Design and Characterization of a Cell-Penetrating Peptide Derived from the SOX2 Transcription Factor
Abstract
1. Introduction
2. Results and Discussion
2.1. Association between SOX2 Expression and Survival of Patients with Breast Cancer
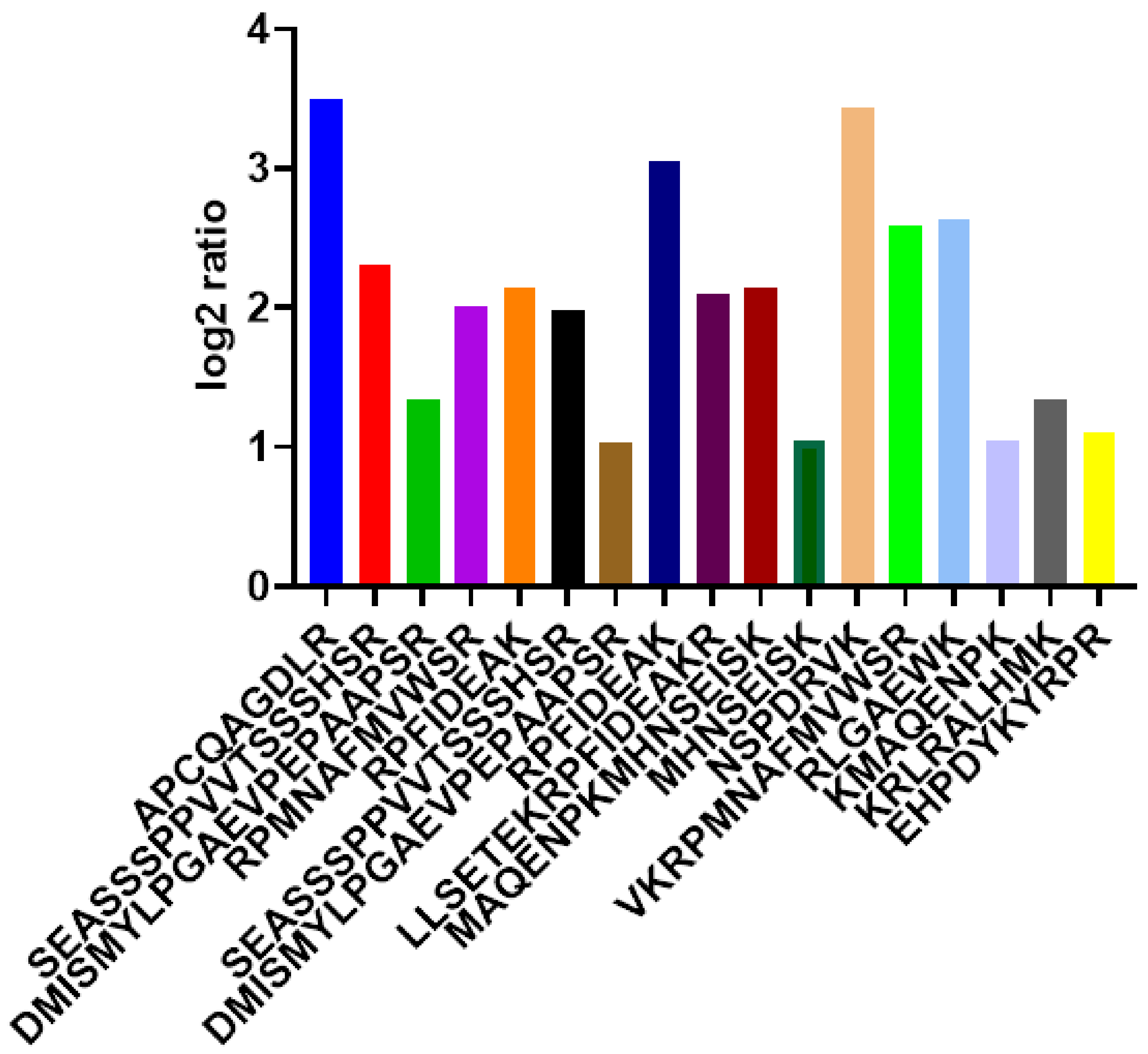
2.2. Peptide Identification and Quantification
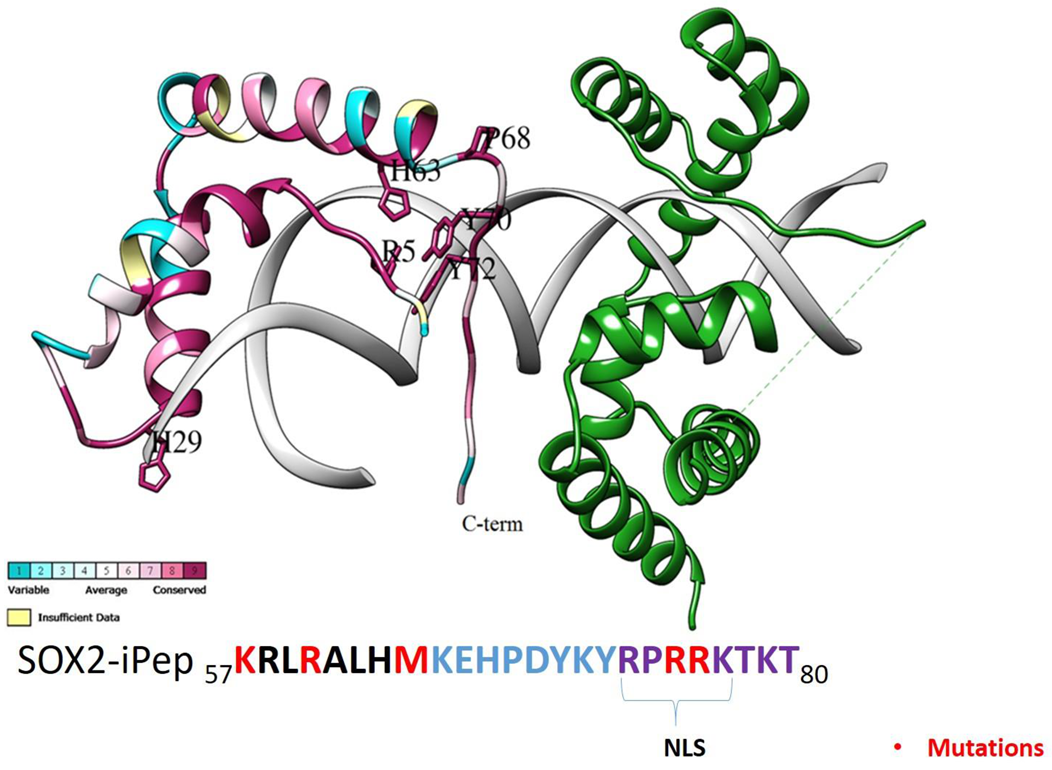
2.3. Design of a SOX2 Interference Peptide (iPep)
2.4. Cellular Internalization and Biological Effects of SOX2 iPep in Basal-like Breast Cancer Cells
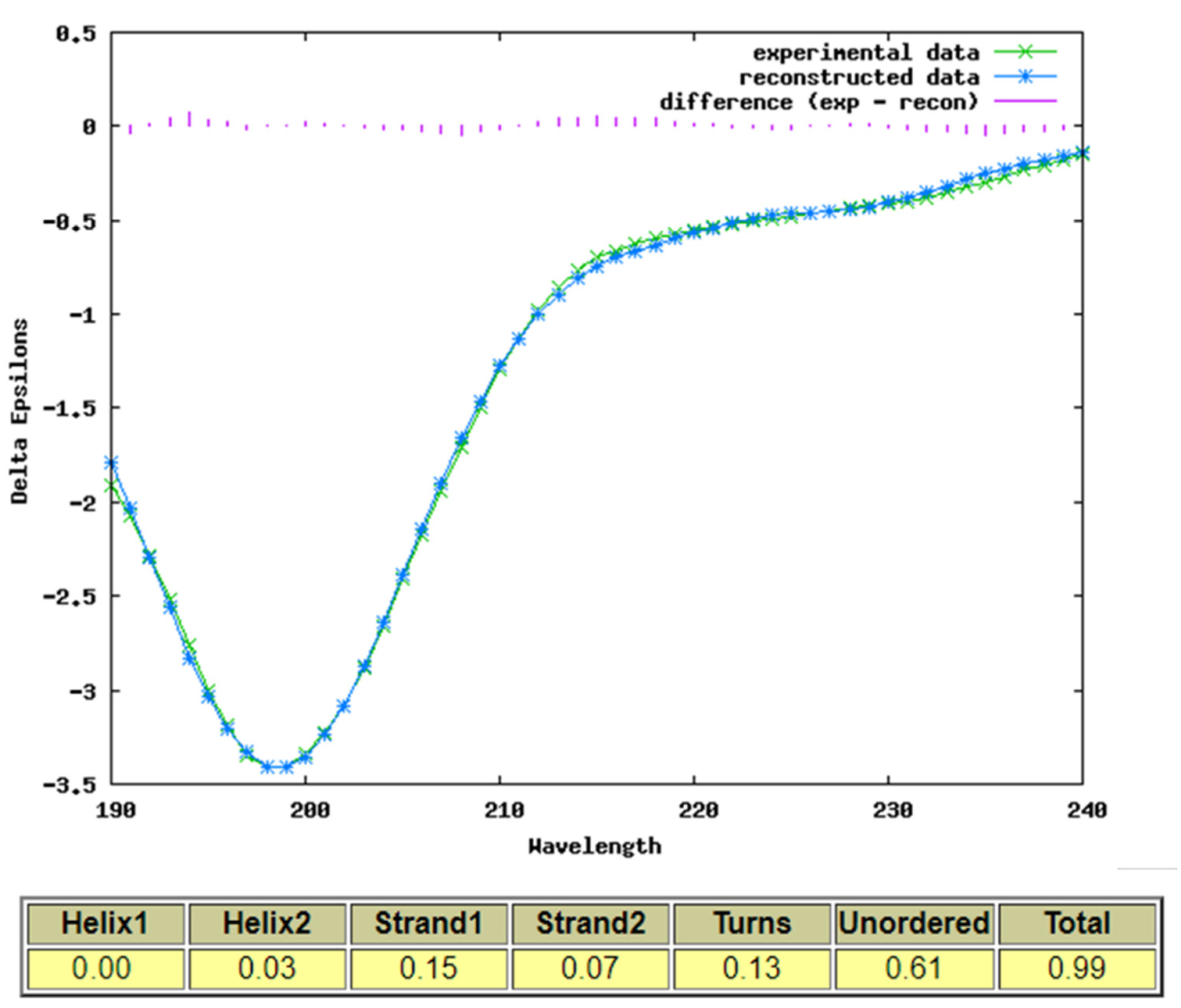
2.5. Secondary Structure of SOX2 iPEP
2.6. MD Simulations
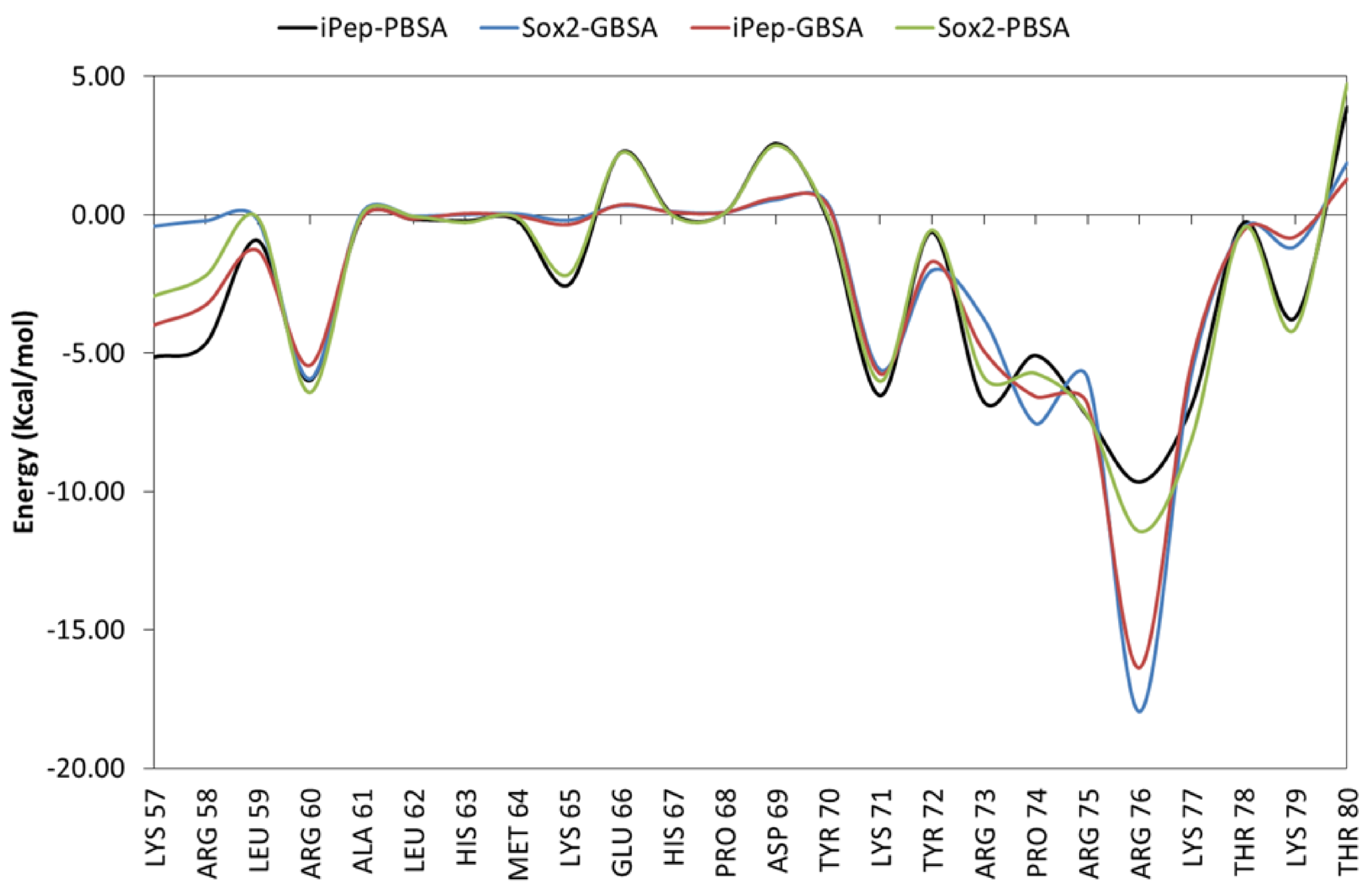
2.6.1. Free Energies of Binding of SOX2 and iPep to DNA in the Presence and Absence of OCT4
2.6.2. Role of Water Molecules in the Interactions of SOX2 with DNA
3. Materials and Methods
3.1. Kaplan Meier Survival Curves
3.2. Identification and Quantification of SOX2 Peptides by MaxQuant
3.3. Cell Line and Culture Conditions
3.4. Cellular Internalisation Assay
3.5. Cell Proliferation and Apoptosis Assay
3.6. Western Blot
3.7. Circular Dichroism (CD) Spectroscopy-
3.8. Molecular Dynamics Simulation of the Interaction of the SOX2 iPep with DNA
3.9. Molecular Dynamics Simulations
3.10. Free Energy of Binding Calculations
3.11. Molecular Entropies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hughes, T.R. Introduction to “A Handbook of Transcription Factors”. In A Handbook of Transcription Factors; Hughes, T.R., Ed.; Subcellular Biochemistry; Springer: Amsterdam, The Netherlands, 2011; Volume 52, pp. 1–6. [Google Scholar]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Luscombe, N.M.; Austin, S.E.; Berman, H.M.; Thornton, J.M. An overview of the structures of protein-DNA complexes. Genome Biol. 2000, 1, 1001. [Google Scholar] [CrossRef]
- Beltran, A.; Rivenbark, A.; Richardson, B.; Yuan, X.; Quian, H.; Hunt, J.; Zimmerman, E.; Graves, L.; Blancafort, P. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res. 2011, 13, R94. [Google Scholar] [CrossRef]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef]
- Raman, V.; Martensen, S.A.; Reisman, D.; Evron, E.; Odenwald, W.F.; Jaffee, E.; Marks, J.; Sukumar, S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 2000, 405, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Gubbay, J.; Collignon, J.; Koopman, P.; Capel, B.; Economou, A.; Munsterberg, A.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 1990, 346, 245–250. [Google Scholar] [CrossRef]
- Wegner, M. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 1999, 27, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cui, W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 2014, 6, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Masui, S.; Nakatake, Y.; Toyooka, Y.; Shimosato, D.; Yagi, R.; Takahashi, K.; Okochi, H.; Okuda, A.; Matoba, R.; Sharov, A.A.; et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007, 9, 625–635. [Google Scholar] [CrossRef]
- Lu, Y.; Futtner, C.; Rock, J.R.; Xu, X.; Whitworth, W.; Hogan, B.L.M.; Onaitis, M.W. Evidence That SOX2 Overexpression Is Oncogenic in the Lung. PLoS ONE 2010, 5, e11022. [Google Scholar] [CrossRef] [PubMed]
- Hussenet, T.; Dali, S.; Exinger, J.; Monga, B.; Jost, B.; Dembelé, D.; Martinet, N.; Thibault, C.; Huelsken, J.; Brambilla, E.; et al. SOX2 Is an Oncogene Activated by Recurrent 3q26.3 Amplifications in Human Lung Squamous Cell Carcinomas. PLoS ONE 2010, 5, e8960. [Google Scholar] [CrossRef]
- Wilson, M.; Koopman, P. Matching SOX: Partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 2002, 12, 441–446. [Google Scholar] [CrossRef]
- Weiss, M.A. Floppy SOX: Mutual Induced Fit in HMG (High-Mobility Group) Box-DNA Recognition. Mol. Endocrinol. 2001, 15, 353–362. [Google Scholar] [CrossRef]
- Reményi, A.; Lins, K.; Nissen, L.J.; Reinbold, R.; Schöler, H.R.; Wilmanns, M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003, 17, 2048–2059. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.C.; Cai, M.; Clore, G.M. Molecular Basis for Synergistic Transcriptional Activation by Oct1 and Sox2 Revealed from the Solution Structure of the 42-kDa Oct1·Sox2·Hoxb1-DNA Ternary Transcription Factor Complex. J. Biol. Chem. 2004, 279, 1449–1457. [Google Scholar] [CrossRef]
- Reményi, A.; Tomilin, A.; Pohl, E.; Lins, K.; Philippsen, A.; Reinbold, R.; Schöler, H.R.; Wilmanns, M. Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping. Mol. Cell 2001, 8, 569–580. [Google Scholar] [CrossRef]
- Beltran, A.S.; Graves, L.M.; Blancafort, P. Novel role of Engrailed 1 as a prosurvival transcription factor in basal-like breast cancer and engineering of interference peptides block its oncogenic function. Oncogene 2014, 33, 4767–4777. [Google Scholar] [CrossRef]
- Sorolla, A.; Wang, E.; Golden, E.; Duffy, C.; Henriques, S.T.; Redfern, A.D.; Blancafort, P. Precision medicine by designer interference peptides: Applications in oncology and molecular therapeutics. Oncogene 2020, 39, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Sorolla, A.; Ho, D.; Wang, E.; Evans, C.W.; Ormonde, C.F.G.; Rashwan, R.; Singh, R.; Iyer, K.S.; Blancafort, P. Sensitizing basal-like breast cancer to chemotherapy using nanoparticles conjugated with interference peptide. Nanoscale 2016, 8, 9343–9353. [Google Scholar] [CrossRef]
- Sorolla, A.; Wang, E.; Clemons, T.D.; Evans, C.W.; Plani-Lam, J.H.; Golden, E.; Dessauvagie, B.; Redfern, A.D.; Swaminathan-Iyer, K.; Blancafort, P. Triple-hit therapeutic approach for triple negative breast cancers using docetaxel nanoparticles, EN1-iPeps and RGD peptides. Nanomed. Nanotechnol. Biol. Med. 2019, 20, 102003. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Blancafort, P.; Mancera, R.L. Atomistic molecular dynamics simulations of bioactive engrailed 1 interference peptides (EN1-iPeps). Oncotarget 2018, 9, 22383–22397. [Google Scholar] [CrossRef]
- Morris, M.C.; Deshayes, S.; Heitz, F.; Divita, G. Cell-penetrating peptides: From molecular mechanisms to therapeutics. Biol. Cell 2008, 100, 201–217. [Google Scholar] [CrossRef]
- Harreither, E.; Rydberg, H.A.; Amand, H.L.; Jadhav, V.; Fliedl, L.; Benda, C.; Esteban, M.A.; Pei, D.; Borth, N.; Grillari-Voglauer, R.; et al. Characterization of a novel cell penetrating peptide derived from human Oct4. Cell Regen. 2014, 3, 2. [Google Scholar] [CrossRef]
- Park, S.B.; Seo, K.W.; So, A.Y.; Seo, M.S.; Yu, K.R.; Kang, S.K.; Kang, K.S. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2012, 19, 534–545. [Google Scholar] [CrossRef]
- Gangemi, R.M.; Griffero, F.; Marubbi, D.; Perera, M.; Capra, M.C.; Malatesta, P.; Ravetti, G.L.; Zona, G.L.; Daga, A.; Corte, G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells Dayt. Ohio 2009, 27, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Stolzenburg, S.; Rots, M.G.; Beltran, A.S.; Rivenbark, A.G.; Yuan, X.; Qian, H.; Strahl, B.D.; Blancafort, P. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res 2012, 40, 6725–6740. [Google Scholar] [CrossRef] [PubMed]
- Duffy, C.; Sorolla, A.; Wang, E.; Golden, E.; Woodward, E.; Davern, K.; Ho, D.; Johnstone, E.; Pfleger, K.; Redfern, A.; et al. Honeybee venom and melittin suppress growth factor receptor activation in HER2-enriched and triple-negative breast cancer. NPJ Precis. Oncol. 2020, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Rai, Y.; Pathak, R.; Kumari, N.; Sah, D.K.; Pandey, S.; Kalra, N.; Soni, R.; Dwarakanath, B.S.; Bhatt, A.N. Mitochondrial biogenesis and metabolic hyperactivation limits the application of MTT assay in the estimation of radiation induced growth inhibition. Sci. Rep. 2018, 8, 1531. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pinilla, S.M.; Sarrio, D.; Moreno-Bueno, G.; Rodriguez-Gil, Y.; Martinez, M.A.; Hernandez, L.; Hardisson, D.; Reis-Filho, J.S.; Palacios, J. Sox2: A possible driver of the basal-like phenotype in sporadic breast cancer. Mod. Pathol. 2007, 20, 474–481. [Google Scholar] [CrossRef]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, F.; Wang, J.; Yin, F.; Xu, Z.; Qi, D.; Wu, X.; Cao, Y.; Liang, W.; Liu, Y.; et al. Pluripotent Stem Cell Protein Sox2 Confers Sensitivity to LSD1 Inhibition in Cancer Cells. Cell Rep. 2013, 5, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Tonelli, M.; Sahu, S.C.; Singarapu, K.K.; Eghbalnia, H.R.; Markley, J.L. Robust, Integrated Computational Control of NMR Experiments to Achieve Optimal Assignment by ADAPT-NMR. PLoS ONE 2012, 7, e33173. [Google Scholar] [CrossRef]
- Jagga, B.; Edwards, M.; Pagin, M.; Wagstaff, K.M.; Aragão, D.; Roman, N.; Nanson, J.D.; Raidal, S.R.; Dominado, N.; Stewart, M.; et al. Structural basis for nuclear import selectivity of pioneer transcription factor SOX2. Nat. Commun. 2021, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.K.; Grand, R.S.; Isbel, L.; Cavadini, S.; Kozicka, Z.; Kempf, G.; Bunker, R.D.; Schenk, A.D.; Graff-Meyer, A.; Pathare, G.R.; et al. Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science 2020, 368, 1460. [Google Scholar] [CrossRef]
- Dodonova, S.O.; Zhu, F.; Dienemann, C.; Taipale, J.; Cramer, P. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 2020, 580, 669–672. [Google Scholar] [CrossRef]
- Milletti, F. Cell-penetrating peptides: Classes, origin, and current landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Joliot, A.; Prochiantz, A. Homeoproteins as natural Penetratin cargoes with signaling properties. Adv. Drug Deliv. Rev. 2008, 60, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Giorello, L.; Clerico, L.; Pescarolo, M.P.; Vikhanskaya, F.; Salmona, M.; Colella, G.; Bruno, S.; Mancuso, T.; Bagnasco, L.; Russo, P.; et al. Inhibition of Cancer Cell Growth and c-Myc Transcriptional Activity by a c-Myc Helix 1-Type Peptide Fused to an Internalization Sequence. Cancer Res. 1998, 58, 3654–3659. [Google Scholar]
- Li, L.; Sun, W.; Zhang, Z.; Huang, Y. Time-staggered delivery of docetaxel and H1-S6A, F8A peptide for sequential dual-strike chemotherapy through tumor priming and nuclear targeting. J. Control. Release 2016, 232, 62–74. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Z.; Li, L.; Chen, B.-C.; Revyakin, A.; Hajj, B.; Legant, W.; Dahan, M.; Lionnet, T.; Betzig, E.; et al. Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell 2014, 156, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Lodato, M.A.; Ng, C.W.; Wamstad, J.A.; Cheng, A.W.; Thai, K.K.; Fraenkel, E.; Jaenisch, R.; Boyer, L.A. SOX2 Co-Occupies Distal Enhancer Elements with Distinct POU Factors in ESCs and NPCs to Specify Cell State. PLoS Genet. 2013, 9, e1003288. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, Y.; Uchikawa, M.; Kondoh, H. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. 2000, 16, 182–187. [Google Scholar] [CrossRef]
- Ambrosetti, D.C.; Basilico, C.; Dailey, L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997, 17, 6321–6329. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, S.; Peng, B.; Ye, Y.; Deng, X.; Yao, K. Embryonic Stem Cells Markers SOX2, OCT4 and Nanog Expression and Their Correlations with Epithelial-Mesenchymal Transition in Nasopharyngeal Carcinoma. PLoS ONE 2013, 8, e56324. [Google Scholar] [CrossRef]
- Tayal, N.; Choudhary, P.; Pandit, S.B.; Sandhu, K.S. Evolutionarily conserved and conformationally constrained short peptides might serve as DNA recognition elements in intrinsically disordered regions. Mol. Biosyst. 2014, 10, 1469–1480. [Google Scholar] [CrossRef]
- Narasimhan, K.; Pillay, S.; Bin, A.N.R.; Bikadi, Z.; Hazai, E.; Yan, L.; Kolatkar, P.R.; Pervushin, K.; Jauch, R. Identification of a Polyoxometalate Inhibitor of the DNA Binding Activity of Sox2. ACS Chem. Biol. 2011, 6, 573–581. [Google Scholar] [CrossRef]
- Scaffidi, P.; Bianchi, M.E. Spatially Precise DNA Bending Is an Essential Activity of the Sox2 Transcription Factor. J. Biol. Chem. 2001, 276, 47296–47302. [Google Scholar] [CrossRef]
- Zhu, Y.-L.; Beroza, P.; Artis, D.R. Including Explicit Water Molecules as Part of the Protein Structure in MM/PBSA Calculations. J. Chem. Inf. Modeling 2014, 54, 462–469. [Google Scholar] [CrossRef]
- Wong, S.; Amaro, R.E.; McCammon, J.A. MM-PBSA Captures Key Role of Intercalating Water Molecules at a Protein−Protein Interface. J. Chem. Theory Comput. 2009, 5, 422–429. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Rueda, O.M.; Sammut, S.-J.; Seoane, J.A.; Chin, S.-F.; Caswell-Jin, J.L.; Callari, M.; Batra, R.; Pereira, B.; Bruna, A.; Ali, H.R.; et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019, 567, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Case, D.; Darden, T.; Cheatham, T.E., III; Simmerling, C.; Wang, J.; Duke, R.; Luo, R.; Walker, R.; Zhang, W.; Merz, K. AMBER 12; University of California: San Francisco, CA, USA, 2012; p. 142. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. How to obtain statistically converged MM/GBSA results. J. Comput. Chem. 2010, 31, 837–846. [Google Scholar] [CrossRef] [PubMed]
- MacKerell, A.D., Jr.; Nilsson, L. Molecular dynamics simulations of nucleic acid–protein complexes. Curr. Opin. Struct. Biol. 2008, 18, 194–199. [Google Scholar] [CrossRef]
- Merino, F.N.; Calista, K.L.; Veerapandian, V.; Schöler, H.R.; Jauch, R.; Cojocaru, V. Structural Basis for the SOX-Dependent Genomic Redistribution of OCT4 in Stem Cell Differentiation. Structure 2014, 22, 1274–1286. [Google Scholar] [CrossRef][Green Version]
- Srinivasan, J.; Cheatham, T.E.; Cieplak, P.; Kollman, P.A.; Case, D.A. Continuum Solvent Studies of the Stability of DNA, RNA, and Phosphoramidate−DNA Helices. J. Am. Chem. Soc. 1998, 120, 9401–9409. [Google Scholar] [CrossRef]
- Tan, C.; Yang, L.; Luo, R. How Well Does Poisson−Boltzmann Implicit Solvent Agree with Explicit Solvent? A Quantitative Analysis. J. Phys. Chem. B 2006, 110, 18680–18687. [Google Scholar] [CrossRef] [PubMed]
- Tsui, V.; Case, D.A. Theory and applications of the generalized born solvation model in macromolecular simulations. Biopolymers 2000, 56, 275–291. [Google Scholar] [CrossRef]
- Tan, C.; Tan, Y.-H.; Luo, R. Implicit Nonpolar Solvent Models. J. Phys. Chem. B 2007, 111, 12263–12274. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins: Struct. Funct. Bioinform. 2004, 55, 383–394. [Google Scholar] [CrossRef]
- Massova, I.; Kollman, P.A. Computational Alanine Scanning To Probe Protein−Protein Interactions: A Novel Approach To Evaluate Binding Free Energies. J. Am. Chem. Soc. 1999, 121, 8133–8143. [Google Scholar] [CrossRef]
- Gohlke, H.; Kiel, C.; Case, D.A. Insights into Protein–Protein Binding by Binding Free Energy Calculation and Free Energy Decomposition for the Ras–Raf and Ras–RalGDS Complexes. J. Mol. Biol. 2003, 330, 891–913. [Google Scholar] [CrossRef]
- Carrington, B.J.; Mancera, R.L. Comparative estimation of vibrational entropy changes in proteins through normal modes analysis. J. Mol. Graph. Model. 2004, 23, 167–174. [Google Scholar] [CrossRef]
- Rodgers, T.; Burnell, D.; Townsend, P.; Pohl, E.; Cann, M.; Wilson, M.; McLeish, T. ΔΔPT: A comprehensive toolbox for the analysis of protein motion. BMC Bioinform. 2013, 14, 183. [Google Scholar] [CrossRef][Green Version]
- Schlitter, J. Estimation of absolute and relative entropies of macromolecules using the covariance matrix. Chem. Phys. Lett. 1993, 215, 617–621. [Google Scholar] [CrossRef]
- Hill, T.L. An Introduction to Statistical Thermodynamics (Dover Books on Physics), 2nd ed.; Courier Corporation: North Chelmsford, MA, USA, 2012; p. 544. [Google Scholar]


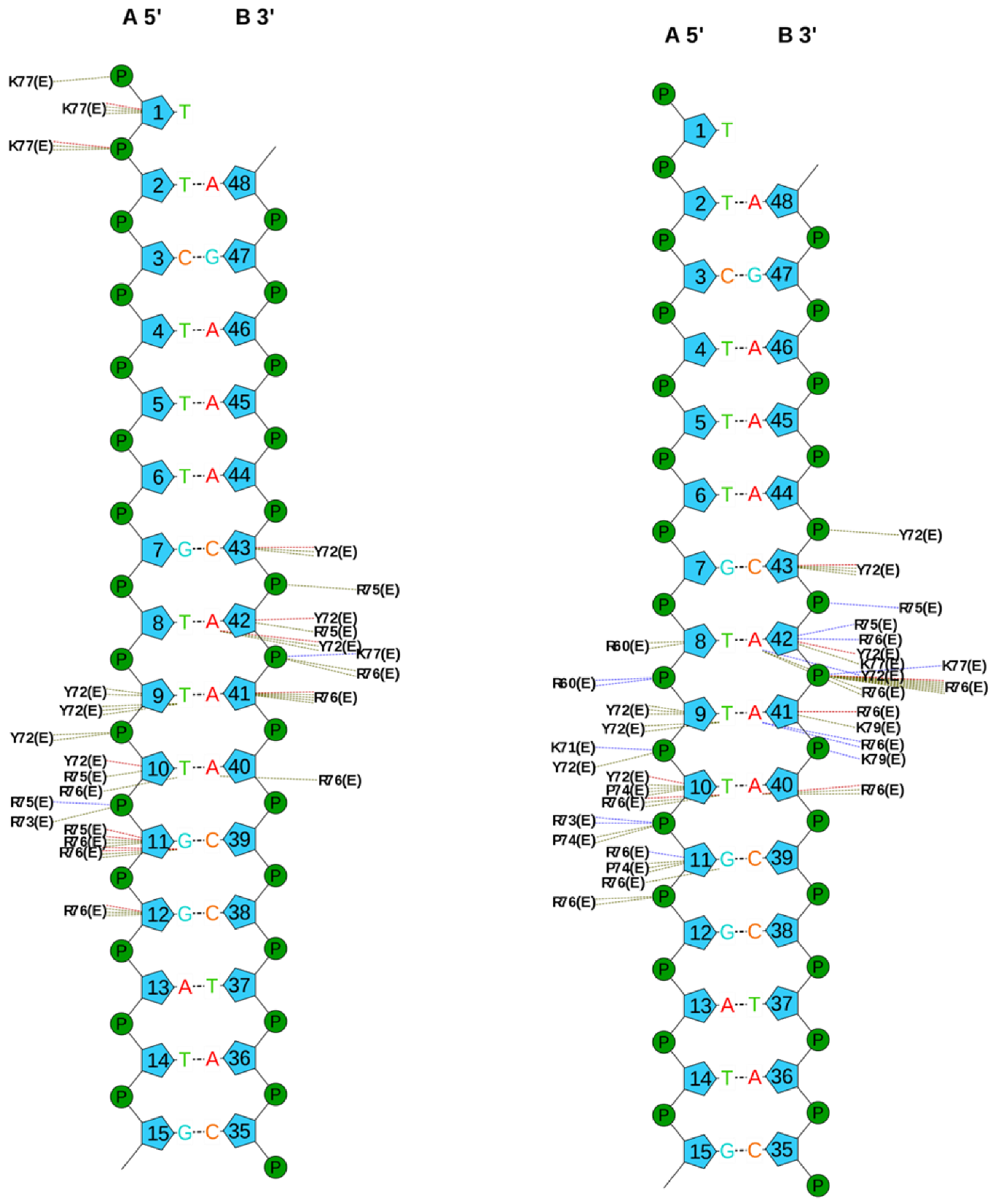
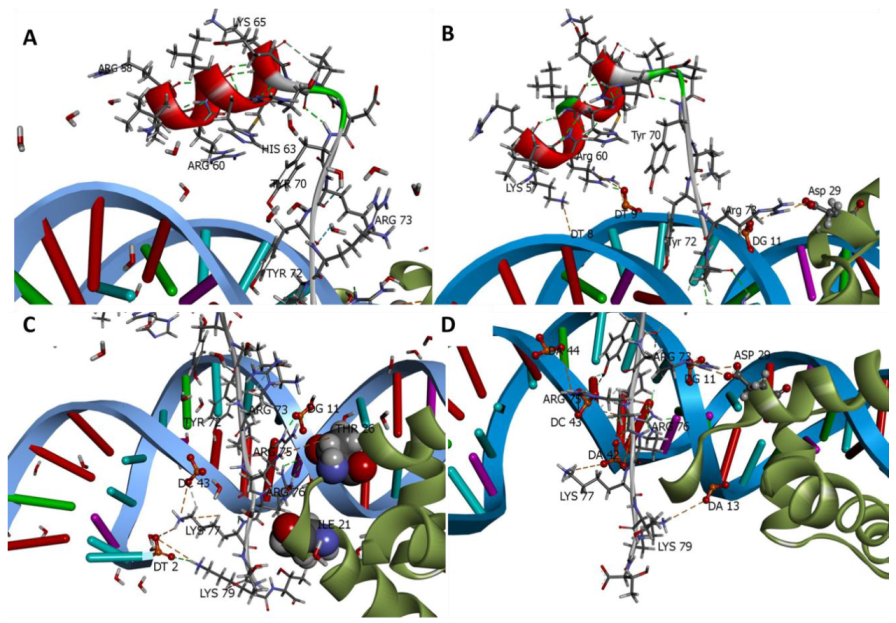

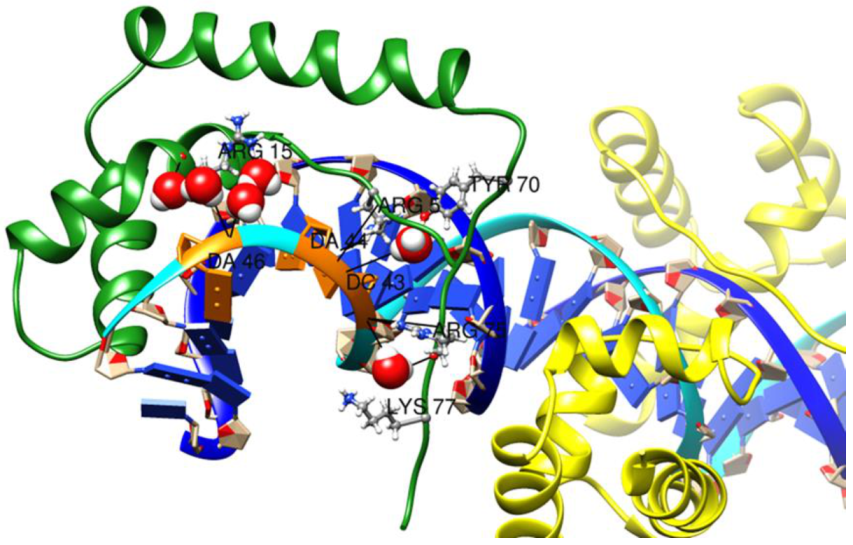
| Energy Term | MM/GBSA | MM/PBSA | ||
|---|---|---|---|---|
| SOX2 | iPep | SOX2 | iPep | |
| ΔEvdw | −197.2 | −84.5 | −197.2 | −85.5 |
| ΔEelec | −8423.8 | −4552.3 | −8423.8 | −4552.3 |
| ΔGgas (ΔEvdw + ΔEelec) | −8620.9 | −4636.9 | −8620.9 | −4636.9 |
| ΔGGB | 8407.9 | 4555.1 | ||
| ΔGPB | 8376.2 | 4532.0 | ||
| ΔGsurface | −23.9 | −11.3 | ||
| ΔGnon-polar | −127.7 | −60.7 | ||
| ΔEdispersion | 254.0 | 117.0 | ||
| ΔEelec + ΔGGB/ΔGPB | −15.9 | 2.8 | −47.6 | −20.4 |
| ΔGsolvated | 8383.9 | 4543.8 | 8502.5 | 4588.2 |
| ΔG | −237.0 | −93.1 | −118.4 | −48.7 |
| TΔS | −44.3 | −37.3 | −44.3 | −37.3 |
| ΔGbinding | −192.7 | −55.8 | −74.1 | −11.4 |
| Hydrogen Bonds with Sox2 | H2O Mediated Hydrogen Bond | Without Crystallographic Water Molecules | With Crystallographic Water Molecules |
|---|---|---|---|
| DC3-Ser31 | No | 7.5% | 0% |
| DT4-Ser34 | No | 82.8% | 99.1% |
| DG7-Trp41 | No | 53.9% | 58.3% |
| DT9-Arg5 | No | 1.8% | 2.0% |
| DG11-Arg75 | No | 15.6% | 13.9% |
| DA42-Tyr72 | No | 67.3% | 88.5% |
| DC43-Arg5 | No | 85.4% | 88.2–90.0% * |
| DC43-Arg75 | Yes | 69.8% | 30.3% |
| DA44-Val3 | Yes | 16.9% | 18.5% |
| DA44-Tyr70 | Yes | 12.8% | 8.7% |
| DA44-Val 3-Tyr70 | Yes | 61.9% | 64.4% |
| DA46-Arg15 | No | 43.7–47.5% | 65.0% |
| DG47-Asn30 | No | 48.6% | 65.4% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gandhi, N.S.; Wang, E.; Sorolla, A.; Kan, Y.J.; Malik, A.; Batra, J.; Young, K.A.; Tie, W.J.; Blancafort, P.; Mancera, R.L. Design and Characterization of a Cell-Penetrating Peptide Derived from the SOX2 Transcription Factor. Int. J. Mol. Sci. 2021, 22, 9354. https://doi.org/10.3390/ijms22179354
Gandhi NS, Wang E, Sorolla A, Kan YJ, Malik A, Batra J, Young KA, Tie WJ, Blancafort P, Mancera RL. Design and Characterization of a Cell-Penetrating Peptide Derived from the SOX2 Transcription Factor. International Journal of Molecular Sciences. 2021; 22(17):9354. https://doi.org/10.3390/ijms22179354
Chicago/Turabian StyleGandhi, Neha S., Edina Wang, Anabel Sorolla, Yu Jie Kan, Adil Malik, Jyotsna Batra, Kimberly A. Young, Wan Jun Tie, Pilar Blancafort, and Ricardo L. Mancera. 2021. "Design and Characterization of a Cell-Penetrating Peptide Derived from the SOX2 Transcription Factor" International Journal of Molecular Sciences 22, no. 17: 9354. https://doi.org/10.3390/ijms22179354
APA StyleGandhi, N. S., Wang, E., Sorolla, A., Kan, Y. J., Malik, A., Batra, J., Young, K. A., Tie, W. J., Blancafort, P., & Mancera, R. L. (2021). Design and Characterization of a Cell-Penetrating Peptide Derived from the SOX2 Transcription Factor. International Journal of Molecular Sciences, 22(17), 9354. https://doi.org/10.3390/ijms22179354








