The Role of Hexokinase and Hexose Transporters in Preferential Use of Glucose over Fructose and Downstream Metabolic Pathways in the Yeast Yarrowia lipolytica
Abstract
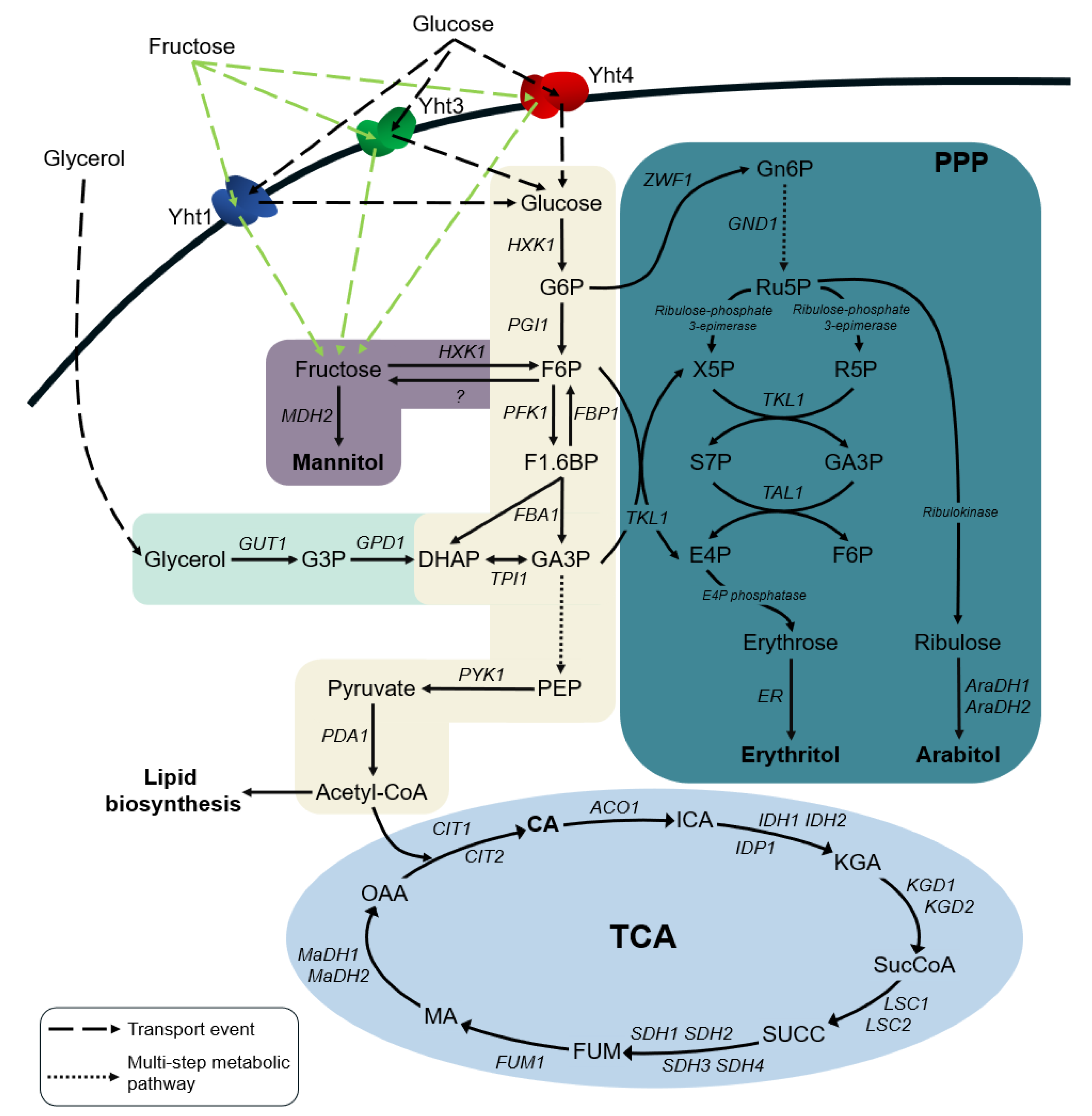
:1. Introduction
2. Results
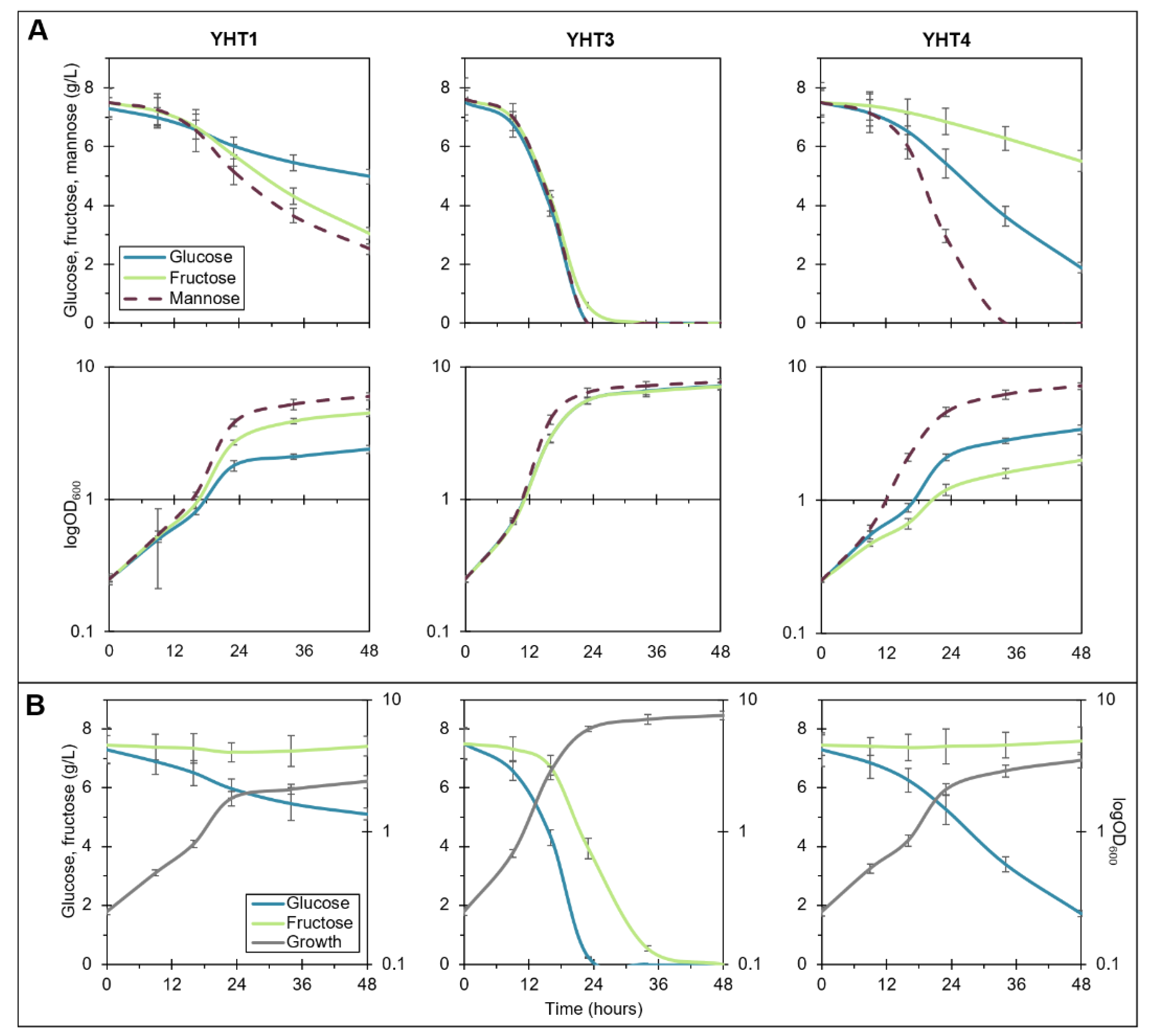
2.1. Expression of Y. lipolytica Hexose Transporters in Heterologous Host Induce Diverse Monosaccharide Utilization Phenotypes
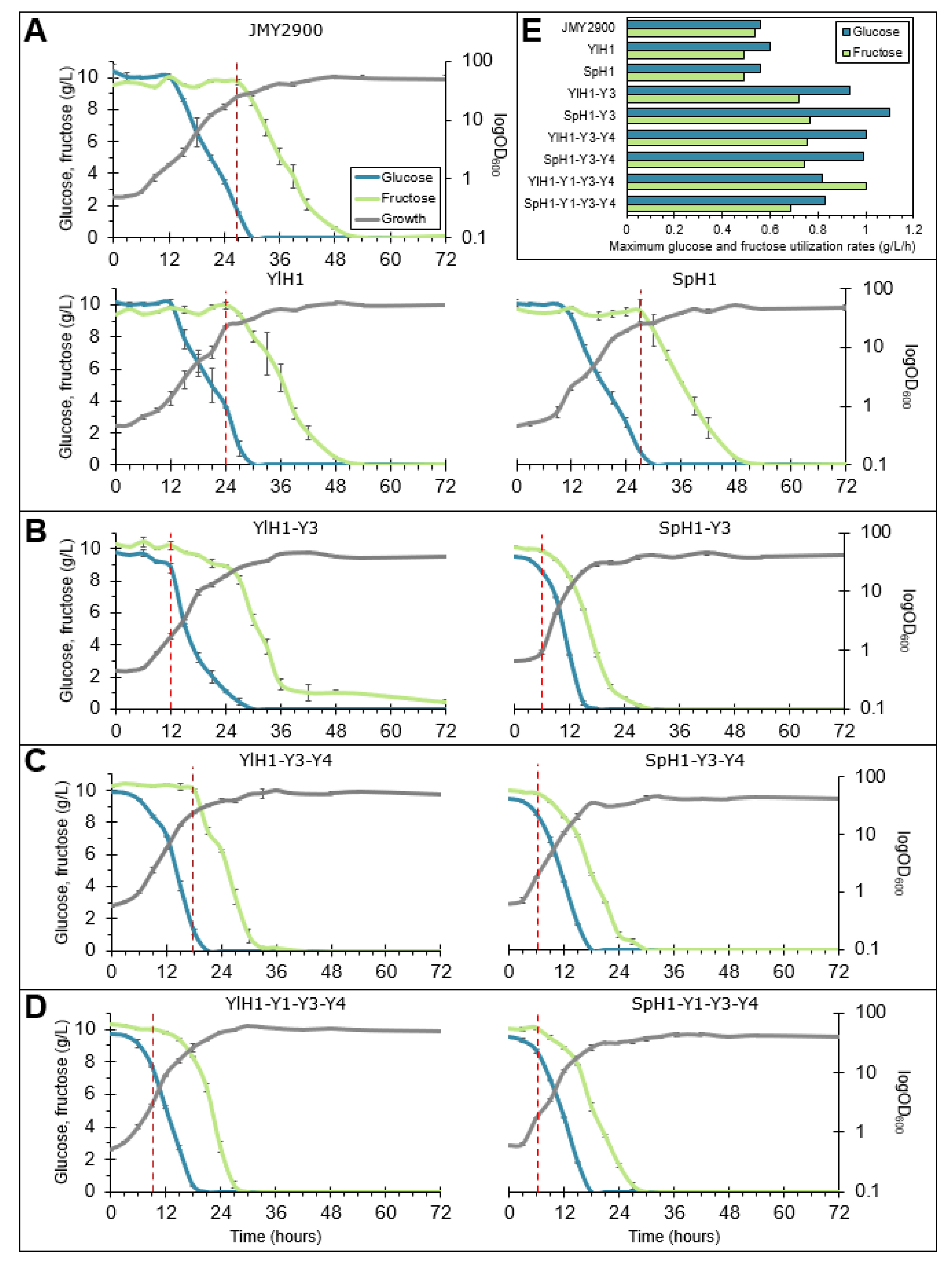
2.2. Construction and Growth Profiling of Y. lipolytica Strains Overexpressing Hexose Transporters and Hexokinases
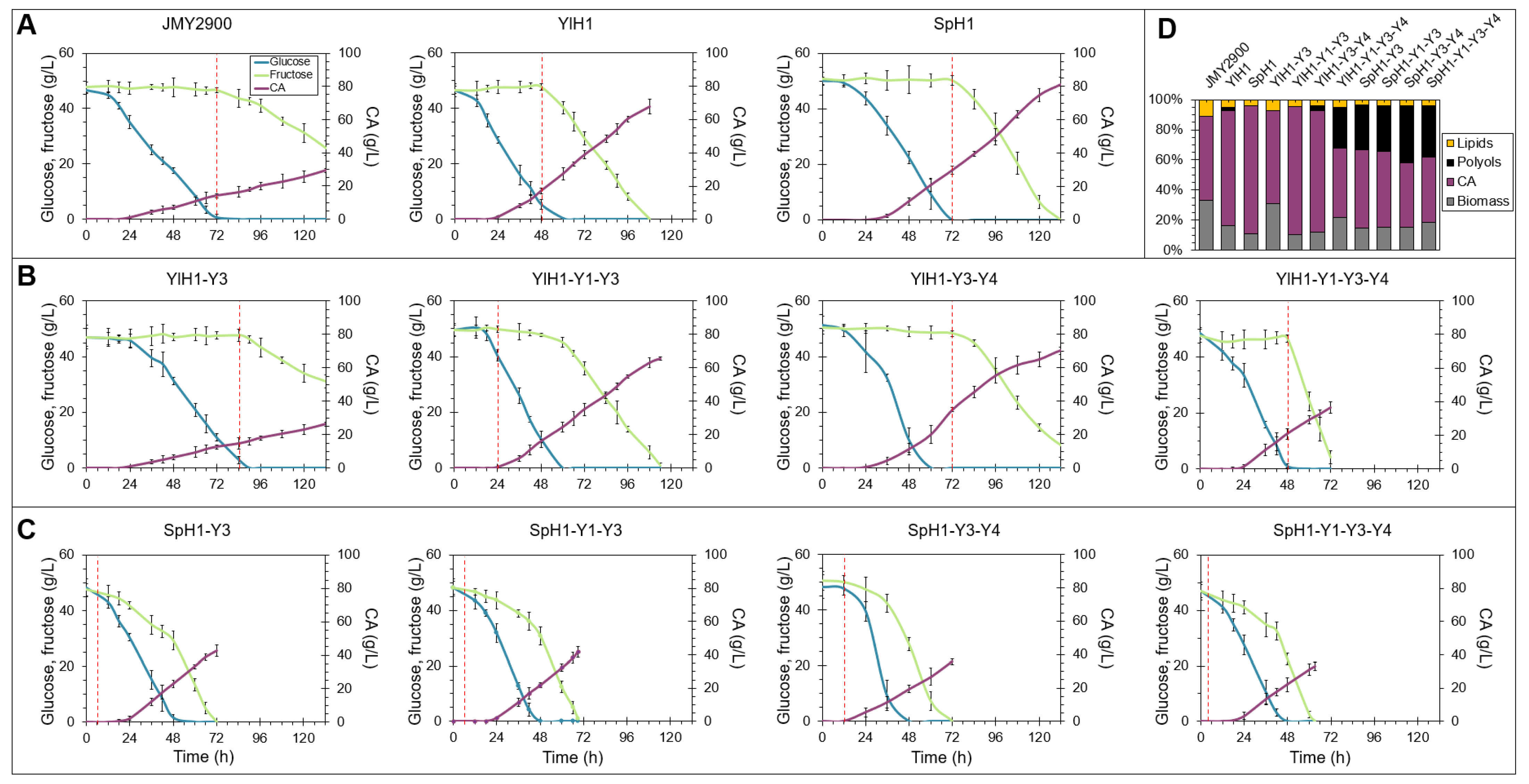
2.3. Rearrangement of Sugar Utilization Profile through Overexpression of Hexose Transporters in Combination with Hexokinases in Y. lipolytica
2.4. The Input Flux of Hexoses Impacts Downstream Metabolic Pathways
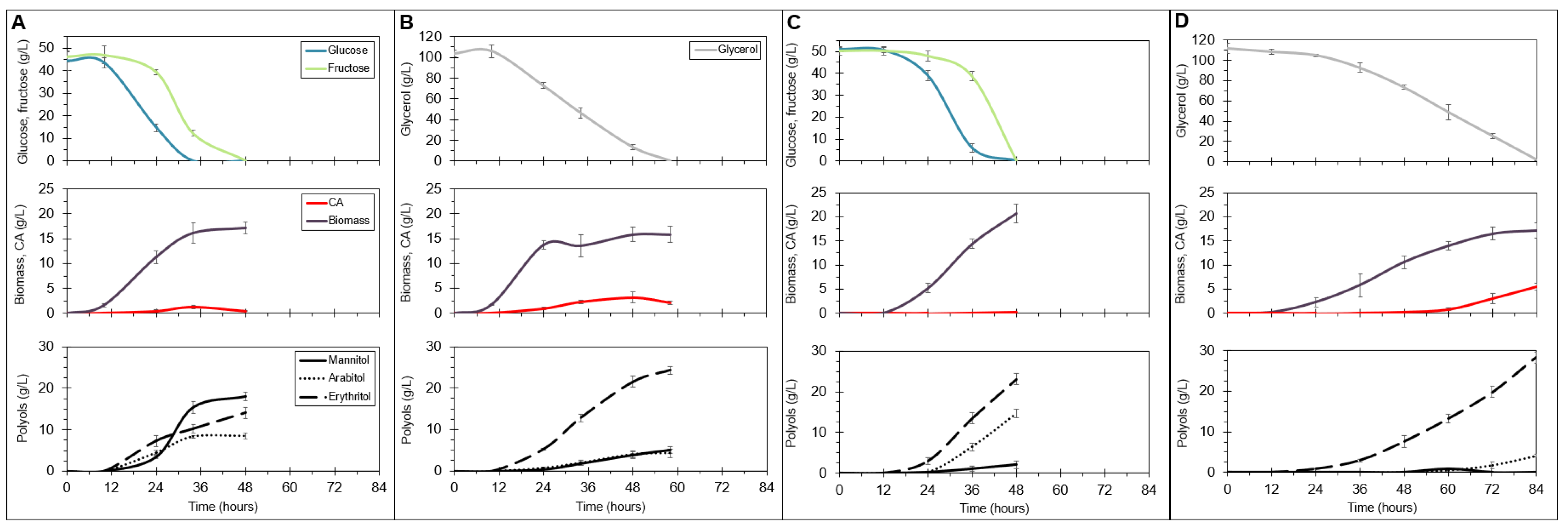
2.5. Overexpression of a Fructophilic Hexokinase and Three Hexose Transporters Induces Rapid Biosynthesis of Polyols from Glucose and Fructose
3. Discussion
4. Materials and Methods
4.1. Culture Media
4.2. Strain Construction
4.3. Microcultivations
4.4. Substrate Utilization Kinetics
4.5. Bioreactor Cultivations
4.6. Analytical Methods
4.7. Gene Expression Analysis
4.8. Calculations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ledesma-Amaro, R.; Nicaud, J.M. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Szczepańska, P.; Hapeta, P.; Lazar, Z. Advances in production of high-value lipids by oleaginous yeasts. Crit. Rev. Biotechnol. 2021, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulermo, R.; Brunel, F.; Dulermo, T.; Ledesma-Amaro, R.; Vion, J.; Trassaert, M.; Thomas, S.; Nicaud, J.M.; Leplat, C. Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microb. Cell Fact. 2017, 16, 31. [Google Scholar] [CrossRef]
- Gasmi, N.; Ayed, A.; Nicaud, J.M.; Kallel, H. Design of an efficient medium for heterologous protein production in Yarrowia lipolytica: Case of human interferon alpha 2b. Microb. Cell Fact. 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rymowicz, W.; Fatykhova, A.R.; Kamzolova, S.V.; Rywińska, A.; Morgunov, I.G. Citric acid production from glycerol-containing waste of biodiesel industry by Yarrowia lipolytica in batch, repeated batch, and cell recycle regimes. Appl. Microbiol. Biotechnol. 2010, 87, 971–979. [Google Scholar] [CrossRef]
- Rakicka, M.; Biegalska, A.; Rymowicz, W.; Dobrowolski, A.; Mirończuk, A.M. Polyol production from waste materials by genetically modified Yarrowia lipolytica. Bioresour. Technol. 2017, 243, 393–399. [Google Scholar] [CrossRef]
- Tomaszewska, L.; Rywińska, A.; Gładkowski, W. Production of erythritol and mannitol by Yarrowia lipolytica yeast in media containing glycerol. J. Ind. Microbiol. Biotechnol. 2012, 39, 1333–1343. [Google Scholar] [CrossRef] [Green Version]
- Hapeta, P.; Kerkhoven, E.J.; Lazar, Z. Nitrogen as the major factor influencing gene expression in Yarrowia lipolytica. Biotechnol. Rep. 2020, 27, e00521. [Google Scholar] [CrossRef]
- Lubuta, P.; Workman, M.; Kerkhoven, E.J.; Workman, C.T. Investigating the influence of glycerol on the utilization of glucose in Yarrowia lipolytica using RNA-Seq-based transcriptomics. G3-Genes Genom Genet. 2019, 9, 4059–4071. [Google Scholar] [CrossRef] [Green Version]
- Rywińska, A.; Rymowicz, W.; Żarowska, B.; Skrzypiński, A. Comparison of citric acid production from glycerol and glucose by different strains of Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2010, 26, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.B.; Zhan, X.B.; Zheng, Z.Y.; Wu, J.R.; Gao, M.J.; Lin, C.C. A novel osmotic pressure control fed-batch fermentation strategy for improvement of erythritol production by Yarrowia lipolytica from glycerol. Bioresour. Technol. 2014, 151, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Barth, G.; Gaillardin, C. Yarrowia lipolytica. In Nonconventional Yeasts in Biotechnology; Springer: Berlin/Heidelberg, Germany, 1996; pp. 313–388. [Google Scholar]
- Lazar, Z.; Gamboa-Meléndez, H.; Crutz-Le Coq, A.M.; Neuvéglise, C.; Nicaud, J.M. Awakening the endogenous Leloir pathway for efficient galactose utilization by Yarrowia lipolytica. Biotechnol. Biofuels 2015, 8, 185. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Lazar, Z.; Rakicka, M.; Guo, Z.; Fouchard, F.; Crutz-Le Coq, A.M.; Nicaud, J.M. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 2016, 38, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Mano, J.; Liu, N.; Hammond, J.H.; Currie, D.H.; Stephanopoulos, G. Engineering Yarrowia lipolytica for the utilization of acid whey. Metab. Eng. 2020, 57, 43–50. [Google Scholar] [CrossRef]
- Lazar, Z.; Walczak, E.; Robak, M. Simultaneous production of citric acid and invertase by Yarrowia lipolytica SUC+ transformants. Bioresour. Technol. 2011, 102, 6982–6989. [Google Scholar] [CrossRef]
- Berthels, N.J.; Otero, R.R.C.; Bauer, F.F.; Pretorius, I.S.; Thevelein, J.M. Correlation between glucose/fructose discrepancy and hexokinase kinetic properties in different Saccharomyces cerevisiae wine yeast strains. Appl. Microbiol. Biotechnol. 2008, 77, 1083–1091. [Google Scholar] [CrossRef]
- Förster, A.; Aurich, A.; Mauersberger, S.; Barth, G. Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2007, 75, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Lazar, Z.; Neuvéglise, C.; Rossignol, T.; Devillers, H.; Morin, N.; Robak, M.; Nicaud, J.M.; Crutz-Le Coq, A.M. Characterization of hexose transporters in Yarrowia lipolytica reveals new groups of sugar porters involved in yeast growth. Fungal Genet. Biol. 2017, 100, 1–12. [Google Scholar] [CrossRef]
- Petit, T.; Gancedo, C. Molecular cloning and characterization of the gene HXK1 encoding the hexokinase from Yarrowia lipolytica. Yeast 1999, 15, 1573–1584. [Google Scholar] [CrossRef]
- Hapeta, P.; Szczepańska, P.; Neuvéglise, C.; Lazar, Z. A 37-amino acid loop in the Yarrowia lipolytica hexokinase impacts its activity and affinity and modulates gene expression. Sci. Rep. 2021, 11, 6412. [Google Scholar] [CrossRef]
- Qiang, S.; Wang, J.; Xiong, X.C.; Qu, Y.L.; Liu, L.; Hu, C.Y.; Meng, Y.H. Promoting the synthesis of precursor substances by overexpressing Hexokinase (Hxk) and Hydroxymethylglutaryl-CoA Synthase (Erg13) to elevate β-carotene production in engineered Yarrowia lipolytica. Front. Microbiol. 2020, 11, 1346. [Google Scholar] [CrossRef]
- Lazar, Z.; Dulermo, T.; Neuvéglise, C.; Crutz-Le Coq, A.M.; Nicaud, J.M. Hexokinase—A limiting factor in lipid production from fructose in Yarrowia lipolytica. Metab. Eng. 2014, 26, 89–99. [Google Scholar] [CrossRef]
- Petit, T.; Blázquez, M.A.; Gancedo, C. Schizosaccharomyces pombe possesses an unusual and a conventional hexokinase: Biochemical and molecular characterization of both hexokinases. FEBS Lett. 1996, 378, 185–189. [Google Scholar] [CrossRef] [Green Version]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Hayes, M.A.; Davies, C.; Dry, I.B. Isolation, functional characterization, and expression analysis of grapevine (Vitis vinifera L.) hexose transporters: Differential roles in sink and source tissues. J. Exp. Bot. 2007, 58, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Schüßler, A.; Martin, H.; Cohen, D.; Fitz, M.; Wipf, D. Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 2006, 444, 933–936. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Muniglia, L.; Chevalot, I.; Aggelis, G.; Marc, I. Yarrowia lipolytica as a potential producer of citric acid from raw glycerol. J. Appl. Microbiol. 2002, 92, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef]
- Rakicka, M.; Kieroń, A.; Hapeta, P.; Neuvéglise, C.; Lazar, Z. Sweet and sour potential of yeast from the Yarrowia clade. Biomass Bioenerg. 2016, 92, 48–54. [Google Scholar] [CrossRef]
- Workman, M.; Holt, P.; Thykaer, J. Comparing cellular performance of Yarrowia lipolytica during growth on glucose and glycerol in submerged cultivations. Amb Express 2013, 3, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulermo, T.; Lazar, Z.; Dulermo, R.; Rakicka, M.; Haddouche, R.; Nicaud, J.M. Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. BBA-Mol. Cell Biol. Lipids 2015, 1851, 1107–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Chi, P.; Zou, Y.; Xu, Y.; Xu, S.; Bilal, M.; Fickers, P.; Cheng, H. Metabolic engineering of Yarrowia lipolytica for thermoresistance and enhanced erythritol productivity. Biotechnol. Biofuels 2020, 13, 176. [Google Scholar] [CrossRef]
- Mirończuk, A.M.; Furgała, J.; Rakicka, M.; Rymowicz, W. Enhanced production of erythritol by Yarrowia lipolytica on glycerol in repeated batch cultures. J. Ind. Microbiol. Biotechnol. 2014, 41, 57–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quarterman, J.; Slininger, P.J.; Kurtzman, C.P.; Thompson, S.R.; Dien, B.S. A survey of yeast from the Yarrowia clade for lipid production in dilute acid pretreated lignocellulosic biomass hydrolysate. Appl. Microbiol. Biotechnol. 2017, 101, 3319–3334. [Google Scholar] [CrossRef]
- Carly, F.; Steels, S.; Telek, S.; Vandermies, M.; Nicaud, J.M.; Fickers, P. Identification and characterization of EYD1, encoding an erythritol dehydrogenase in Yarrowia lipolytica and its application to bioconvert erythritol into erythrulose. Bioresour. Technol. 2018, 247, 963–969. [Google Scholar] [CrossRef]
- Carly, F.; Vandermies, M.; Telek, S.; Steels, S.; Thomas, S.; Nicaud, J.M.; Fickers, P. Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering. Metab. Eng. 2017, 42, 19–24. [Google Scholar] [CrossRef]
- Janek, T.; Dobrowolski, A.; Biegalska, A.; Mirończuk, A.M. Characterization of erythrose reductase from Yarrowia lipolytica and its influence on erythritol synthesis. Microb. Cell Fact. 2017, 16, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirończuk, A.M.; Biegalska, A.; Dobrowolski, A. Functional overexpression of genes involved in erythritol synthesis in the yeast Yarrowia lipolytica. Biotechnol. Biofuels 2017, 10, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carly, F.; Gamboa-Melendez, H.; Vandermies, M.; Damblon, C.; Nicaud, J.M.; Fickers, P. Identification and characterization of EYK1, a key gene for erythritol catabolism in Yarrowia lipolytica. Appl. Microbiol. 2017, 101, 6587–6596. [Google Scholar]
- Lazar, Z.; Rossignol, T.; Verbeke, J.; Crutz-Le Coq, A.M.; Nicaud, J.M.; Robak, M. Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. J. Ind. Microbiol. Biotechnol. 2013, 40, 1273–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fickers, P.; Le Dall, M.T.; Gaillardin, C.; Thonart, P.; Nicaud, J.M. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods 2003, 55, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Le Dall, M.T.; Nicaud, J.M.; Gaillardin, C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 1994, 26, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Trassaert, M.; Vandermies, M.; Carly, F.; Denies, O.; Thomas, S.; Fickers, P.; Nicaud, J.M. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microb. Cell Fact. 2017, 16, 141. [Google Scholar] [CrossRef]
- Dulermo, R.; Dulermo, T.; Gamboa-Meléndez, H.; Thevenieau, F.; Nicaud, J.M. Role of Pex11p in lipid homeostasis in Yarrowia lipolytica. Eukaryot. Cell 2015, 14, 511–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, M.J.; Van Hille, R.P.; Harrison, S.T.L. Selection of direct transesterification as the preferred method for assay of fatty acid content of microalgae. Lipids 2010, 45, 1053–1060. [Google Scholar] [CrossRef]

| Parameter | Unit | JMY2900 | YlH1 | SpH1 | YlH1-Y3 | YlH1-Y1-Y3 | YlH1-Y3-Y4 | YlH1-Y1-Y3-Y4 | SpH1-Y3 | SpH1-Y1-Y3 | SpH1-Y3-Y4 | SpH1-Y1-Y3-Y4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rxmax | g/L/h | 0.13 | 0.14 | 0.08 | 0.10 | 0.07 | 0.08 | 0.24 | 0.17 | 0.19 | 0.18 | 0.22 |
| rglc | 0.73 | 1.04 | 1.03 | 1.09 | 1.26 | 1.86 | 1.35 | 1.16 | 1.53 | 2.51 | 1.35 | |
| rfru | 0.43 | 0.84 | 0.77 | 0.41 | 0.91 | 0.89 | 1.77 | 1.34 | 1.43 | 1.70 | 1.67 | |
| X | g/L | 17.4 ± 1.2 | 14.5 ± 1.0 | 10.5 ± 0.9 | 13.5 ± 0.3 | 8.0 ± 1.0 | 10.5 ± 1.4 | 17.2 ± 2.4 | 11.9 ± 0.8 | 13.0 ± 1.3 | 12.6 ± 1.2 | 14.0 ± 1.8 |
| CA | 29.39 ± 2.33 | 67.90 ± 3.51 | 81.09 ± 4.55 | 26.45 ± 1.24 | 65.50 ± 4.33 | 70.19 ± 2.22 | 36.59 ± 4.22 | 42.84 ± 1.83 | 41.87 ± 1.97 | 35.78 ± 1.76 | 33.19 ± 2.40 | |
| Lipids | 5.74 ± 1.35 | 4.42 ± 1.00 | 4.02 ± 0.98 | 3.23 ± 1.33 | 3.46 ± 1.03 | 3.61 ± 1.22 | 3.97 ± 1.27 | 2.83 ± 0.66 | 3.51 ± 0.83 | 3.51 ± 0.97 | 3.08 ± 1.56 | |
| Man | ND | 1.34 ± 0.27 | ND | ND | ND | 1.93 ± 0.06 | 8.47 ± 1.03 | 8.47 ± 0.98 | 13.26 ± 0.56 | 13.56 ± 0.96 | 13.42 ± 1.88 | |
| Ara | ND | 0.90 ± 0.03 | ND | ND | ND | 0.78 ± 0.13 | 4.98 ± 0.52 | 4.10 ± 0.08 | 3.02 ± 0.22 | 4.08 ± 1.03 | 3.20 ± 0.06 | |
| Ery | ND | ND | ND | ND | ND | ND | 7.87 ± 1.12 | 11.94 ± 1.16 | 9.03 ± 1.09 | 13.84 ± 1.69 | 9.34 ± 0.97 | |
| Sum of polyols | ND | 2.24 | ND | ND | ND | 2.71 | 21.32 | 24.51 | 25.31 | 31.48 | 25.96 | |
| YCA/S | g/g | 0.427 | 0.728 | 0.804 | 0.420 | 0.667 | 0.751 | 0.398 | 0.447 | 0.438 | 0.361 | 0.353 |
| QCA | g/L/h | 0.111 | 0.314 | 0.307 | 0.100 | 0.287 | 0.266 | 0.254 | 0.298 | 0.303 | 0.248 | 0.263 |
| qCA | g/g/h | 0.013 | 0.043 | 0.060 | 0.015 | 0.072 | 0.051 | 0.030 | 0.050 | 0.047 | 0.039 | 0.038 |
| Parameter | Unit | Polyol Media | Erythritol Media | ||
|---|---|---|---|---|---|
| Glucose Fructose | Glycerol | Glucose Fructose | Glycerol | ||
| rxmax | g/L/h | 0.693 | 0.857 | 0.767 | 0.400 |
| rglc | 2.048 | - | 2.738 | - | |
| rfru | 2.698 | - | 3.229 | - | |
| rgly | - | 2.432 | - | 2.013 | |
| Man | g/L | 18.03 ± 1.37 | 5.04 ± 0.90 | 2.09 ± 0.66 | 0.11 ± 0.01 |
| Ara | 8.48 ± 0.68 | 4.29 ± 1.02 | 14.66 ± 1.06 | 4.07 ± 0.32 | |
| Ery | 14.07 ± 1.29 | 24.28 ± 0.99 | 23.12 ± 1.43 | 28.31 ± 1.33 | |
| YMan/S | g/g | 0.199 | 0.049 | 0.021 | 0.001 |
| YAra/S | 0.094 | 0.041 | 0.145 | 0.037 | |
| YEry/S | 0.155 | 0.235 | 0.229 | 0.257 | |
| YMan/X | 1.050 | 0.319 | 0.101 | 0.007 | |
| YAra/X | 0.493 | 0.271 | 0.708 | 0.238 | |
| YEry/X | 0.819 | 1.534 | 1.117 | 1.655 | |
| qMan | g/g/h | 0.022 | 0.012 | 0.002 | 0.000 |
| qAra | 0.010 | 0.006 | 0.015 | 0.003 | |
| qEry | 0.017 | 0.018 | 0.023 | 0.020 | |
| QMan | g/L/h | 0.188 | 0.043 | 0.022 | 0.000 |
| QAra | 0.088 | 0.037 | 0.153 | 0.024 | |
| QEry | 0.147 | 0.209 | 0.241 | 0.169 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hapeta, P.; Szczepańska, P.; Witkowski, T.; Nicaud, J.-M.; Crutz-Le Coq, A.-M.; Lazar, Z. The Role of Hexokinase and Hexose Transporters in Preferential Use of Glucose over Fructose and Downstream Metabolic Pathways in the Yeast Yarrowia lipolytica. Int. J. Mol. Sci. 2021, 22, 9282. https://doi.org/10.3390/ijms22179282
Hapeta P, Szczepańska P, Witkowski T, Nicaud J-M, Crutz-Le Coq A-M, Lazar Z. The Role of Hexokinase and Hexose Transporters in Preferential Use of Glucose over Fructose and Downstream Metabolic Pathways in the Yeast Yarrowia lipolytica. International Journal of Molecular Sciences. 2021; 22(17):9282. https://doi.org/10.3390/ijms22179282
Chicago/Turabian StyleHapeta, Piotr, Patrycja Szczepańska, Tadeusz Witkowski, Jean-Marc Nicaud, Anne-Marie Crutz-Le Coq, and Zbigniew Lazar. 2021. "The Role of Hexokinase and Hexose Transporters in Preferential Use of Glucose over Fructose and Downstream Metabolic Pathways in the Yeast Yarrowia lipolytica" International Journal of Molecular Sciences 22, no. 17: 9282. https://doi.org/10.3390/ijms22179282
APA StyleHapeta, P., Szczepańska, P., Witkowski, T., Nicaud, J.-M., Crutz-Le Coq, A.-M., & Lazar, Z. (2021). The Role of Hexokinase and Hexose Transporters in Preferential Use of Glucose over Fructose and Downstream Metabolic Pathways in the Yeast Yarrowia lipolytica. International Journal of Molecular Sciences, 22(17), 9282. https://doi.org/10.3390/ijms22179282






