Endocytic Protein Defects in the Neural Crest Cell Lineage and Its Pathway Are Associated with Congenital Heart Defects
Abstract
1. Introduction
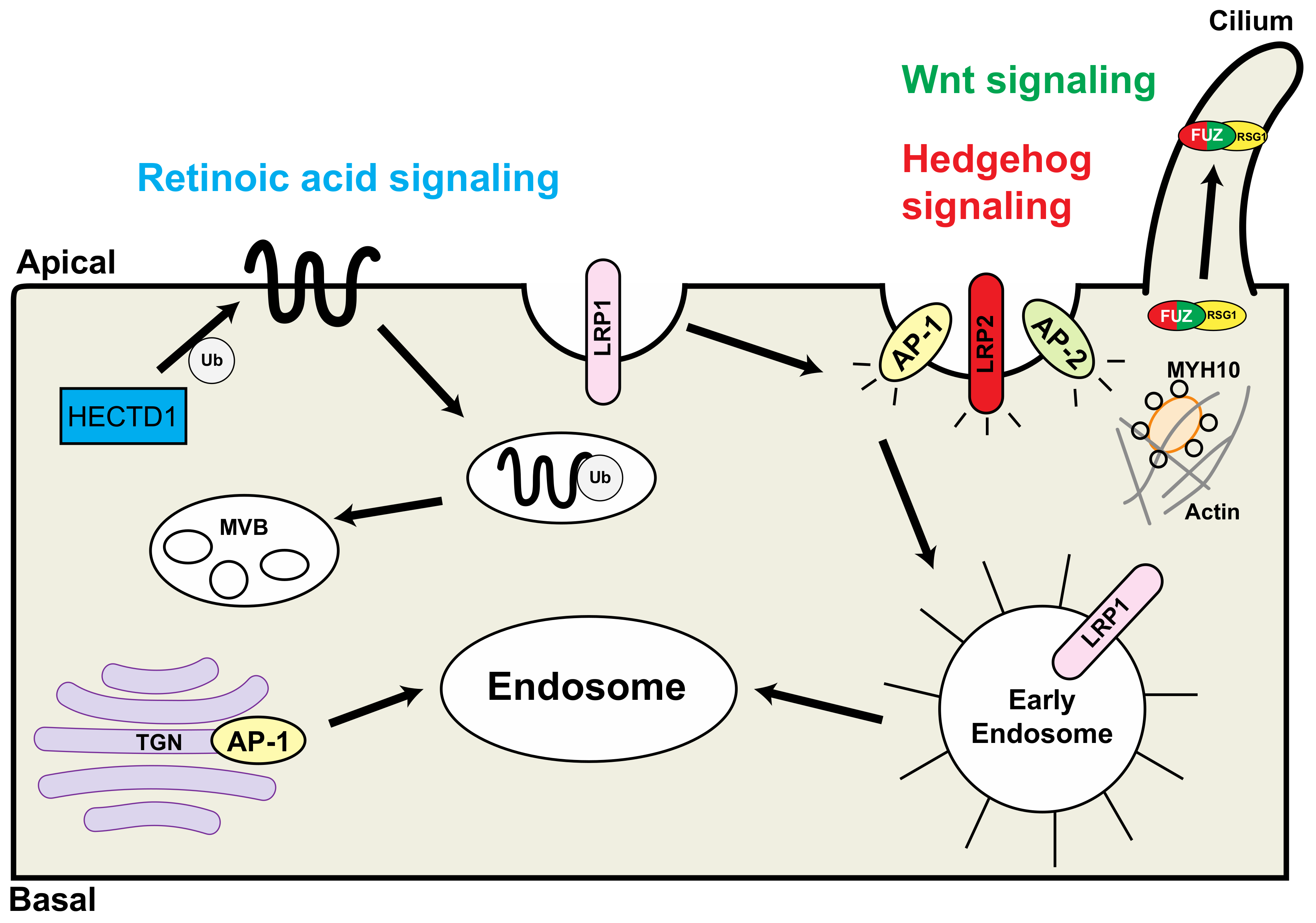
2. LRP1 (Low Density Lipoprotein Receptor-Related Protein 1)
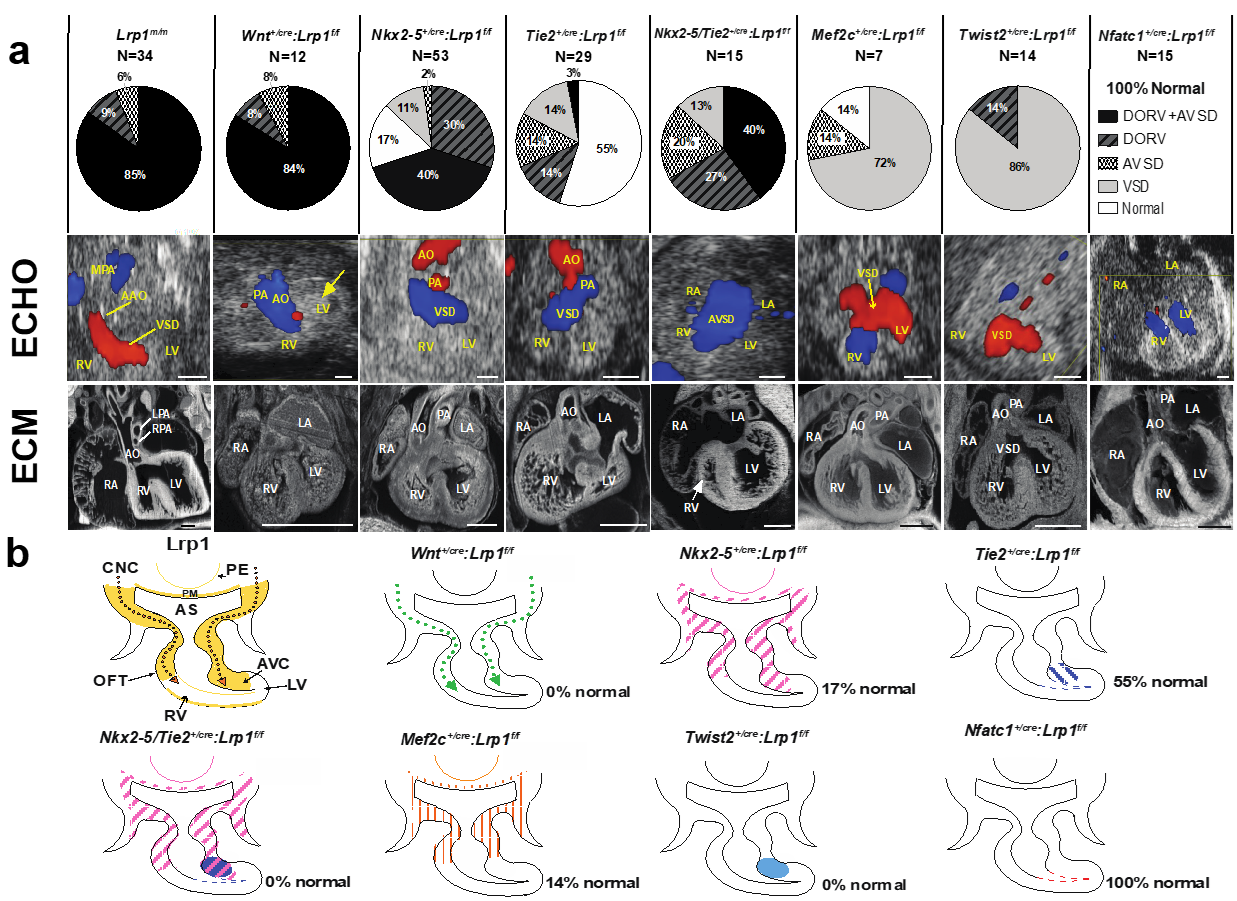
2.1. Lrp1 Mutation Is Associated with DORV and AVSD
2.2. Lineage-Specific Roles of Lrp1 in Cardiac Development
2.3. LRP1 Is Required for Cardiac Neural Crest Cell Migration
2.4. LRP1 Is Essential for Cardiac Outflow Tract Development and Endocardial Cushion Maturation
3. LRP2 (Low-Density Lipoprotein Receptor-Related Protein 2)
4. AP1B1 (Adaptor Related Protein Complex 1 Subunit Beta 1)
5. AP2B1 (Adaptor-Related Protein Complex 2 Beta1)
6. Fuz (Fuzzy Planar Cell Polarity Protein)
7. Myh10 (Myosin Heavy Chain 10)
8. HECTD1 (HECT Domain E3 Ubiquitin Protein Ligase 1)
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Srivastava, D.; Olson, E.N. A genetic blueprint for cardiac development. Nature 2000, 407, 221–226. [Google Scholar] [CrossRef]
- Li, Y.; Lui, K.O.; Zhou, B. Reassessing endothelial-to-mesenchymal transition in cardiovascular diseases. Nat. Rev. Cardiol. 2018, 15, 445–456. [Google Scholar] [CrossRef]
- Combs, M.D.; Yutzey, K.E. Heart valve development: Regulatory networks in development and disease. Circ. Res. 2009, 105, 408–421. [Google Scholar] [CrossRef]
- Keyte, A.; Hutson, M.R. The neural crest in cardiac congenital anomalies. Differentiation 2012, 84, 25–40. [Google Scholar] [CrossRef]
- Kirby, M.L.; Gale, T.F.; Stewart, D.E. Neural crest cells contribute to normal aorticopulmonary septation. Science 1983, 220, 1059–1061. [Google Scholar] [CrossRef]
- Kirby, M.L.; Hutson, M.R. Factors controlling cardiac neural crest cell migration. Cell Adhes. Migr. 2010, 4, 609–621. [Google Scholar] [CrossRef]
- Jiang, X.; Rowitch, D.H.; Soriano, P.; McMahon, A.P.; Sucov, H.M. Fate of the mammalian cardiac neural crest. Development 2000, 127, 1607–1616. [Google Scholar] [CrossRef]
- Hutson, M.R.; Kirby, M.L. Neural crest and cardiovascular development: A 20-year perspective. Birth Defects Res. C Embryo Today 2003, 69, 2–13. [Google Scholar] [CrossRef]
- Waldo, K.L.; Lo, C.W.; Kirby, M.L. Connexin 43 expression reflects neural crest patterns during cardiovascular development. Dev. Biol. 1999, 208, 307–323. [Google Scholar] [CrossRef]
- Ma, P.; Gu, S.; Karunamuni, G.H.; Jenkins, M.W.; Watanabe, M.; Rollins, A.M. Cardiac neural crest ablation results in early endocardial cushion and hemodynamic flow abnormalities. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H1150–H1159. [Google Scholar] [CrossRef]
- Yelbuz, T.M.; Waldo, K.L.; Kumiski, D.H.; Stadt, H.A.; Wolfe, R.R.; Leatherbury, L.; Kirby, M.L. Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation 2002, 106, 504–510. [Google Scholar] [CrossRef]
- Etchevers, H.C.; Dupin, E.; Le Douarin, N.M. The diverse neural crest: From embryology to human pathology. Development 2019, 146, 169821. [Google Scholar] [CrossRef]
- Driessen, A.K.; Farrell, M.J.; Mazzone, S.B.; McGovern, A.E. Multiple neural circuits mediating airway sensations: Recent advances in the neurobiology of the urge-to-cough. Respir. Physiol. Neurobiol. 2016, 226, 115–120. [Google Scholar] [CrossRef]
- Arima, Y.; Miyagawa-Tomita, S.; Maeda, K.; Asai, R.; Seya, D.; Minoux, M.; Rijli, F.M.; Nishiyama, K.; Kim, K.S.; Uchijima, Y.; et al. Preotic neural crest cells contribute to coronary artery smooth muscle involving endothelin signalling. Nat. Commun. 2012, 3, 1267. [Google Scholar] [CrossRef]
- Ebadi, A.; Spicer, D.E.; Backer, C.L.; Fricker, F.J.; Anderson, R.H. Double-outlet right ventricle revisited. J. Thorac. Cardiovasc. Surg. 2017, 154, 598–604. [Google Scholar] [CrossRef]
- Neeb, Z.; Lajiness, J.D.; Bolanis, E.; Conway, S.J. Cardiac outflow tract anomalies. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 499–530. [Google Scholar] [CrossRef]
- Bajolle, F.; Zaffran, S.; Kelly, R.G.; Hadchouel, J.; Bonnet, D.; Brown, N.A.; Buckingham, M.E. Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circ. Res. 2006, 98, 421–428. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, H.; Correa, A.; Devine, O.; Mathews, T.J.; Honein, M.A. Racial differences in infant mortality attributable to birth defects in the United States, 1989–2002. Birth Defects Res. A Clin. Mol. Teratol. 2006, 76, 706–713. [Google Scholar] [CrossRef]
- Gilboa, S.M.; Devine, O.J.; Kucik, J.E.; Oster, M.E.; Riehle-Colarusso, T.; Nembhard, W.N.; Xu, P.; Correa, A.; Jenkins, K.; Marelli, A.J. Congenital Heart Defects in the United States: Estimating the Magnitude of the Affected Population in 2010. Circulation 2016, 134, 101–109. [Google Scholar] [CrossRef]
- Li, Y.; Klena, N.T.; Gabriel, G.C.; Liu, X.; Kim, A.J.; Lemke, K.; Chen, Y.; Chatterjee, B.; Devine, W.; Damerla, R.R.; et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 2015, 521, 520–524. [Google Scholar] [CrossRef]
- Hobert, O. The impact of whole genome sequencing on model system genetics: Get ready for the ride. Genetics 2010, 184, 317–319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammes, A.; Andreassen, T.K.; Spoelgen, R.; Raila, J.; Hubner, N.; Schulz, H.; Metzger, J.; Schweigert, F.J.; Luppa, P.B.; Nykjaer, A.; et al. Role of endocytosis in cellular uptake of sex steroids. Cell 2005, 122, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Willnow, T.E.; Christ, A.; Hammes, A. Endocytic receptor-mediated control of morphogen signaling. Development 2012, 139, 4311–4319. [Google Scholar] [CrossRef]
- Strickland, D.K.; Au, D.T.; Cunfer, P.; Muratoglu, S.C. Low-density lipoprotein receptor-related protein-1: Role in the regulation of vascular integrity. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 487–498. [Google Scholar] [CrossRef]
- May, P.; Herz, J.; Bock, H.H. Molecular mechanisms of lipoprotein receptor signalling. Cell. Mol. Life Sci. 2005, 62, 2325–2338. [Google Scholar] [CrossRef]
- Rudenko, G.; Henry, L.; Henderson, K.; Ichtchenko, K.; Brown, M.S.; Goldstein, J.L.; Deisenhofer, J. Structure of the LDL receptor extracellular domain at endosomal pH. Science 2002, 298, 2353–2358. [Google Scholar] [CrossRef]
- Lin, J.I.; Feinstein, T.N.; Jha, A.; McCleary, J.T.; Xu, J.; Arrigo, A.B.; Rong, G.; Maclay, L.M.; Ridge, T.; Xu, X. Mutation of LRP1 in cardiac neural crest cells causes congenital heart defects by perturbing outflow lengthening. Commun. Biol. 2020, 3, 312. [Google Scholar] [CrossRef]
- Herz, J.; Clouthier, D.E.; Hammer, R.E. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 1992, 71, 411–421. [Google Scholar] [CrossRef]
- Rohlmann, A.; Gotthardt, M.; Willnow, T.E.; Hammer, R.E.; Herz, J. Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat. Biotechnol. 1996, 14, 1562–1565. [Google Scholar] [CrossRef] [PubMed]
- Danielian, P.S.; Muccino, D.; Rowitch, D.H.; Michael, S.K.; McMahon, A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998, 8, 1323–1326. [Google Scholar] [CrossRef]
- Brewer, S.; Feng, W.; Huang, J.; Sullivan, S.; Williams, T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev. Biol. 2004, 267, 135–152. [Google Scholar] [CrossRef]
- Verzi, M.P.; McCulley, D.J.; De Val, S.; Dodou, E.; Black, B.L. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 2005, 287, 134–145. [Google Scholar] [CrossRef]
- Kisanuki, Y.Y.; Hammer, R.E.; Miyazaki, J.; Williams, S.C.; Richardson, J.A.; Yanagisawa, M. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001, 230, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, Z.; Lui, W.; Chen, X.; Wang, Y.; Chamberlain, A.A.; Moreno-Rodriguez, R.A.; Markwald, R.R.; O’Rourke, B.P.; Sharp, D.J.; et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 2012, 151, 1083–1096. [Google Scholar] [CrossRef]
- Šošić, D.; Richardson, J.A.; Yu, K.; Ornitz, D.M.; Olson, E.N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell 2003, 112, 169–180. [Google Scholar] [CrossRef]
- Goddard, L.M.; Duchemin, A.L.; Ramalingan, H.; Wu, B.; Chen, M.; Bamezai, S.; Yang, J.; Li, L.; Morley, M.P.; Wang, T.; et al. Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis. Dev. Cell 2017, 43, 274–289.e5. [Google Scholar] [CrossRef] [PubMed]
- Stanley, E.G.; Biben, C.; Elefanty, A.; Barnett, L.; Koentgen, F.; Robb, L.; Harvey, R.P. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 2002, 46, 431–439. [Google Scholar]
- Maretto, S.; Cordenonsi, M.; Dupont, S.; Braghetta, P.; Broccoli, V.; Hassan, A.B.; Volpin, D.; Bressan, G.M.; Piccolo, S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 3299–3304. [Google Scholar] [CrossRef]
- Liebner, S.; Cattelino, A.; Gallini, R.; Rudini, N.; Iurlaro, M.; Piccolo, S.; Dejana, E. Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J. Cell Biol. 2004, 166, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999, 21, 70–71. [Google Scholar] [CrossRef]
- Theret, L.; Jeanne, A.; Langlois, B.; Hachet, C.; David, M.; Khrestchatisky, M.; Devy, J.; Hervé, E.; Almagro, S.; Dedieu, S. Identification of LRP-1 as an endocytosis and recycling receptor for beta1-integrin in thyroid cancer cells. Oncotarget 2017, 8, 78614–78632. [Google Scholar] [CrossRef]
- Liang, D.; Wang, X.; Mittal, A.; Dhiman, S.; Hou, S.Y.; Degenhardt, K.; Astrof, S. Mesodermal expression of integrin alpha5beta1 regulates neural crest development and cardiovascular morphogenesis. Dev. Biol. 2014, 395, 232–244. [Google Scholar] [CrossRef]
- Hakim, Z.S.; DiMichele, L.A.; Doherty, J.T.; Homeister, J.W.; Beggs, H.E.; Reichardt, L.F.; Schwartz, R.J.; Brackhan, J.; Smithies, O.; Mack, C.P.; et al. Conditional deletion of focal adhesion kinase leads to defects in ventricular septation and outflow tract alignment. Mol. Cell. Biol. 2007, 27, 5352–5364. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Bu, G.; Mars, W.M.; Reeves, W.B.; Tanaka, S.; Hu, K. tPA activates LDL receptor-related protein 1-mediated mitogenic signaling involving the p90RSK and GSK3beta pathway. Am. J. Pathol. 2010, 177, 1687–1696. [Google Scholar] [CrossRef]
- Hu, K.; Wu, C.; Mars, W.M.; Liu, Y. Tissue-type plasminogen activator promotes murine myofibroblast activation through LDL receptor-related protein 1-mediated integrin signaling. J. Clin. Investig. 2007, 117, 3821–3832. [Google Scholar] [CrossRef]
- Zhang, H.; von Gise, A.; Liu, Q.; Hu, T.; Tian, X.; He, L.; Pu, W.; Huang, X.; He, L.; Cai, C.L.; et al. Yap1 is required for endothelial to mesenchymal transition of the atrioventricular cushion. J. Biol. Chem. 2014, 289, 18681–18692. [Google Scholar] [CrossRef]
- Xiong, H.; Yu, Q.; Gong, Y.; Chen, W.; Tong, Y.; Wang, Y.; Xu, H.; Shi, Y. Yes-Associated Protein (YAP) Promotes Tumorigenesis in Melanoma Cells Through Stimulation of Low-Density Lipoprotein Receptor-Related Protein 1 (LRP1). Sci. Rep. 2017, 7, 15528. [Google Scholar] [CrossRef]
- Takeda, T.; Yamazaki, H.; Farquhar, M.G. Identification of an apical sorting determinant in the cytoplasmic tail of megalin. Am. J. Physiol. Cell Physiol. 2003, 284, C1105–C1113. [Google Scholar] [CrossRef]
- Christ, A.; Christa, A.; Klippert, J.; Eule, J.C.; Bachmann, S.; Wallace, V.A.; Hammes, A.; Willnow, T.E. LRP2 Acts as SHH Clearance Receptor to Protect the Retinal Margin from Mitogenic Stimuli. Dev. Cell 2015, 35, 36–48. [Google Scholar] [CrossRef]
- Christ, A.; Christa, A.; Kur, E.; Lioubinski, O.; Bachmann, S.; Willnow, T.E.; Hammes, A. LRP2 is an auxiliary SHH receptor required to condition the forebrain ventral midline for inductive signals. Dev. Cell 2012, 22, 268–278. [Google Scholar] [CrossRef]
- McCarthy, R.A.; Argraves, W.S. Megalin and the neurodevelopmental biology of sonic hedgehog and retinol. J. Cell Sci. 2003, 116, 955–960. [Google Scholar] [CrossRef]
- Kerjaschki, D.; Farquhar, M.G. The pathogenic antigen of Heymann nephritis is a membrane glycoprotein of the renal proximal tubule brush border. Proc. Natl. Acad. Sci. USA 1982, 79, 5557–5561. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Pietromonaco, S.; Loo, A.K.; Farquhar, M.G. Complete cloning and sequencing of rat gp330/“megalin,” a distinctive member of the low density lipoprotein receptor gene family. Proc. Natl. Acad. Sci. USA 1994, 91, 9725–9729. [Google Scholar] [CrossRef] [PubMed]
- Willnow, T.E.; Hilpert, J.; Armstrong, S.A.; Rohlmann, A.; Hammer, R.E.; Burns, D.K.; Herz, J. Defective forebrain development in mice lacking gp330/megalin. Proc. Natl. Acad. Sci. USA 1996, 93, 8460–8464. [Google Scholar] [CrossRef] [PubMed]
- Gomes, J.R.; Lobo, A.; Nogueira, R.; Terceiro, A.F.; Costelha, S.; Lopes, I.M.; Magalhães, A.; Summavielle, T.; Saraiva, M.J. Neuronal megalin mediates synaptic plasticity-a novel mechanism underlying intellectual disabilities in megalin gene pathologies. Brain Commun. 2020, 2, 135. [Google Scholar] [CrossRef]
- Flemming, J.; Marczenke, M.; Rudolph, I.M.; Nielsen, R.; Storm, T.; Erik, I.C.; Diecke, S.; Emma, F.; Willnow, T.E. Induced pluripotent stem cell-based disease modeling identifies ligand-induced decay of megalin as a cause of Donnai-Barrow syndrome. Kidney Int. 2020, 98, 159–167. [Google Scholar] [CrossRef]
- Storm, T.; Burgoyne, T.; Dunaief, J.L.; Christensen, E.I.; Futter, C.; Nielsen, R. Selective Ablation of Megalin in the Retinal Pigment Epithelium Results in Megaophthalmos, Macromelanosome Formation and Severe Retina Degeneration. Investig. Ophthalmol. Vis. Sci. 2019, 60, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Baardman, M.E.; Zwier, M.V.; Wisse, L.J.; Gittenberger-de Groot, A.C.; Kerstjens-Frederikse, W.S.; Hofstra, R.M.; Jurdzinski, A.; Hierck, B.P.; Jongbloed, M.R.; Berger, R.M.; et al. Common arterial trunk and ventricular non-compaction in Lrp2 knockout mice indicate a crucial role of LRP2 in cardiac development. Dis. Models Mech. 2016, 9, 413–425. [Google Scholar]
- Christ, A.; Marczenke, M.; Willnow, T.E. LRP2 controls sonic hedgehog-dependent differentiation of cardiac progenitor cells during outflow tract formation. Hum. Mol. Genet. 2020, 29, 3183–3196. [Google Scholar] [CrossRef]
- Goddeeris, M.M.; Schwartz, R.; Klingensmith, J.; Meyers, E.N. Independent requirements for Hedgehog signaling by both the anterior heart field and neural crest cells for outflow tract development. Development 2007, 134, 1593–1604. [Google Scholar] [CrossRef]
- Alsaif, H.S.; Al-Owain, M.; Barrios-Llerena, M.E.; Gosadi, G.; Binamer, Y.; Devadason, D.; Ravenscroft, J.; Suri, M.; Alkuraya, F.S. Homozygous Loss-of-Function Mutations in AP1B1, Encoding Beta-1 Subunit of Adaptor-Related Protein Complex 1, Cause MEDNIK-like Syndrome. Am. J. Hum. Genet. 2019, 105, 1016–1022. [Google Scholar] [CrossRef]
- Li, P.; Merrill, S.A.; Jorgensen, E.M.; Shen, K. Two Clathrin Adaptor Protein Complexes Instruct Axon-Dendrite Polarity. Neuron 2016, 90, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Guardia, C.M.; De Pace, R.; Mattera, R.; Bonifacino, J.S. Neuronal functions of adaptor complexes involved in protein sorting. Curr. Opin. Neurobiol. 2018, 51, 103–110. [Google Scholar] [CrossRef]
- Brooks, E.R.; Wallingford, J.B. Control of vertebrate intraflagellar transport by the planar cell polarity effector Fuz. J. Cell Biol. 2012, 198, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.S.; Abitua, P.B.; Wlodarczyk, B.J.; Szabo-Rogers, H.L.; Blanchard, O.; Lee, I.; Weiss, G.S.; Liu, K.J.; Marcotte, E.M.; Wallingford, J.B.; et al. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat. Cell Biol. 2009, 11, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wlodarczyk, B.J.; Niederreither, K.; Venugopalan, S.; Florez, S.; Finnell, R.H.; Amendt, B.A. Fuz regulates craniofacial development through tissue specific responses to signaling factors. PLoS ONE 2011, 6, e24608. [Google Scholar] [CrossRef]
- Tabler, J.M.; Barrell, W.B.; Szabo-Rogers, H.L.; Healy, C.; Yeung, Y.; Perdiguero, E.G.; Schulz, C.; Yannakoudakis, B.Z.; Mesbahi, A.; Wlodarczyk, B.; et al. Fuz mutant mice reveal shared mechanisms between ciliopathies and FGF-related syndromes. Dev. Cell 2013, 25, 623–635. [Google Scholar] [CrossRef]
- Seo, J.H.; Zilber, Y.; Babayeva, S.; Liu, J.; Kyriakopoulos, P.; De Marco, P.; Merello, E.; Capra, V.; Gros, P.; Torban, E. Mutations in the planar cell polarity gene, Fuzzy, are associated with neural tube defects in humans. Hum. Mol. Genet. 2011, 20, 4324–4333. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, I.; Goeckeler, Z.M.; Turney, S.G.; Wang, P.; Wysolmerski, R.B.; Adelstein, R.S.; Bridgman, P.C. Nonmuscle myosin II is a critical regulator of clathrin-mediated endocytosis. Traffic 2014, 15, 418–432. [Google Scholar] [CrossRef]
- Ma, X.; Adelstein, R.S. A point mutation in Myh10 causes major defects in heart development and body wall closure. Circ. Cardiovasc. Genet. 2014, 7, 257–265. [Google Scholar] [CrossRef]
- Rosenquist, T.H.; Bennett, G.D.; Brauer, P.R.; Stewart, M.L.; Chaudoin, T.R.; Finnell, R.H. Microarray analysis of homocysteine-responsive genes in cardiac neural crest cells in vitro. Dev. Dyn. 2007, 236, 1044–1054. [Google Scholar] [CrossRef]
- Zohn, I.E.; Anderson, K.V.; Niswander, L. The Hectd1 ubiquitin ligase is required for development of the head mesenchyme and neural tube closure. Dev. Biol. 2007, 306, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, K.F.; Zohn, I.E. Reduced maternal vitamin A status increases the incidence of normal aortic arch variants. Genesis 2019, 57, e23343. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Pashmforoush, M.; Sucov, H.M. Retinoic acid regulates differentiation of the secondary heart field and TGFbeta-mediated outflow tract septation. Dev. Cell 2010, 18, 480–485. [Google Scholar] [CrossRef] [PubMed]
- El Robrini, N.; Etchevers, H.C.; Ryckebüsch, L.; Faure, E.; Eudes, N.; Niederreither, K.; Zaffran, S.; Bertrand, N. Cardiac outflow morphogenesis depends on effects of retinoic acid signaling on multiple cell lineages. Dev. Dyn. 2016, 245, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Chisaka, O.; Capecchi, M.R. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature 1991, 350, 473–479. [Google Scholar] [CrossRef]
| Gene | Mouse Line ID | MGI ID | Nucleotide Change | Protein Change | Mouse Phenotype | Related Pathways | Human Disease |
|---|---|---|---|---|---|---|---|
| Ap1b1 | 1660 | 5433323 | c.T1094C | p.V365A | Heterotaxy, AVSD, DORV, right arch, micrognathia, craniofacial defects | ||
| Ap2b2 | 2321 | 5552944 | c.T1343A | p.M448K | DORV/Taussig-Bing, AVSD/VSD, aortic arch defects, craniofacial defects | Ataxia telangiectasia, cerebellar degeneration | |
| Fuz | 1273 | 5311392 | c.387+2T>A | This changes splice donor site G-GT to G-GA which is assumed to be much less efficient | Pulmonary atresia, AVSD, MAPCA, right arch, TEF, craniofacial defects, diaphragmatic hernia, limb defects | Hedgehog signaling WNT signaling | Neural tube defects |
| Hectd1 | 327 | 5313700 | c.T3264A | p.Y1088X | Aortic atresia/hypoplasia, dysplastic semilunar valve, VSD, exencephaly, neural tube defects | Retinoic acid signaling | |
| Lrp1 | 1554 | 5437079 | c.T12694C | p.C4232R | DORV, AVSD, pulmonary stenosis, craniofacial defects | Alzheimer disease, Schizophrenia | |
| Lrp2 | 1625 | 5489925 | c.8456-3A>G | Y2204X | PTA, IAA, AVSD, craniofacial defects | Hedgehog signaling | Donnai-Barrow syndrome |
| Myh10 | 2437 | 5552947 | c.T1054C | p.S352P | DORV/OA, exencephaly, micrognathia, hydronephrosis, body wall defects |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arrigo, A.B.; Lin, J.-H.I. Endocytic Protein Defects in the Neural Crest Cell Lineage and Its Pathway Are Associated with Congenital Heart Defects. Int. J. Mol. Sci. 2021, 22, 8816. https://doi.org/10.3390/ijms22168816
Arrigo AB, Lin J-HI. Endocytic Protein Defects in the Neural Crest Cell Lineage and Its Pathway Are Associated with Congenital Heart Defects. International Journal of Molecular Sciences. 2021; 22(16):8816. https://doi.org/10.3390/ijms22168816
Chicago/Turabian StyleArrigo, Angelo B., and Jiuann-Huey Ivy Lin. 2021. "Endocytic Protein Defects in the Neural Crest Cell Lineage and Its Pathway Are Associated with Congenital Heart Defects" International Journal of Molecular Sciences 22, no. 16: 8816. https://doi.org/10.3390/ijms22168816
APA StyleArrigo, A. B., & Lin, J.-H. I. (2021). Endocytic Protein Defects in the Neural Crest Cell Lineage and Its Pathway Are Associated with Congenital Heart Defects. International Journal of Molecular Sciences, 22(16), 8816. https://doi.org/10.3390/ijms22168816






