The Role of Strigolactones in the Regulation of Root System Architecture in Grapevine (Vitis vinifera L.) in Response to Root-Restriction Cultivation
Abstract
1. Introduction
2. Results
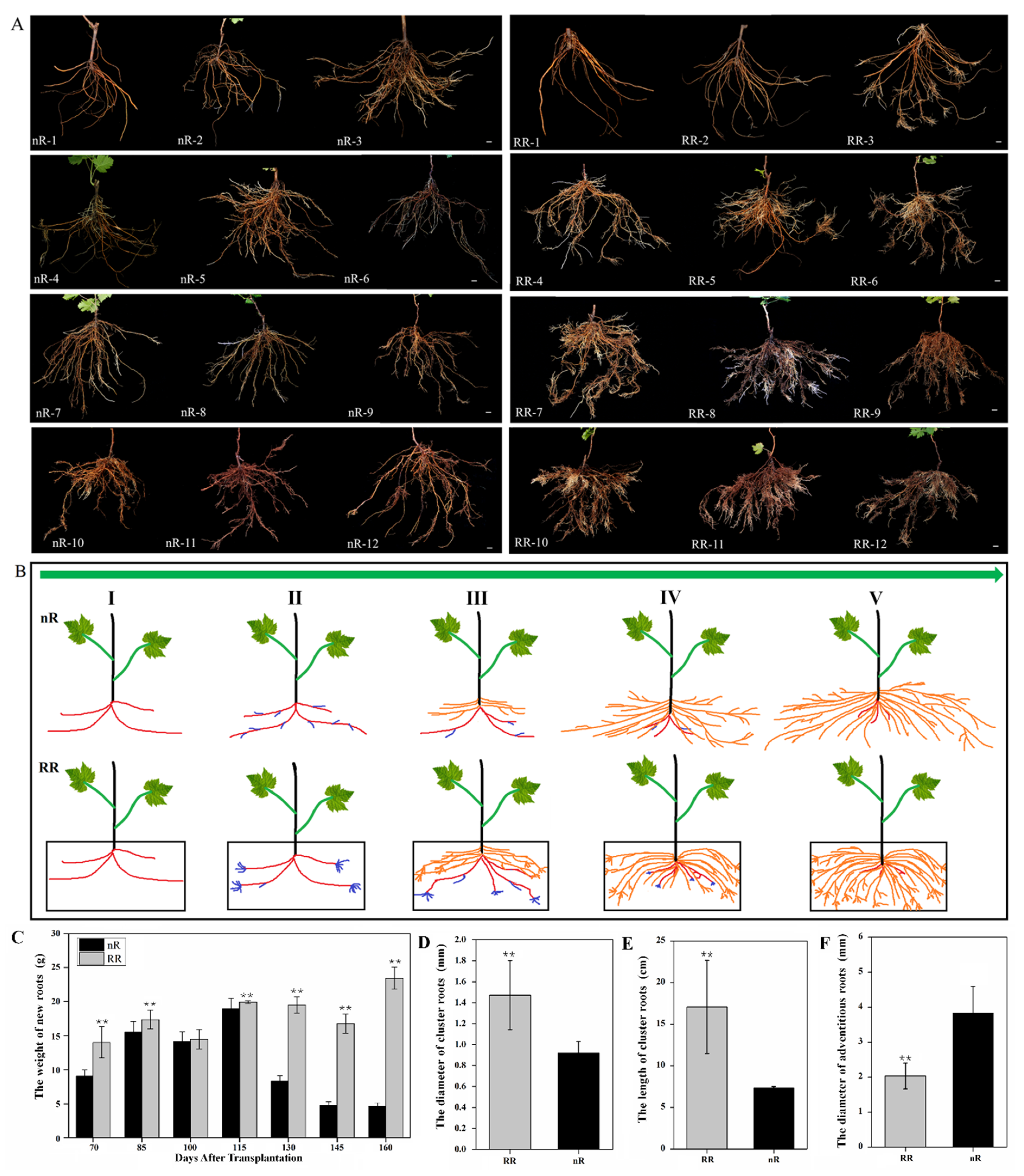
2.1. Characterization and Observation of Grapevine Root Development under Root-Restriction and Non-Root-Restriction Cultivation
2.2. Change in Endogenous SL Content of Grapevine Roots under Root-Restriction and Non-Root-Restriction Cultivation
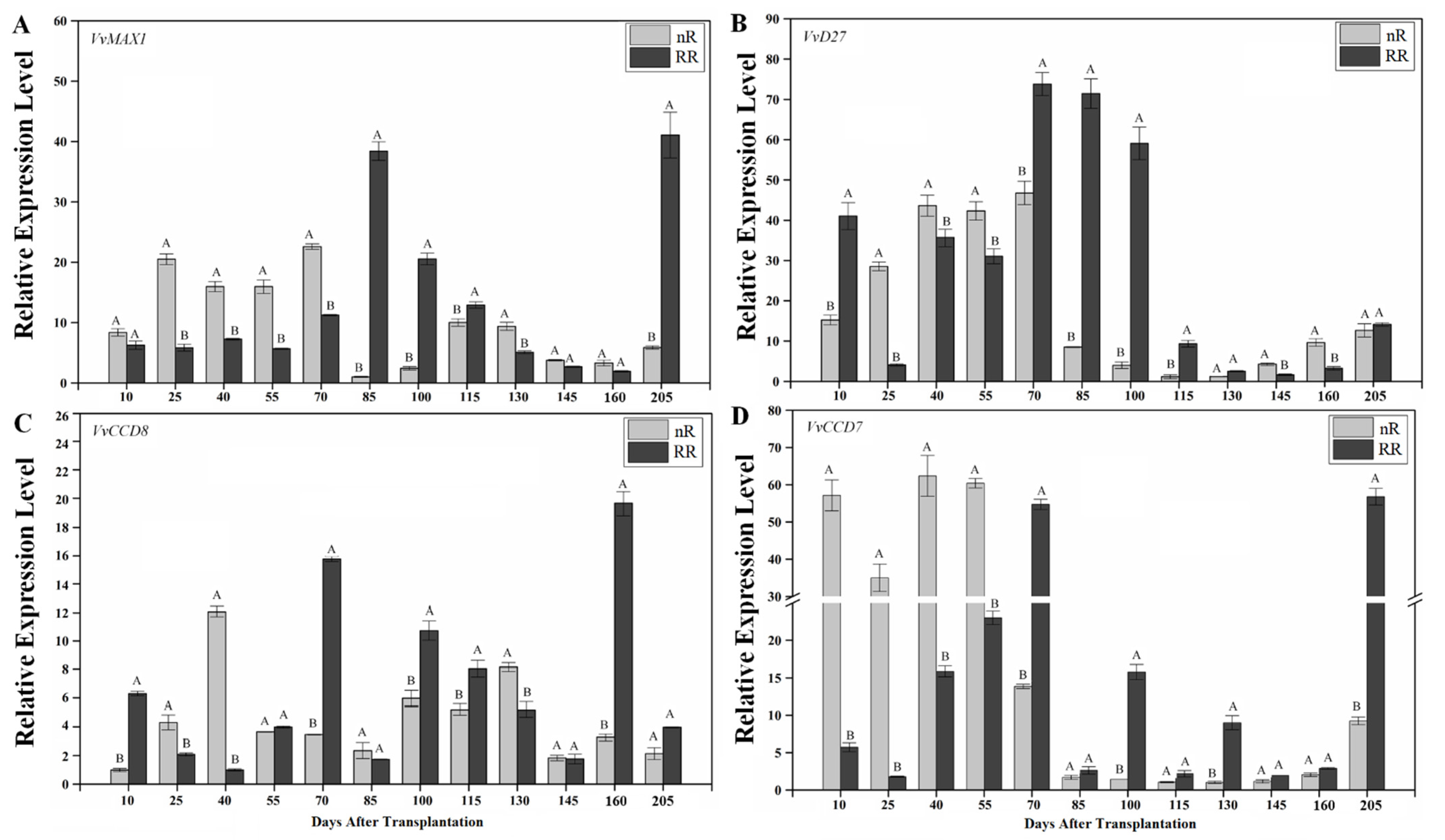
2.3. Expression of SL Biosynthesis Genes in Grapevine Roots under Root-Restriction Cultivation
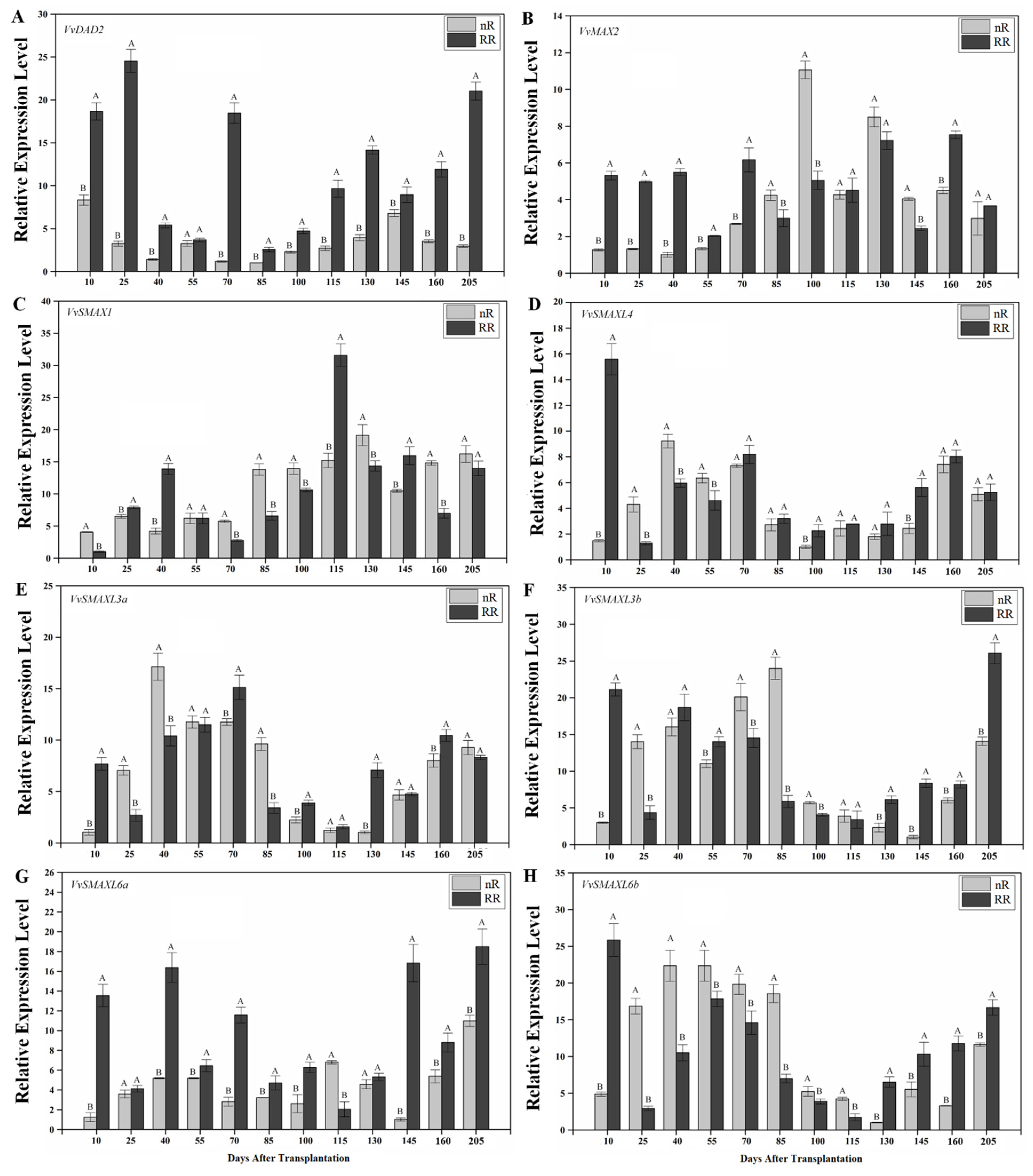
2.4. Expression of SL Signaling Genes in Grapevine Roots under Root-Restriction Cultivation
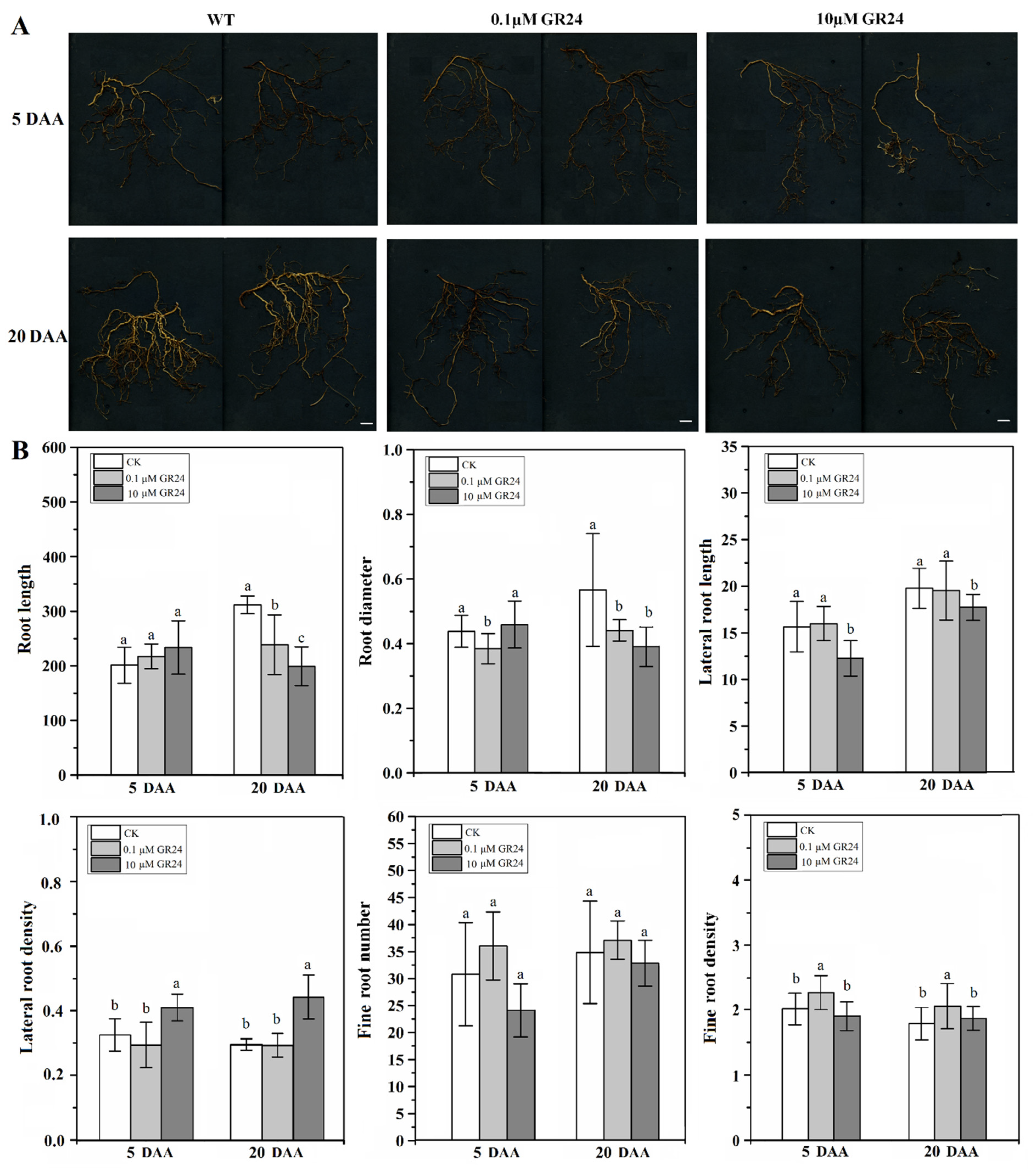
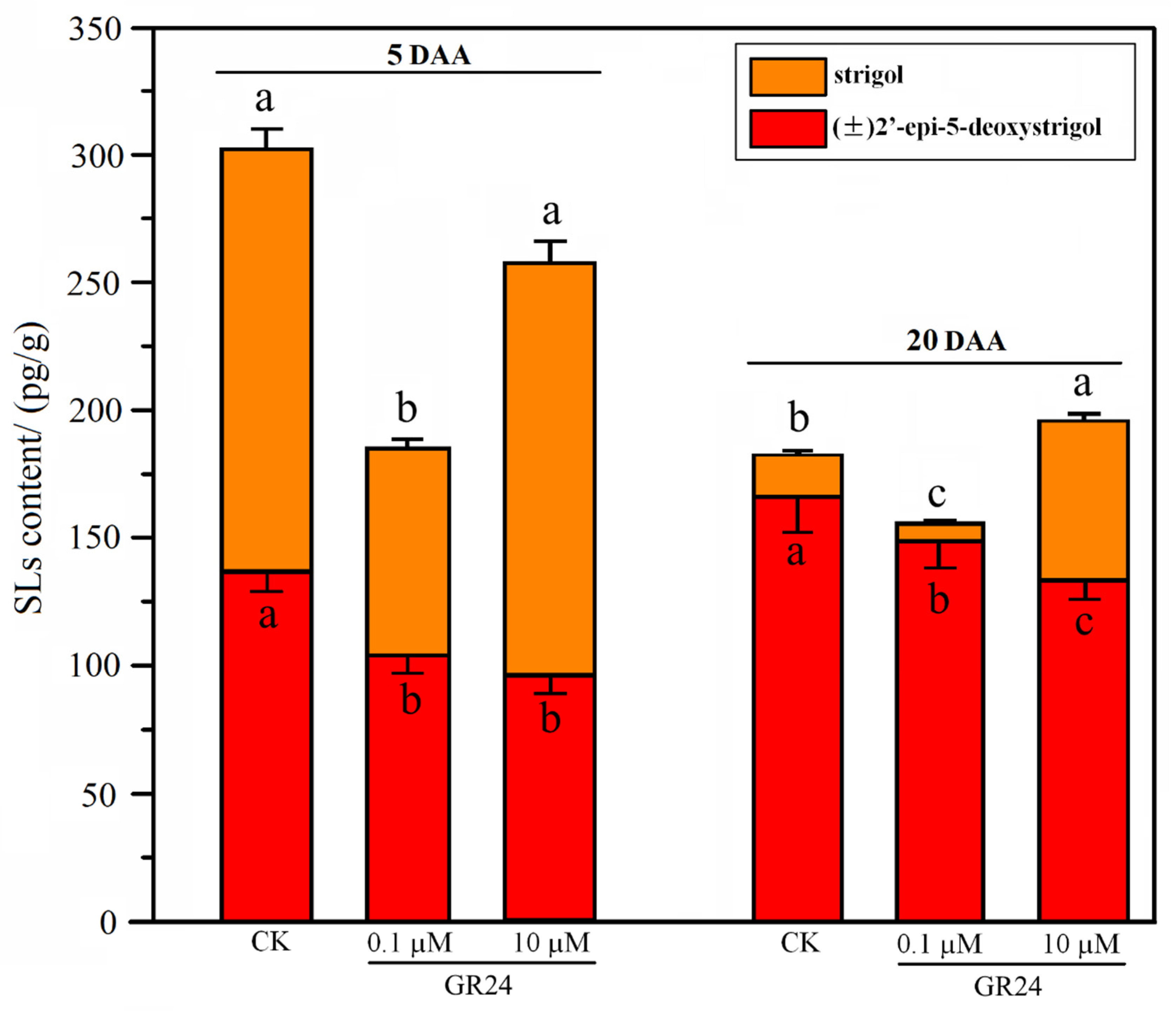
2.5. Effect of SL Analog on Grapevine Root System Architecture
2.6. SL Analog Affects Endogenous Hormone Content of Grapevine Root
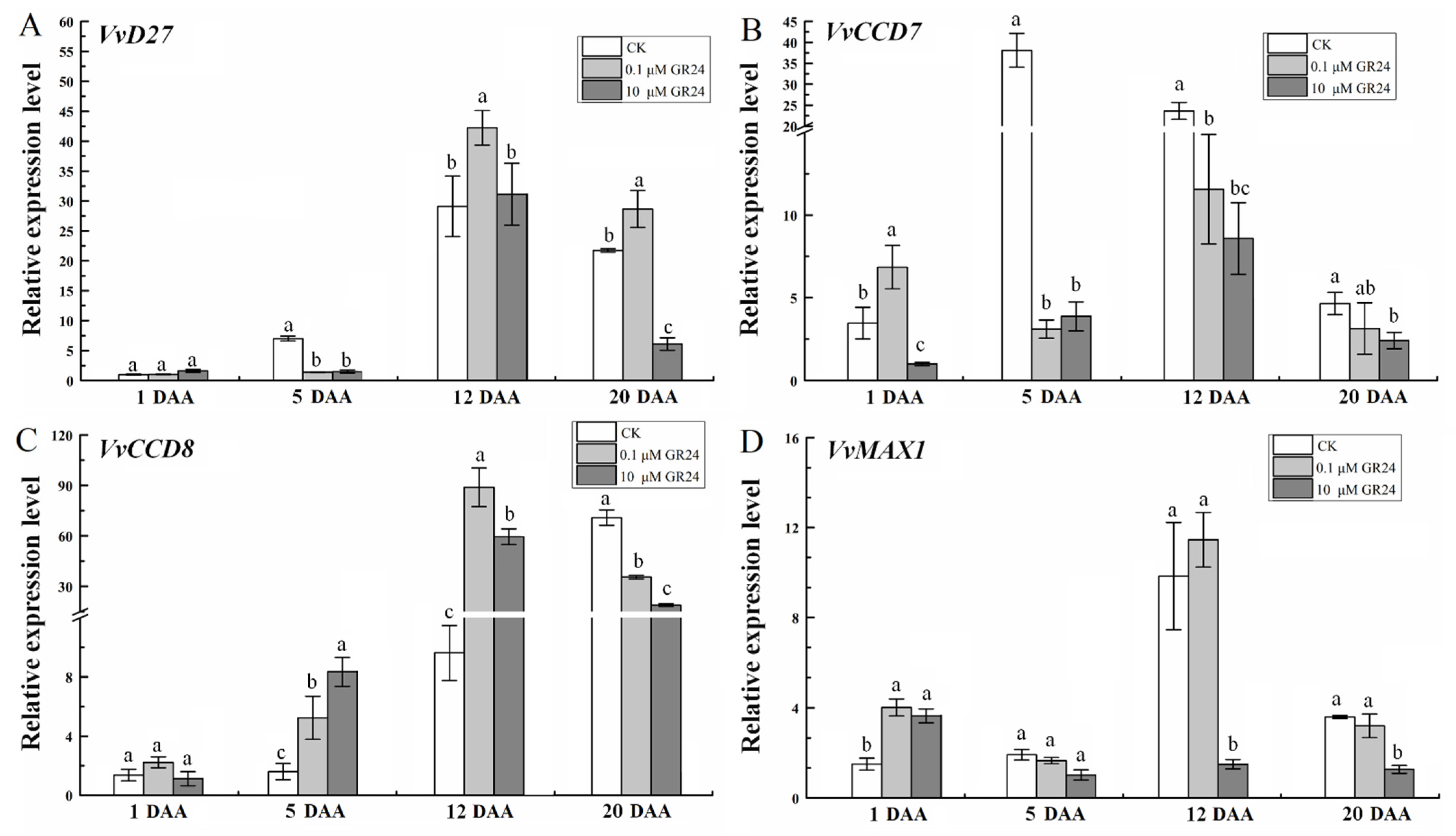
2.7. GR24 Treatment Affects the Expression of SL Biosynthesis Genes
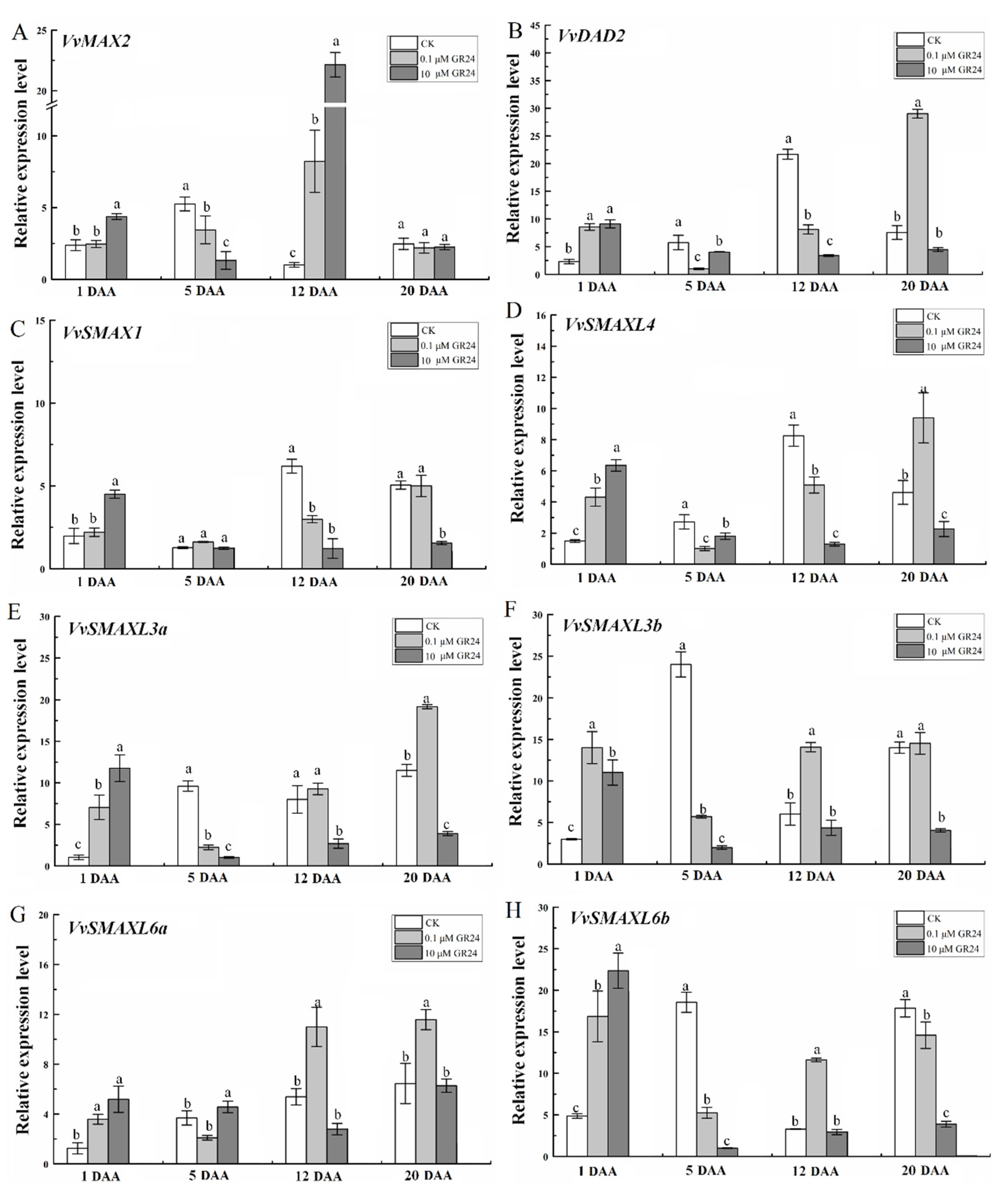
2.8. GR24 Treatment Affects the Expression of SL Signaling-Related Genes
3. Discussion
3.1. Effects of Root Restriction on Grapevine Root Architecture
3.2. Effect of Root Restriction on Hormone Content and SL-Related Gene Expression in Grapevine Roots
3.3. Effect of SLs on the Growth and Development of Grapevine Roots
4. Materials and Methods
4.1. Plant Materials
4.2. Plant Sampling
4.3. Exogenous GR24 Treatment
4.4. Root Morphological Evaluation
4.5. Paraffin Section Sample Preparation
4.6. SL Content Measurement
4.7. RNA Isolation and cDNA Synthesis
4.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Zhang, C.; Luo, J. Advances in research of root-zone restriction fruit growing. J. Fruit Sci. 2002, 19, 298–301. [Google Scholar]
- Stoll, M.; Loveys, B.; Dry, P. Hormonal changes induced by partial rootzone drying of irrigated grapevine. J. Exp. Bot. 2000, 51, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Christmann, A.; Weiler, E.W.; Steudle, E.; Grill, E. A hydraulic signal in root-to-shoot signaling of water shortage. Plant J. 2007, 52, 167–174. [Google Scholar] [CrossRef]
- Upreti, K.K.; Murti, G.S.R. Response of grape rootstocks to salinity: Changes in root growth, polyamines and abscisic acid. Biol. Plant. 2010, 54, 730–734. [Google Scholar] [CrossRef]
- Ren, Y.Z.; Xu, Y.H.; Ding, J.P. Regulation of abiotic factors on the plasticity of plant root development. Chin. Agric. Sci. Bull. 2011, 27, 34–38. [Google Scholar]
- Yakushiji, H.; Nonami, H.; Fukuyama, T.; Ono, S.; Takagi, N.; Yasushi, H. Sugar accumulation enhanced by osmoregulation in Satsuma mandarin fruit. J. Am. Soc. Hortic. Sci. 1996, 121, 466–472. [Google Scholar] [CrossRef]
- Wang, S.; Okamoto, G.; Hirano, K. Effects of rooting-zone restriction on the changes in carbohydrates and nitrogenous compounds in ’Kyoho’ grapevines during winter dormancy and early shoot growth. J. Jpn. Soc. Hortic. Sci. 1998, 67, 577–582. [Google Scholar] [CrossRef][Green Version]
- Wang, L.R.; Zheng, J.T.; Xu, Y.J.; Fang, C.L.; Xu, S.Q. Effect of rooting-zone restriction on the root growth of Minicure Finger grapevine. Sino-Overseas Grapevine Vine 2008, 2, 17–19. [Google Scholar]
- Jiao, H.J.; Leng, H.; Zhou, L.; Ai, K.B.E. Influence of rooting-zone restriction cultivation on growth and development of ‘Quanqiuhong’ grape. Chin. Hortic. Abstr. 2013, 29, 24–25, 223. [Google Scholar]
- Bar-Yosef, B.; Schwartz, S.; Markovich, T.; Lucas, B.; Assaf, R. Effect of root volume and nitrate solution concentration on growth, fruit yield, and temporal N and water uptake rates by apple trees. Plant Soil 1988, 107, 49–56. [Google Scholar] [CrossRef]
- Wang, B.; He, J.J.; Bai, Y.; Yu, X.M.; Li, J.F.; Zhang, C.X.; Xu, W.P.; Bai, X.J.; Cao, X.J.; Wang, S.P. Root restriction affected anthocyanin composition and up regulated the transcription of their biosynthetic genes during berry development in ‘Summer Black’ grape. Acta Physiol. Plant 2013, 35, 2205–2217. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef]
- Yoneyama, K.; Xie, X.N.; Sekimoto, H.; Takeuchi, Y.; Ogasawara, S.; Akiyama, K.; Hayashi, H.; Yoneyama, K. Strigolactones, host recognition signals for root parasitic plants and Arbuscular mycorrhizal fungi, from Fabaceae plants. New Phytol. 2008, 179, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.; Wegmann, K.; Frischmuth, K.; Samson, E.; Kranz, A.; Weigelt, D.; Koll, P.; Welzel, P. Stimulation of Orobanche crenata seed germination by (+)-strigol and structural analogues dependence on constitution and configuration of the germinatio stimulants. Plant Physiol. 1993, 142, 338–342. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; Zeijl, A.V.; Bezouwen, L.V.; de Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011, 155, 721–734. [Google Scholar] [CrossRef]
- Rasmussen, A.; Mason, M.G.; De Cuyper, C.; Brewer, P.B.; Herold, S.; Agusti, J.; Geelen, D.; Greb, T.; Goormachtig, S.; Beeckman, T.; et al. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 2012, 158, 1976–1987. [Google Scholar] [CrossRef] [PubMed]
- Arite, T.; Kameoka, H.; Kyozuka, J. Strigolactone positively controls crown root elongation in rice. Plant Growth Regul. 2012, 31, 165–172. [Google Scholar] [CrossRef]
- Kapulnik, Y.; Delaux, P.M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Séjalon-Delmas, N.; Combier, J.P.; Bécard, G.; Belausov, E. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H. Strigolactones are regulators of root development. New Phytol. 2011, 190, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Resnick, N.; Mayzlish-Gati, E.; Kaplan, Y.; Wininger, S.; Hershenhorn, J.; Koltai, H. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 2011, 62, 2915–2924. [Google Scholar] [CrossRef]
- Foo, E.; Davies, N.W. Strigolactones promote nodulation in pea. Planta 2011, 234, 1073–1081. [Google Scholar] [CrossRef]
- Soto, M.J.; Fernández-Aparicio, M.; Castellanos-Morales, V.; García-Garrido, J.M.; Ocampo, J.A.; Delgado, M.J.; Vierheilig, H. First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biol. Biochem. 2010, 42, 383–385. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, X.; Wang, Y.; Ruyterlogpira, C. The tomato MAX1 homolog, SlMAX1, is involved in the biosynthesis of tomato strigolactones from carlactone. New Phytol. 2018, 219, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Kagiyama, M.; Hirano, Y.; Mori, T.; Kim, S.Y.; Kyozuka, J.; Seto, Y.; Yamaguchi, S.; Hakoshima, T. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 2013, 18, 147–160. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.L.; Zhou, K.N.; Shabek, N.; Wu, F.Q.; Mao, H.B.; Dong, W.; Gan, L.; et al. D14–SCF D3-dependent degradation of D53 regulates strigolactone signaling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.H.; Chen, F.L.; Wang, L.; Meng, X.B.; Liu, G.F.; Yu, H.; Yuan, Y.D.; et al. DWARF 53 acts as a repressor of strigolactone signaling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef]
- Chevalier, F.; Nieminen, K.; Sánchez-Ferrero, J.C.; Rodríguez, M.L.; Chagoyen, M.; Hardtke, C.S.; Cubas, P. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 2014, 26, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, S.; Yang, T.; Zhang, C.; Xu, W. Vine growth and nitrogen metabolism of ‘Fujiminori’ grapevines in response to root restriction. Sci. Hortic. 2006, 107, 143–149. [Google Scholar] [CrossRef]
- de Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef]
- Osmont, K.S.; Sibout, R.; Hardtke, C.S. Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 2007, 58, 93–113. [Google Scholar] [CrossRef]
- Schmidt, W. Root systems biology. Front. Plant Sci. 2014, 5, 215. [Google Scholar] [CrossRef]
- Haider, M.S.; Jogaiah, S.; Pervaiz, T.; Zhao, Y.; Khan, N.; Fang, J. Physiological and transcriptional variations inducing complex adaptive mechanisms in grapevine by salt stress. Environ. Exp. Bot. 2019, 162, 455–467. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Yang, Y.X. Grape root growth and management. Nephron Clin. Pract. 2004, 61, 69–80. [Google Scholar]
- Fang, J.; Jogaiah, S.; Guan, L.; Sun, X.; Abdelrahman, M. Coloring biology in grape skin: A prospective strategy for molecular farming. Physiol. Plant. 2018, 164, 429–441. [Google Scholar] [CrossRef]
- Bai, J.; Li, N.; Fu, C.X.; Zhang, Z.Y.; Wang, Y.A.; Shu, H.R. Effect of micro-environment regulation mode around root zone on the soil property and root growth of apple trees. Acta Hortic. Sin. 2016, 43, 829–840. [Google Scholar]
- Li, X. The Effection on Osmanthus fragrans’s Root System and Function under the Control Root Cultivation Mode. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2010. [Google Scholar]
- Zhu, L.; Tang, X.; Lu, C. Effects of root zone restriction on growth, fruit quality and nutrient element content of Fujiminori grape. South China Fruits 2004, 33, 80–82. [Google Scholar]
- Cao, W. Kinetic Study of Plant Hormone Regulation on Root Growth of Arabidopsis thaliana. Ph.D. Thesis, Central China Normal University, Wuhan, China, 2019. [Google Scholar]
- Richards, D.; Rowe, R.N. Root-shoot interactions in peach: The function of the root. Ann. Bot. 1977, 41, 1211–1216. [Google Scholar] [CrossRef]
- Xie, Z.S.; Li, B.; Xu, W.P. Effects of rooting-zone restriction on the development and main nutriment contents in grape (Vitis vinifera L.) berry. Plant Physiol. Commun. 2008, 44, 927–930. [Google Scholar]
- Hawver, G.A. Influence of Root Restriction and Drought Stress on Container-Grown Trees: Impacts on Plant Morphology and Physiology. Master’s Thesis, Cornell University, New York, NY, USA, 1997. [Google Scholar]
- Cui, S.; Hu, W.X.; Xie, Z.Q.; Zhu, X.D. Effects of root restriction cultivation on endogenous hormones of grape fruits. Contemp. Hortic. 2019, 8, 4–5. [Google Scholar]
- Xu, Y.; Jiu, S.T.; Wang, J.Y.; Wang, L.; Xu, W.P.; Zhang, C.X.; Zhao, L.P.; Wang, S.P. Effect of root restriction on IAA content and metabolism-related gene expression in root of Vitis vinifera. Acta Bot. Boreali-Occident. Sin. 2021, 41, 52–62. [Google Scholar]
- Li, R.Y.; Min, Z.; Fang, Y.L. Effects of strigolactones on growth of ‘Cabernet Sauvignon’ seedlings under drought stress. J. Northwest A F Univ. 2019, 47, 73–83. [Google Scholar]
- Ma, N.; Wan, L.; Zhao, W.; Liu, H.; Li, J.; Zhang, C. Exogenous strigolactones promote lateral root growth by reducing the endogenous auxin level in rapeseed. J. Integ. Agric. 2020, 19, 465–482. [Google Scholar] [CrossRef]
- Koltai, H.; Dor, E.; Hershenhorn, J.; Joel, D.M.; Weininger, S.; Lekalla, S.; Shealtiel, H.; Bhattacharya, C.; Eliahu, E.; Resnick, N. Strigolactones effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J. Plant Growth Regul. 2010, 29, 129–136. [Google Scholar] [CrossRef]
- Koltai, H.; Lekkala, S.P.; Bhattacharya, C.; Mayzlish-Gati, E.; Resnick, N.; Wininger, S.; Dor, E.; Yoneyama, K.; Hershenhorn, J.; Joel, D.M.; et al. A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 2010, 61, 1739–1749. [Google Scholar] [CrossRef]
- Shen, X.; Xue, Z.; Sun, L.; Zhao, C.; Sun, S.; Liu, J.; Zhao, Y.; Liu, J. Effect of GR24 concentrations on biogas upgrade and nutrient removal by microalgae- based technology. Bioresour. Technol. 2020, 312, 123563. [Google Scholar] [CrossRef]
- Mitra, D.; Rad, K.V.; Chaudhary, P.; Ruparelia, J.; Panneerselvam, P. Involvement of strigolactone hormone in root development, influence and interaction with mycorrhizal fungi in plant: Mini-review. Curr. Res. Microb. Sci. 2021, 2, 100026. [Google Scholar]
- Min, Z.; Li, R.; Chen, L.; Zhang, Y.; Li, Z.; Liu, M.; Fang, Y. Alleviation of drought stress in grapevine by foliar-applied strigolactones. Plant Physiol. Biochem. 2019, 135, 99–110. [Google Scholar] [CrossRef] [PubMed]
- An, J.C. Molecular Cloning and Functional Characterization of Two Genes (CiNIP5 and CiNIP6) Associated with Boron Transport from Trifoliate Orange [Poncirus trifoliata (L.) RAF.]. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2012. [Google Scholar]
- Rial, C.; Varela, R.M.; Molinillo, J.M.G.; López-Ráez, J.A.; Macías, F.A. A new UHPLC-MS/MS method for the direct determination of strigolactones in root exudates and extracts. Phytochem. Anal. 2018, 30, 110–116. [Google Scholar] [CrossRef]
- Jiu, S.; Zhu, X.; Wang, J.; Zhang, C.; Mu, Q.; Wang, C.; Fang, J.G. Genome-wide mapping and analysis of grapevine microRNAs and their potential target genes. Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Jiu, S.T.; Wang, C.; Zheng, T.; Liu, Z.J.; Leng, X.P.; Pervaiz, T.; Lotfi, A.; Fang, J.G.; Wang, X.M. Characterization of VvPAL-like promoter from grapevine using transgenic tobacco plants. Funct. Integr. Genom. 2016, 16, 595–617. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Jiu, S.; Zhang, C.; Wang, C.; Tariq, P.; Liu, Z.; Wang, B.; Cui, L.; Fang, J. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress ripening transcription factor. Plant Biotechnol. J. 2016, 14, 2045–2065. [Google Scholar] [CrossRef]
- Jiu, S.; Xu, Y.; Wang, J.; Wang, L.; Liu, X.; Sun, W.; Irfan, A.S.; Ma, C.; Xu, W.; Wang, S.; et al. The cytochrome P450 monooxygenase inventory of grapevine (Vitis Vinifera L.): Genome-wide identification, evolutionary characterization and expression analysis. Front. Genet. 2020, 11, 44. [Google Scholar] [CrossRef]
- Jiu, S.; Guan, L.; Leng, X.; Zhang, K.K.; Haider, M.S.; Yu, X.; Zheng, T.; Ge, M.; Wang, C.; Jia, H.; et al. The role of VvMYBA2r and VvMYBA2w alleles of MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 2021, 19, 1216–1239. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, C.; Jogaiah, S.; Zhang, W.; Abdelrahman, M.; Fang, J. Spatio-temporal expression of miRNA159 family members and their GAMYB target gene during the modulation of gibberellin-induced grapevine parthenocarpy. J. Exp. Bot. 2018, 69, 3639–3650. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Wang, J.; Wang, R.; Wang, L.; Zhang, C.; Xu, W.; Wang, S.; Jiu, S. The Role of Strigolactones in the Regulation of Root System Architecture in Grapevine (Vitis vinifera L.) in Response to Root-Restriction Cultivation. Int. J. Mol. Sci. 2021, 22, 8799. https://doi.org/10.3390/ijms22168799
Xu Y, Wang J, Wang R, Wang L, Zhang C, Xu W, Wang S, Jiu S. The Role of Strigolactones in the Regulation of Root System Architecture in Grapevine (Vitis vinifera L.) in Response to Root-Restriction Cultivation. International Journal of Molecular Sciences. 2021; 22(16):8799. https://doi.org/10.3390/ijms22168799
Chicago/Turabian StyleXu, Yan, Jiyuan Wang, Ruiqi Wang, Lei Wang, Caixi Zhang, Wenping Xu, Shiping Wang, and Songtao Jiu. 2021. "The Role of Strigolactones in the Regulation of Root System Architecture in Grapevine (Vitis vinifera L.) in Response to Root-Restriction Cultivation" International Journal of Molecular Sciences 22, no. 16: 8799. https://doi.org/10.3390/ijms22168799
APA StyleXu, Y., Wang, J., Wang, R., Wang, L., Zhang, C., Xu, W., Wang, S., & Jiu, S. (2021). The Role of Strigolactones in the Regulation of Root System Architecture in Grapevine (Vitis vinifera L.) in Response to Root-Restriction Cultivation. International Journal of Molecular Sciences, 22(16), 8799. https://doi.org/10.3390/ijms22168799






