes the Effects of circBTBD7 on Immature Porcine Sertoli Cells
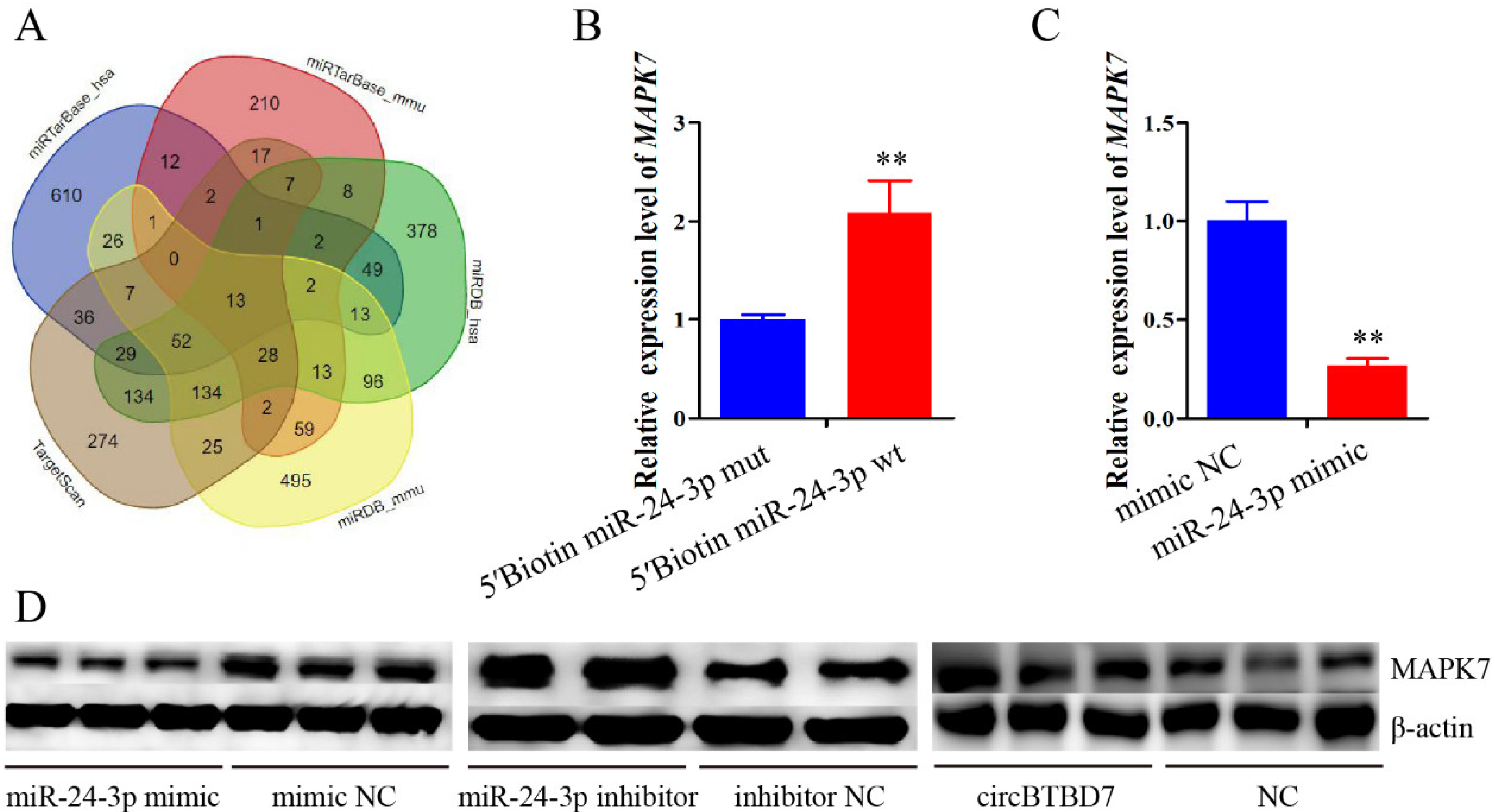
2.6. miR-24-3p Binding to the MAPK7 Gene and Controls Its Expression
2.7. MAPK7 Knockdown Attenuates the Effects of miR-24-3p Inhibition on Immature Porcine Sertoli Cells
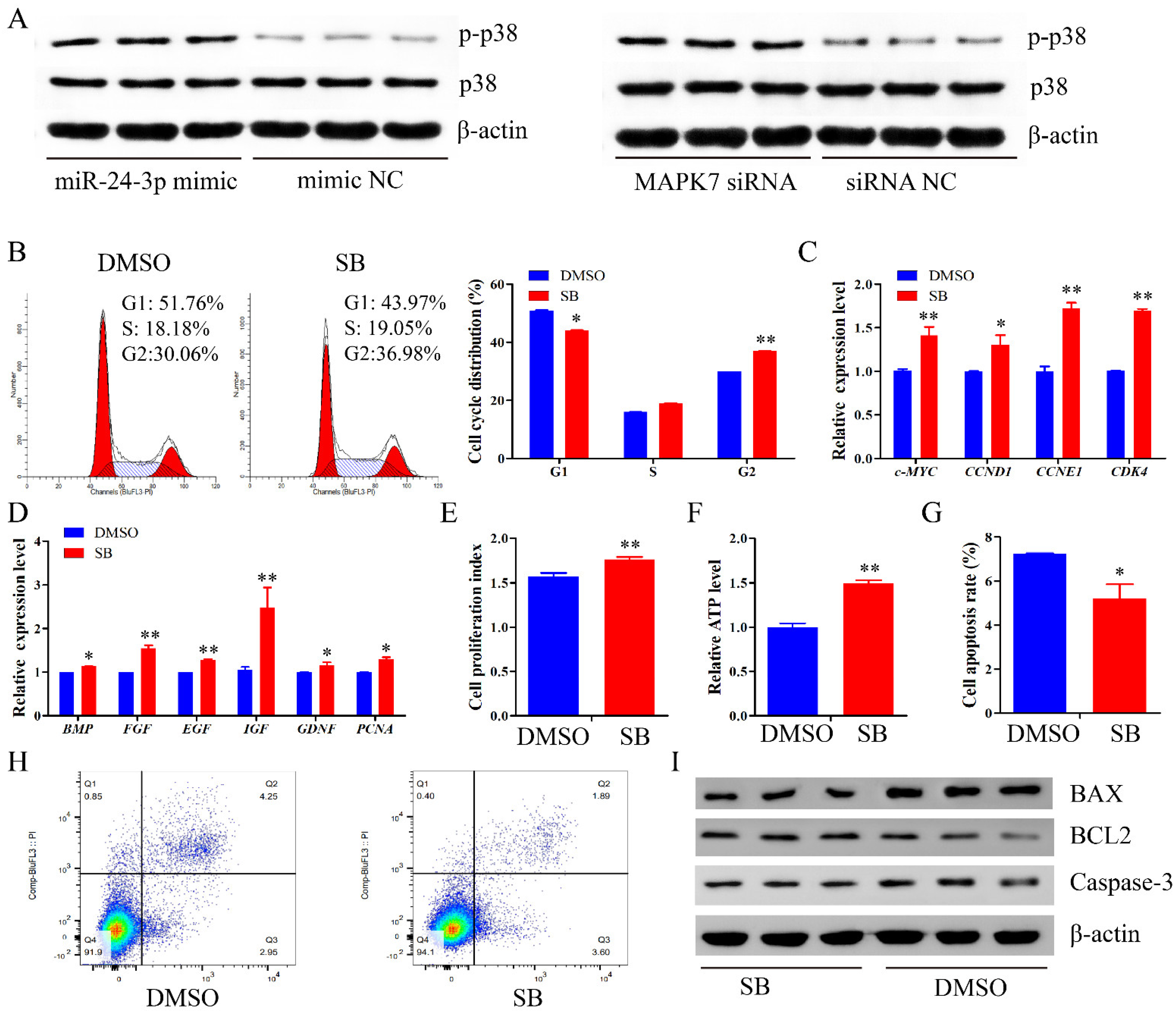
2.8. miR-24-3p Activates the p38 MAPK Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Reverse Transcription PCR
4.3. Fluorescence in Situ Hybridization
4.4. Cell Cycle Assay
4.5. Cell Proliferation Assay
4.6. Cell Apoptosis Assay
4.7. RNA Pull Down Assay
4.8. Real-Time qPCR
4.9. Western Blotting
4.10. Dual-Luciferase Activity Assay
4.11. Statistical Analysis
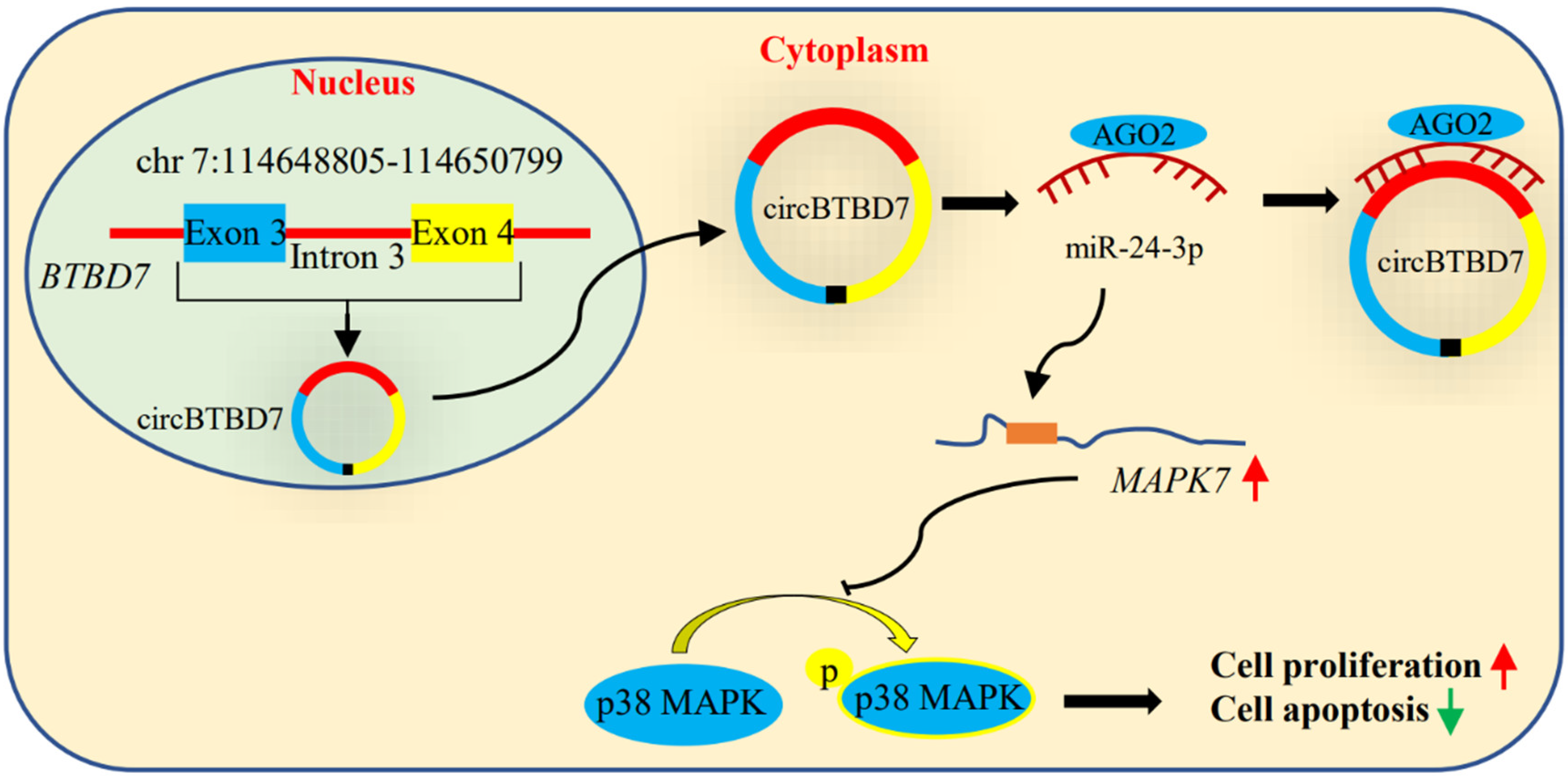
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fayomi, A.P.; Orwig, K.E. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018, 29, 207–214. [Google Scholar] [CrossRef]
- Franca, L.R.; Hess, R.A.; Dufour, J.M.; Hofmann, M.C.; Griswold, M.D. The Sertoli cell: One hundred fifty years of beauty and plasticity. Andrology 2016, 4, 189–212. [Google Scholar] [CrossRef] [Green Version]
- Griswold, M.D. 50 years of spermatogenesis: Sertoli cells and their interactions with germ cells. Biol. Reprod. 2018, 99, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003, 125, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Rebourcet, D.; Darbey, A.; Monteiro, A.; Soffientini, U.; Tsai, Y.T.; Handel, I.; Pitetti, J.L.; Nef, S.; Smith, L.B.; O’Shaughnessy, P.J. Sertoli Cell Number Defines and Predicts Germ and Leydig Cell Population Sizes in the Adult Mouse Testis. Endocrinology 2017, 158, 2955–2969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, L.; Thompson, D.L., Jr.; Varner, D.D. Role of Sertoli cell number and function on regulation of spermatogenesis. Anim. Reprod. Sci. 2008, 105, 23–51. [Google Scholar] [CrossRef]
- Franca, L.R.; Silva, V.A., Jr.; Chiarini-Garcia, H.; Garcia, S.K.; Debeljuk, L. Cell proliferation and hormonal changes during postnatal development of the testis in the pig. Biol. Reprod. 2000, 63, 1629–1636. [Google Scholar] [CrossRef]
- Ma, C.; Song, H.; Yu, L.; Guan, K.; Hu, P.; Li, Y.; Xia, X.; Li, J.; Jiang, S.; Li, F. miR-762 promotes porcine immature Sertoli cell growth via the ring finger protein 4 (RNF4) gene. Sci. Rep. 2016, 6, 32783. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Guan, K.; Feng, Y.; Ma, C.; Song, H.; Li, Y.; Xia, X.; Li, J.; Li, F. miR-638 Inhibits immature Sertoli cell growth by indirectly inactivating PI3K/AKT pathway via SPAG1 gene. Cell Cycle 2017, 16, 2290–2300. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Z.J.; Yi, W.; Rong, Y.W.; Kee, J.D.; Zhong, W.X. MicroRNA-1285 Regulates 17beta-Estradiol-Inhibited Immature Boar Sertoli Cell Proliferation via Adenosine Monophosphate-Activated Protein Kinase Activation. Endocrinology 2015, 156, 4059–4070. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Guo, J.; Liang, M.; Qi, J.; Wang, Z.; Jian, X.; Zhang, Z.; Sun, B.; Li, Z. miR-196a Promotes Proliferation and Inhibits Apoptosis of Immature Porcine Sertoli Cells. DNA Cell Biol. 2019, 38, 41–48. [Google Scholar] [CrossRef]
- Luo, H.; Chen, B.; Weng, B.; Tang, X.; Chen, Y.; Yang, A.; Chu, D.; Zeng, X.; Ran, M. miR-130a promotes immature porcine Sertoli cell growth by activating SMAD5 through the TGF-beta-PI3K/AKT signaling pathway. FASEB J. 2020, 34, 15164–15179. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ran, M.; Luo, H.; Weng, B.; Tang, X.; Chen, Y.; Yang, A.; Chen, B. miR-499 promotes immature porcine Sertoli cell growth by the PI3K/AKT pathway by targeting the PTEN gene. Reproduction 2019, 159, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Ebbesen, K.K.; Hansen, T.B.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045. [Google Scholar] [CrossRef]
- Li, T.; Luo, R.; Wang, X.; Wang, H.; Zhao, X.; Guo, Y.; Jiang, H.; Ma, Y. Unraveling Stage-Dependent Expression Patterns of Circular RNAs and Their Related ceRNA Modulation in Ovine Postnatal Testis Development. Front. Cell Dev. Biol. 2021, 9, 627439. [Google Scholar] [CrossRef]
- Ran, M.L.; Weng, B.; Chen, B.; Wu, M.S.; He, C.Q.; Zhang, S.W. Strand-specific RNA sequencing in pig testes identifies developmentally regulated genes and circular RNAs. Genes Genom. 2017, 39, 1083–1094. [Google Scholar] [CrossRef]
- Liang, G.; Yang, Y.; Niu, G.; Tang, Z.; Li, K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017, 24, 523–535. [Google Scholar] [CrossRef] [Green Version]
- Robic, A.; Cerutti, C.; Kuhn, C.; Faraut, T. Comparative Analysis of the Circular Transcriptome in Muscle, Liver, and Testis in Three Livestock Species. Front. Genet. 2021, 12, 665153. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Patop, I.L.; Wust, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Qin, L.; Lin, J.; Xie, X. CircRNA-9119 suppresses poly I:C induced inflammation in Leydig and Sertoli cells via TLR3 and RIG-I signal pathways. Mol. Med. 2019, 25, 28. [Google Scholar] [CrossRef]
- Ran, M.; Chen, B.; Li, Z.; Wu, M.; Liu, X.; He, C.; Zhang, S.; Li, Z. Systematic Identification of Long Noncoding RNAs in Immature and Mature Porcine Testes. Biol. Reprod. 2016, 94, 77. [Google Scholar] [CrossRef]
- Weng, B.; Ran, M.; Chen, B.; He, C.; Dong, L.; Peng, F. Genome-wide analysis of long non-coding RNAs and their role in postnatal porcine testis development. Genomics 2017, 109, 446–456. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, X.; Ning, W.; Zhang, X.; Ru, Z.; Wang, S.; Sheng, M.; Zhang, J.; Zhang, X.; Luo, H.; et al. Expression Analysis of Circular RNAs in Young and Sexually Mature Boar Testes. Animals 2021, 11, 1430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.A.; Reckman, Y.J.; Aufiero, S.; van den Hoogenhof, M.M.; van der Made, I.; Beqqali, A.; Koolbergen, D.R.; Rasmussen, T.B.; van der Velden, J.; Creemers, E.E.; et al. RBM20 Regulates Circular RNA Production from the Titin Gene. Circ. Res. 2016, 119, 996–1003. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Xing, Y.; Ren, L.; Wang, Y.; Li, Q.; Fu, X.; Yang, Q.; Xu, L.; Willems, L.; Li, J.; et al. Bta-miR-24-3p Controls the Myogenic Differentiation and Proliferation of Fetal, Bovine, Skeletal Muscle-Derived Progenitor Cells by Targeting ACVR1B. Animals 2019, 9, 859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Xue, S.; Feng, Y.; Shen, J.; Zhao, J. MicroRNA-24-3p inhibition prevents cell growth of vascular smooth muscle cells by targeting Bcl-2-like protein 11. Exp. Ther. Med. 2020, 19, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zhong, J.; Li, S.W.; Li, J.Z.; Zhou, M.; Chen, Y.; Sang, Y.; Liu, L. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget 2016, 7, 86755–86765. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.F.; Shan, Z.; Ma, J.Y.; Wang, M.; Zhang, C.X.; Liu, R.M.; Wu, W.B.; Shi, Y.W.; Li, W.; Wang, S.M. Investigating the Role of the Posttranscriptional Gene Regulator MiR-24-3p in the Proliferation, Migration and Apoptosis of Human Arterial Smooth Muscle Cells in Arteriosclerosis Obliterans. Cell Physiol. Biochem. 2015, 36, 1359–1370. [Google Scholar] [CrossRef]
- Ran, M.L.; Chen, B.; Wu, M.S.; Liu, X.C.; He, C.Q.; Yang, A.Q.; Li, Z.; Xiang, Y.J.; Li, Z.H.; Zhang, S.W. Integrated analysis of miRNA and mRNA expression profiles in development of porcine testes. RSC Adv. 2015, 5, 63439–63449. [Google Scholar] [CrossRef]
- Lian, C.; Sun, B.; Niu, S.; Yang, R.; Liu, B.; Lu, C.; Meng, J.; Qiu, Z.; Zhang, L.; Zhao, Z. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J. 2012, 279, 964–975. [Google Scholar] [CrossRef]
- Papaioannou, M.D.; Pitetti, J.L.; Ro, S.; Park, C.; Aubry, F.; Schaad, O.; Vejnar, C.E.; Kuhne, F.; Descombes, P.; Zdobnov, E.M.; et al. Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 2009, 326, 250–259. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zheng, Y.; Li, X.; Gao, Q.; Feng, T.; Zhang, P.; Liao, M.; Tian, X.; Lu, H.; Zeng, W. Profiling of miRNAs in porcine Sertoli cells. J. Anim. Sci. Biotechnol. 2020, 11, 85. [Google Scholar] [CrossRef]
- Tesser-Gamba, F.; Lopes, L.J.; Petrilli, A.S.; Toledo, S.R. MAPK7 gene controls proliferation, migration and cell invasion in osteosarcoma. Mol. Carcinog. 2016, 55, 1700–1713. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Mohanmed, M.M.; Li, Z.; Zhu, L.; Zhang, Q.; Huang, Q.; Shen, W. Capn4 promotes colorectal cancer cell proliferation by increasing MAPK7 through activation of the Wnt/beta-Catenin pathway. Exp. Cell Res. 2018, 363, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.D.; Hao, S.L.; Yang, W.X. Multiple signaling pathways in Sertoli cells: Recent findings in spermatogenesis. Cell Death Dis. 2019, 10, 541. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.H.; Cheng, C.Y. Mitogen-activated protein kinases, adherens junction dynamics, and spermatogenesis: A review of recent data. Dev. Biol. 2005, 286, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Gautam, M.; Bhattacharya, I.; Rai, U.; Majumdar, S.S. Hormone induced differential transcriptome analysis of Sertoli cells during postnatal maturation of rat testes. PLoS ONE 2018, 13, e0191201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.; Tang, X.; Wei, Y.; Long, C.; Tan, B.; Wu, S.; Sun, M.; Zhou, Y.; Cao, X.; Wei, G. Vitamin E and vitamin C attenuate Di-(2-ethylhexyl) phthalate-induced blood-testis barrier disruption by p38 MAPK in immature SD rats. Reprod. Toxicol. 2018, 81, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, L.; Wei, J.; Zhang, J.; Zhu, Y.; Li, X.; Jing, L.; Duan, J.; Zhou, X.; Sun, Z. Fine particle matter disrupts the blood-testis barrier by activating TGF-beta3/p38 MAPK pathway and decreasing testosterone secretion in rat. Environ. Toxicol. 2018, 33, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Qian, Y.; Liu, Z.; Wang, C.; Qu, J.; Wang, X.; Wang, S. Perfluorooctane sulfonate (PFOS) disrupts blood-testis barrier by down-regulating junction proteins via p38 MAPK/ATF2/MMP9 signaling pathway. Toxicology 2016, 373, 1–12. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, Q.; Chen, B.; Weng, B.; Chu, D.; Tang, X.; Yan, S.; Yin, Y.; Ran, M. circBTBD7 Promotes Immature Porcine Sertoli Cell Growth through Modulating miR-24-3p/MAPK7 Axis to Inactivate p38 MAPK Signaling Pathway. Int. J. Mol. Sci. 2021, 22, 9385. https://doi.org/10.3390/ijms22179385
Bian Q, Chen B, Weng B, Chu D, Tang X, Yan S, Yin Y, Ran M. circBTBD7 Promotes Immature Porcine Sertoli Cell Growth through Modulating miR-24-3p/MAPK7 Axis to Inactivate p38 MAPK Signaling Pathway. International Journal of Molecular Sciences. 2021; 22(17):9385. https://doi.org/10.3390/ijms22179385
Chicago/Turabian StyleBian, Qiao, Bin Chen, Bo Weng, Dan Chu, Xiangwei Tang, Saina Yan, Yanfei Yin, and Maoliang Ran. 2021. "circBTBD7 Promotes Immature Porcine Sertoli Cell Growth through Modulating miR-24-3p/MAPK7 Axis to Inactivate p38 MAPK Signaling Pathway" International Journal of Molecular Sciences 22, no. 17: 9385. https://doi.org/10.3390/ijms22179385
APA StyleBian, Q., Chen, B., Weng, B., Chu, D., Tang, X., Yan, S., Yin, Y., & Ran, M. (2021). circBTBD7 Promotes Immature Porcine Sertoli Cell Growth through Modulating miR-24-3p/MAPK7 Axis to Inactivate p38 MAPK Signaling Pathway. International Journal of Molecular Sciences, 22(17), 9385. https://doi.org/10.3390/ijms22179385




