Conditional Inactivation of Limbic Neuropeptide Y-1 Receptors Increases Vulnerability to Diet-Induced Obesity in Male Mice
Abstract
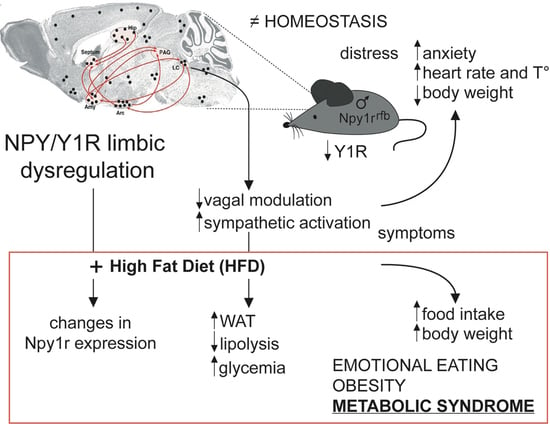
:1. Introduction
2. Results
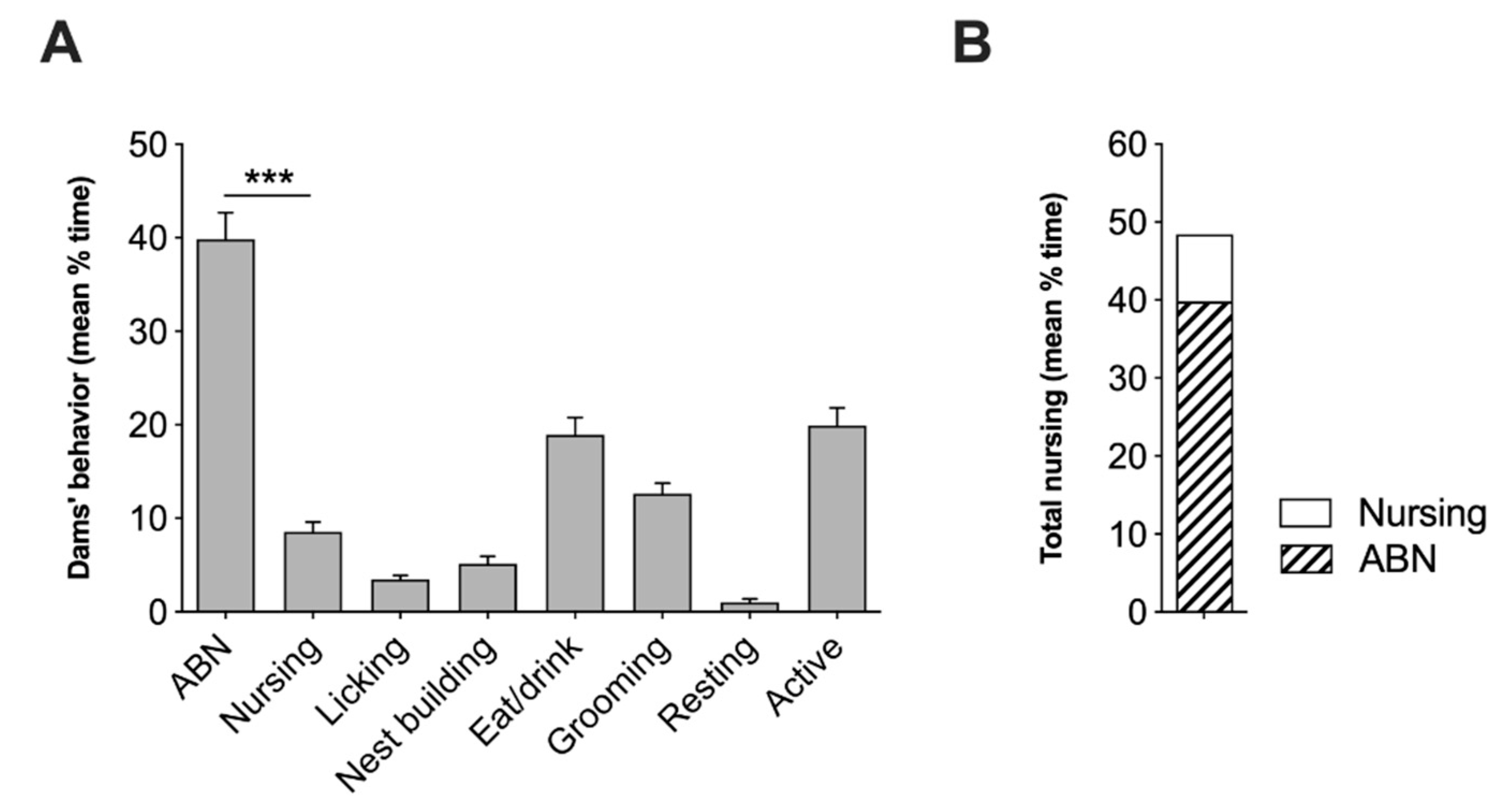
2.1. Analysis of Spontaneous Maternal Behavior of Swiss CD1 Foster Dams
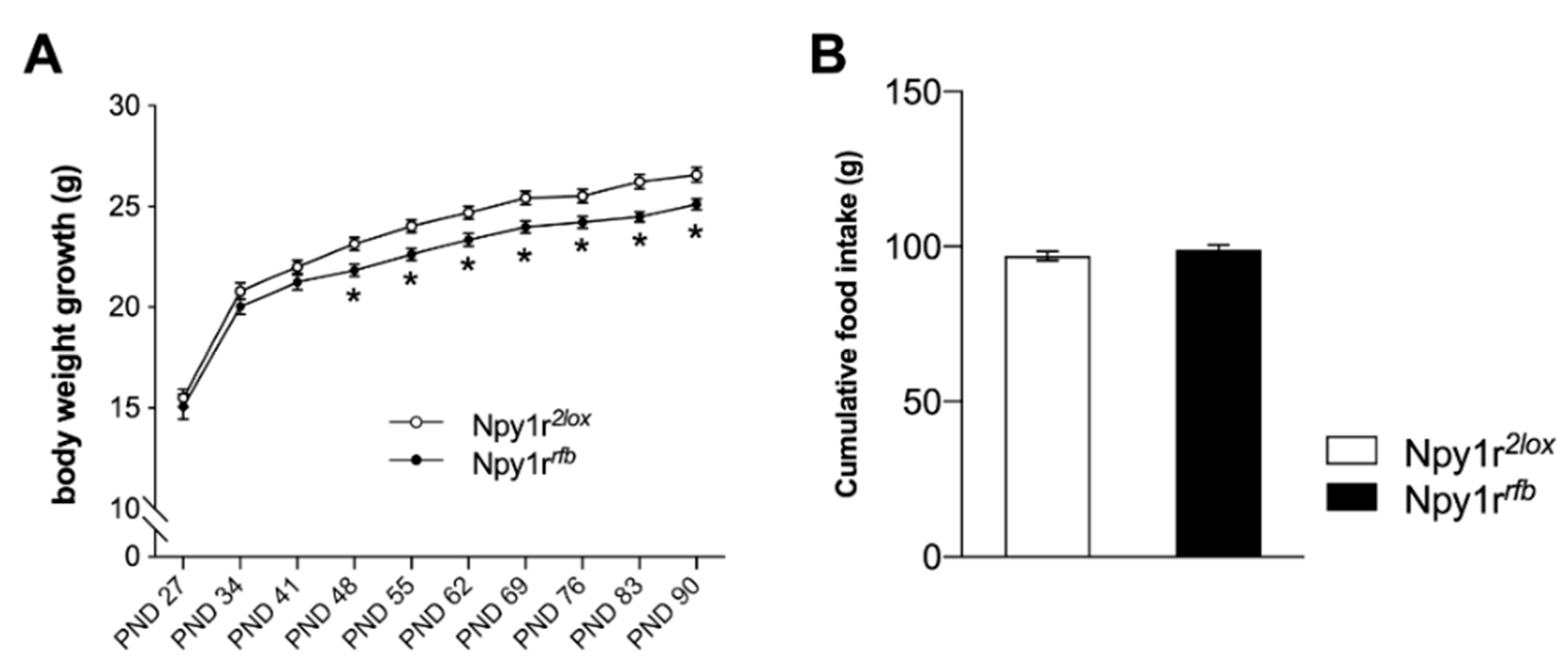
2.2. Body Weight Growth
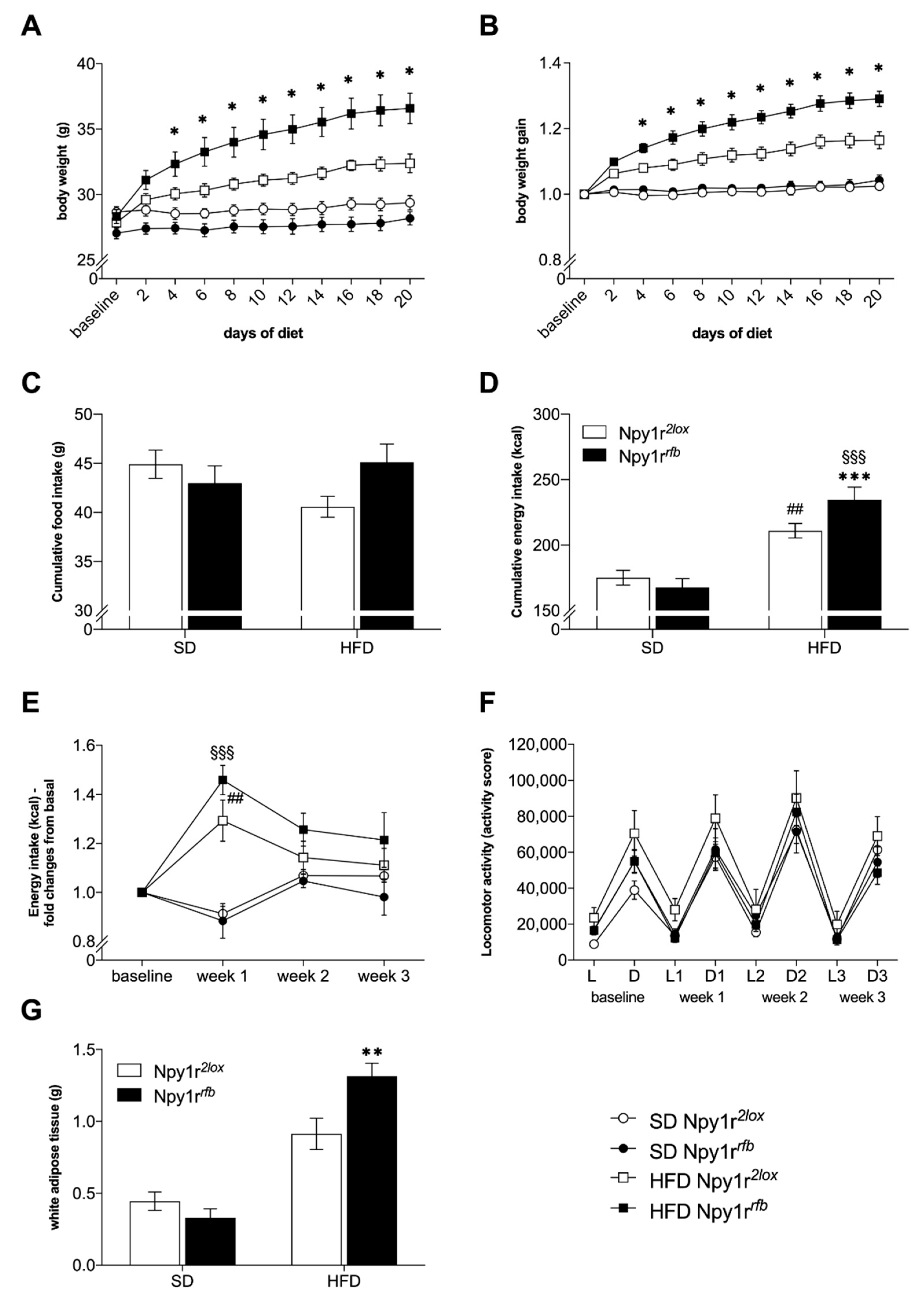
2.3. Metabolic Consequences of Diet Exposure
2.3.1. Body Weight and Food Intake
2.3.2. Locomotor Activity
2.3.3. Visceral Adiposity
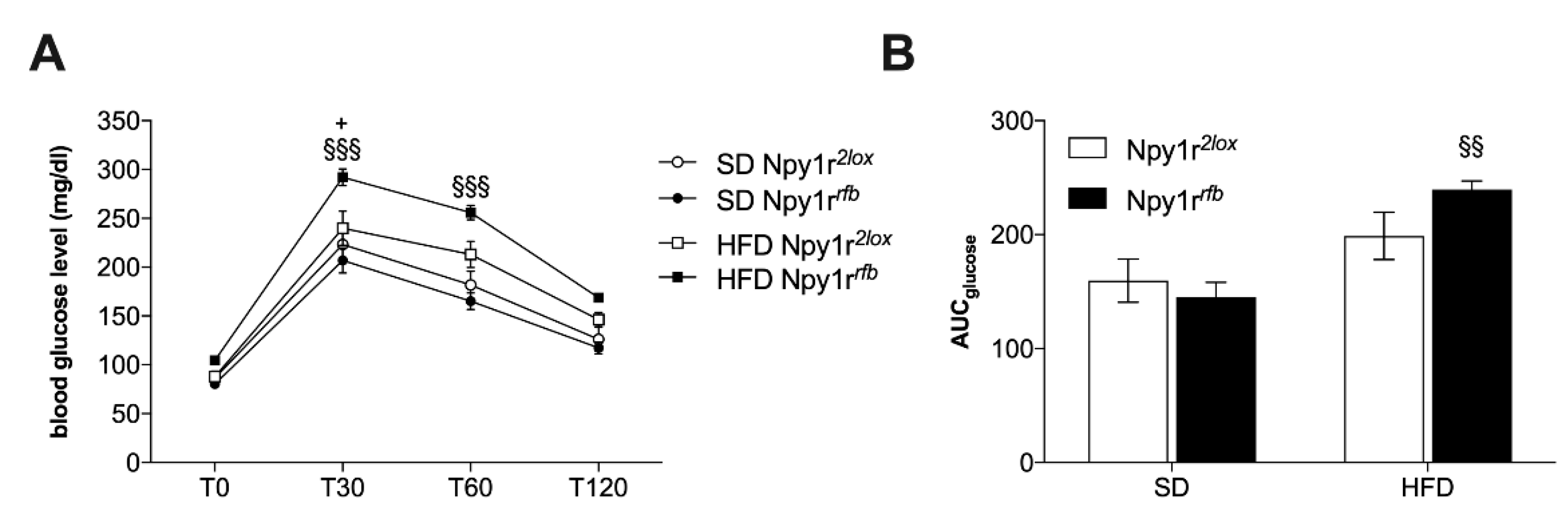
2.3.4. Blood Glucose Level
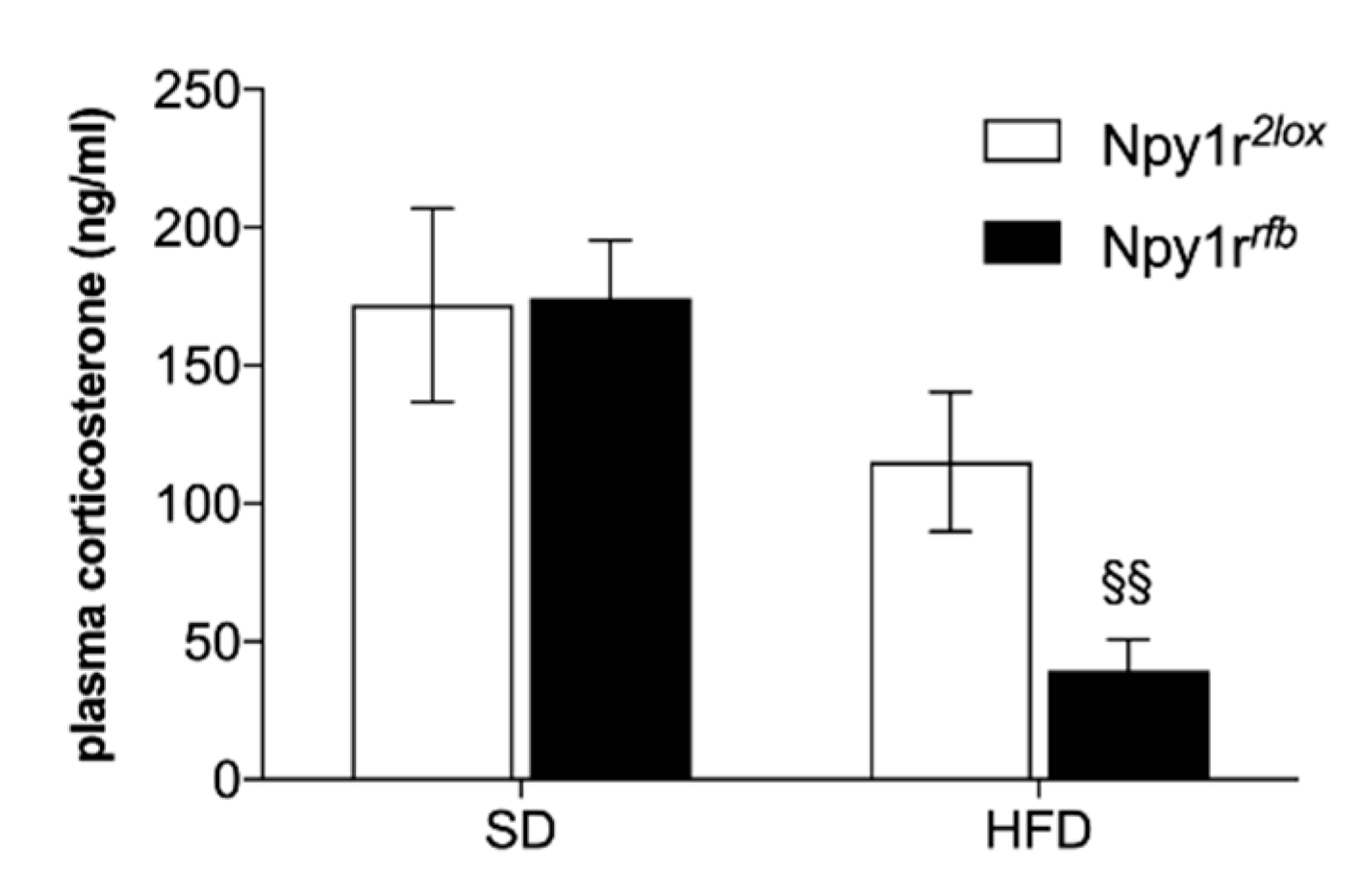
2.4. Plasma Analysis
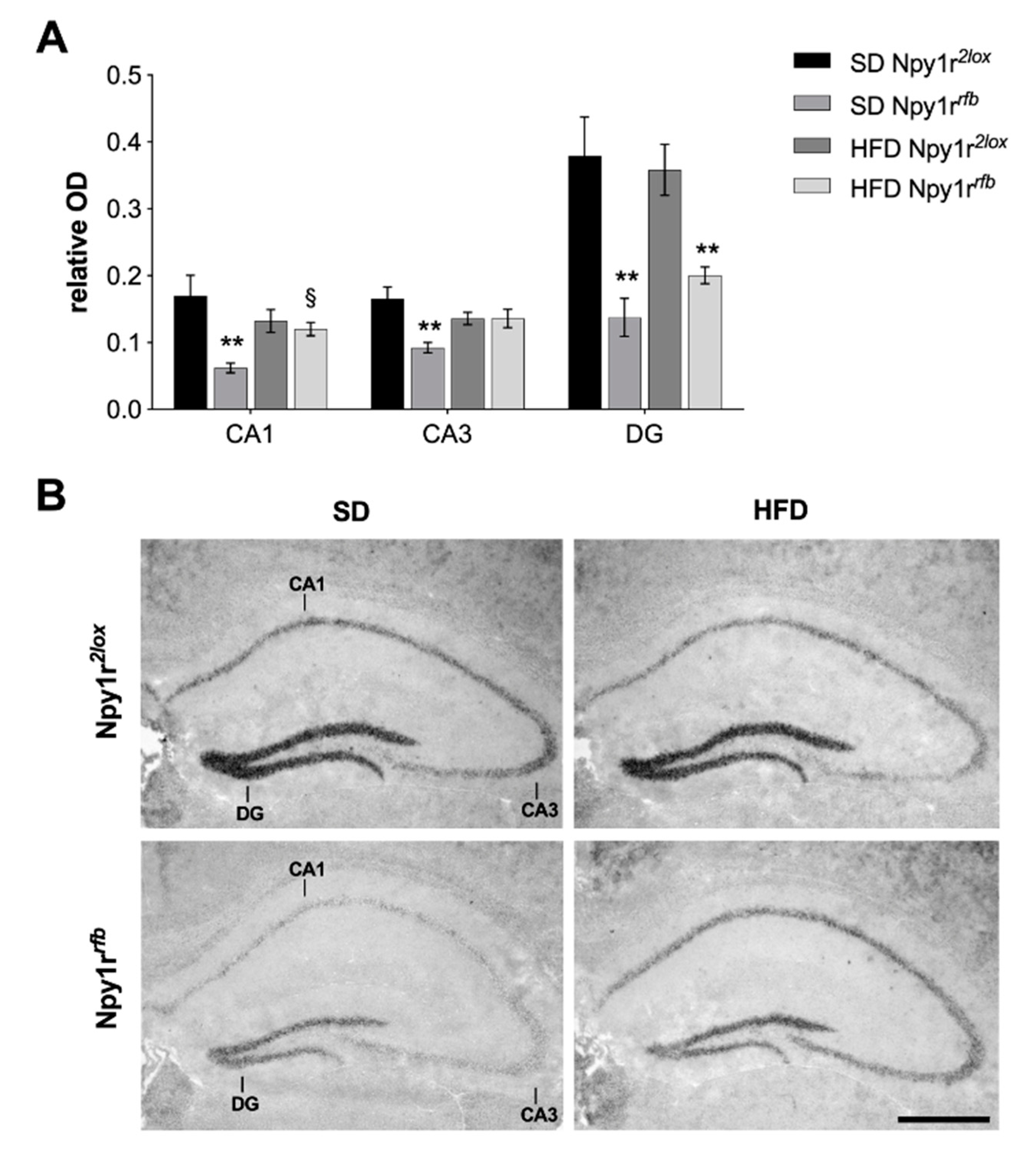
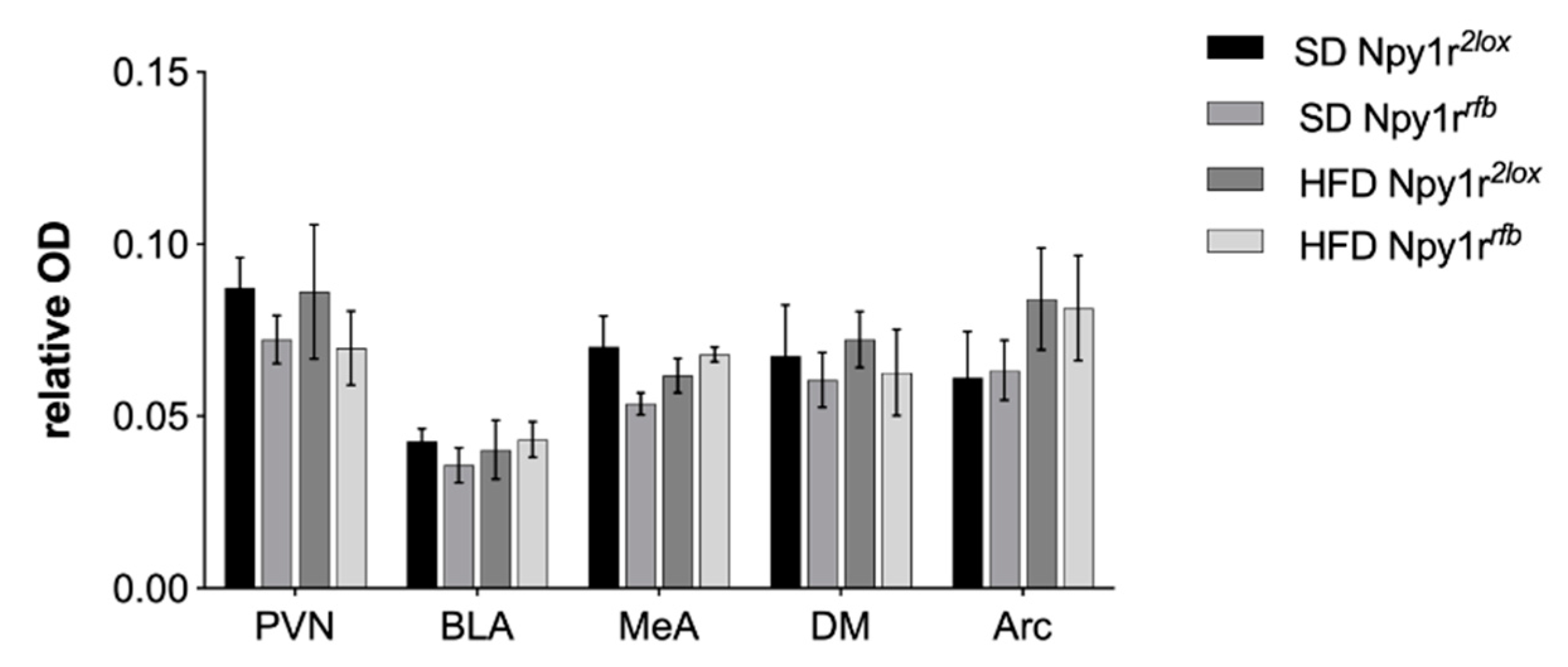
2.5. Npy1r mRNA Expression
3. Discussion
4. Materials and Methods
4.1. Experimental Outline
4.2. Animals and Housing
4.3. Maternal Behavior
4.4. Body Weight Growth
4.5. Diet Exposure
4.5.1. Body Weight and Food Intake
4.5.2. Home Cage Locomotor Activity
4.5.3. Glucose Tolerance Test (GTT)
4.5.4. Blood and Organ Collection
4.5.5. Plasma Analysis
4.6. In Situ Hybridization for Npy1r mRNA
4.7. Statistical Analysis
5. Conclusions
- No data were collected on the components of energy metabolism (e.g., energy expenditure and respiratory exchange ratio).
- Anxiety-like behavior was not measured in mice used in this study.
- Only males were used in this study, their female siblings were used in a different parallel study (Paterlini et al. in preparation).
- Limbic NPY-Y1R system is involved in energy balance and emotional behavior.
- High fat diet increases body weight, food and calory intake, glycemia and fat depots in male mice with limbic Y1R gene deletion.
- Limbic Npy1r gene deletion increases susceptibility to diet-induced obesity in male mice.
- Selective inactivation of limbic NPY1r gene might be involved in emotional eating disorders.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| Arc | arcuate hypothalamic nucleus |
| AUC | area under the curve |
| BLA | basolateral amygdala |
| CA1 | cornu ammonis 1 hippocampal subregion |
| CA3 | cornu ammonis 3 hippocampal subregion |
| CamKII | calcium/calmodulin dependent protein kinase II |
| Cre | cre recombinase |
| DM | dorsomedial hypothalamic nucleus |
| DG | dentate gyrus |
| HFD | high fat diet |
| MeA | medial amygdala |
| NPY | neuropeptide Y |
| Npy1r | gene encoding the Y1 receptor for NPY |
| PND | postnatal day |
| PVN | paraventricular hypothalamic nucleus |
| SD | standard diet |
| Y1R | Y1 receptor |
References
- Eva, C.; Oberto, A.; Longo, A.; Palanza, P.; Bertocchi, I. Sex differences in behavioral and metabolic effects of gene inactivation: The neuropeptide Y and Y receptors in the brain. Neurosci. Biobehav. Rev. 2020, 119, 333–347. [Google Scholar] [CrossRef]
- Eva, C.; Serra, M.; Mele, P.; Panzica, G.; Oberto, A. Physiology and gene regulation of the brain NPY Y1 receptor. Front. Neuroendocrinol. 2006, 27, 308–339. [Google Scholar] [CrossRef]
- Hökfelt, T.; Broberger, C.; Zhang, X.; Diez, M.; Kopp, J.; Xu, Z.Q.; Landry, M.; Bao, L.; Schalling, M.; Koistinaho, J.; et al. Neuropeptide Y: Some viewpoints on a multifaceted peptide in the normal and diseased nervous system. Brain Res. Brain Res. Rev. 1998, 26, 154–166. [Google Scholar] [CrossRef]
- Tatemoto, K.; Carlquist, M.; Mutt, V. Neuropeptide Y—A novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature 1982, 296, 659–660. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.T.; Kalra, P.S.; Crowley, W.R.; Kalra, S.P. Neuropeptide γ and human pancreatic polypeptide stimulate feeding behavior in rats. Endocrinology 1984, 115, 427–429. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.S.; Morley, J.E. Neuropeptide Y: A potent inducer of consummatory behavior in rats. Peptides 1984, 5, 1025–1029. [Google Scholar] [CrossRef]
- Rasmusson, A.M.; Schnurr, P.P.; Zukowska, Z.; Scioli, E.; Forman, D.E. Adaptation to extreme stress: Post-traumatic stress disorder, neuropeptide Y and metabolic syndrome. Exp. Biol. Med. 2010, 235, 1150–1162. [Google Scholar] [CrossRef]
- Gumbs, M.C.R.; van den Heuvel, J.K.; la Fleur, S.E. The effect of obesogenic diets on brain neuropeptide Y. Physiol. Behav. 2016, 162, 161–173. [Google Scholar] [CrossRef]
- Guan, X.M.; Yu, H.; Trumbauer, M.; Frazier, E.; Van Der Ploeg, L.H.T.; Chen, H. Induction of neuropeptide Y expression in dorsomedial hypothalamus of diet-induced obese mice. Neuroreport 1998, 9, 3415–3419. [Google Scholar] [CrossRef]
- Kesterson, R.A.; Huszar, D.; Lynch, C.A.; Simerly, R.B.; Cone, R.D. Induction of neuropeptide Y gene expression in the dorsal medial hypothalamic nucleus in two models of the agouti obesity syndrome. Mol. Endocrinol. 1997, 11, 630–637. [Google Scholar] [CrossRef]
- Wilding, J.P.H.; Gilbey, S.G.; Bailey, C.J.; Batt, R.A.L.; Williams, G.; Ghatei, M.A.; Bloom, S.R. Increased neuropeptide-y messenger ribonucleic acid (mRNA) and decreased neurotensin mrna in the hypothalamus of the obese (ob/ob) mouse. Endocrinology 1993, 132, 1939–1944. [Google Scholar] [CrossRef]
- Sanacora, G.; Kershaw, M.; Judith, J.A.; White, J.D. Increased hypothalamic content of preproneuropeptide y messenger ribonucleic acid in genetically obese zucker rats and its regulation by food deprivation. Endocrinology 1990, 127, 730–737. [Google Scholar] [CrossRef]
- Dryden, S.; Pickavance, L.; Frankish, H.M.; Williams, G. Increased neuropeptide Y secretion in the hypothalamic paraventricular nucleus of obese (fa/fa) Zucker rats. Brain Res. 1995, 690, 185–188. [Google Scholar] [CrossRef]
- Li, A.J.; Ritter, S. Glucoprivation increases expression of neuropeptide Y mRNA in hindbrain neurons that innervate the hypothalamus. Eur. J. Neurosci. 2004, 19, 2147–2154. [Google Scholar] [CrossRef]
- Mitchell, G.C.; Wang, Q.; Ramamoorthy, P.; Whim, M.D. A common single nucleotide polymorphism alters the synthesis and secretion of neuropeptide Y. J. Neurosci. 2008, 28, 14428–14434. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Kull, B.; Liu, Z.; Mottagui-Tabar, S.; Thonberg, H.; Gu, H.F.; Brookes, A.J.; Grundemar, L.; Karlsson, C.; Hamsten, A.; et al. Human neuropeptide Y signal peptide gain-of-function polymorphism is associated with increased body mass index: Possible mode of function. Regul. Pept. 2005, 127, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Nordman, S.; Ding, B.; Ostenson, C.-G.; Karvestedt, L.; Brismar, K.; Efendic, S.; Gu, H.F. Leu7Pro polymorphism in the neuropeptide Y (NPY) gene is associated with impaired glucose tolerance and type 2 diabetes in Swedish men. Exp. Clin. Endocrinol. Diabetes 2005, 113, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Ukkola, O.; Kesaniemi, Y.A. Leu7Pro polymorphism of PreproNPY associated with an increased risk for type II diabetes in middle-aged subjects. Eur. J. Clin. Nutr. 2007, 61, 1102–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karvonen, M.K.; Pesonen, U.; Koulu, M.; Niskanen, L.; Laakso, M.; Rissanen, A.; Dekker, J.M.; Hart, L.M.; Valve, R.; Uusitupa, M.I. Association of a leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y with high serum cholesterol and LDL cholesterol levels. Nat. Med. 1998, 4, 1434–1437. [Google Scholar] [CrossRef]
- Wallerstedt, S.M.; Skrtic, S.; Eriksson, A.-L.; Ohlsson, C.; Hedner, T. Association analysis of the polymorphism T1128C in the signal peptide of neuropeptide Y in a Swedish hypertensive population. J. Hypertens. 2004, 22, 1277–1281. [Google Scholar] [CrossRef]
- Kallio, J.; Pesonen, U.; Kaipio, K.; Karvonen, M.K.; Jaakkola, U.; Heinonen, O.J.; Uusitupa, M.I.; Koulu, M. Altered intracellular processing and release of neuropeptide Y due to leucine 7 to proline 7 polymorphism in the signal peptide of preproneuropeptide Y in humans. FASEB J. 2001, 15, 1242–1244. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, F.; Holzer, P. Neuropeptide Y: A stressful review. Neuropeptides 2016, 55, 99–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zammaretti, F.; Panzica, G.; Eva, C. Sex-dependent regulation of hypothalamic neuropeptide Y-Y1 receptor gene expression in moderate/high fat, high-energy diet-fed mice. J. Physiol. 2007, 583, 445–454. [Google Scholar] [CrossRef]
- Herzog, H. Neuropeptide Y and energy homeostasis: Insights from Y receptor knockout models. Eur. J. Pharmacol. 2003, 480, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.S.B.; Wilding, J.P.H.; Pinkney, J.H. Gut peptides and the regulation of appetite. Obes. Rev. 2006, 7, 163–182. [Google Scholar] [CrossRef]
- Loh, K.; Zhang, L.; Brandon, A.; Wang, Q.; Begg, D.; Qi, Y.; Fu, M.; Kulkarni, R.; Teo, J.; Baldock, P.; et al. Insulin controls food intake and energy balance via NPY neurons. Mol. Metab. 2017, 6, 574–584. [Google Scholar] [CrossRef]
- Renshaw, D.; Batterham, R.L. Peptide YY: A potential therapy for obesity. Curr. Drug Targets 2005, 6, 171–179. [Google Scholar] [CrossRef]
- Bertocchi, I.; Oberto, A.; Longo, A.; Mele, P.; Sabetta, M.; Bartolomucci, A.; Palanza, P.; Sprengel, R.; Eva, C. Regulatory functions of limbic Y1 receptors in body weight and anxiety uncovered by conditional knockout and maternal care. Proc. Natl. Acad. Sci. USA 2011, 108, 19395–19400. [Google Scholar] [CrossRef] [Green Version]
- Bertocchi, I.; Oberto, A.; Longo, A.; Palanza, P.; Eva, C. Conditional inactivation of Npy1r gene in mice induces sex-related differences of metabolic and behavioral functions. Horm. Behav. 2020, 125, 104824. [Google Scholar] [CrossRef]
- Champagne, F.A.; Curley, J.P.; Keverne, E.B.; Bateson, P.P.G. Natural variations in postpartum maternal care in inbred and outbred mice. Physiol. Behav. 2007, 91, 325–334. [Google Scholar] [CrossRef]
- Liu, D.; Diorio, J.; Tannenbaum, B.; Caldji, C.; Francis, D.; Freedman, A.; Sharma, S.; Pearson, D.; Plotsky, P.M.; Meaney, M.J. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 1997, 277, 1659–1662. [Google Scholar] [CrossRef] [Green Version]
- Statello, R.; Carnevali, L.; Paterlini, S.; Gioiosa, L.; Bertocchi, I.; Oberto, A.; Eva, C.; Palanza, P.; Sgoifo, A. Reduced NPY Y1 receptor hippocampal expression and signs of decreased vagal modulation of heart rate in mice. Physiol. Behav. 2017, 172, 31–39. [Google Scholar] [CrossRef]
- Grundy, S.M. Metabolic syndrome pandemic. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 629–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallman, M.F.; Pecoraro, N.; Akana, S.F.; La Fleur, S.E.; Gomez, F.; Houshyar, H.; Bell, M.E.; Bhatnagar, S.; Laugero, K.D.; Manalo, S. Chronic stress and obesity: A new view of “comfort food”. Proc. Natl. Acad. Sci. USA 2003, 100, 11696–11701. [Google Scholar] [CrossRef] [Green Version]
- Singh, M. Mood, food and obesity. Front. Psychol. 2014, 5, 1–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecoraro, N.; Gomez, F.; Dallman, M.F. Glucocorticoids dose-dependently remodel energy stores and amplify incentive relativity effects. Psychoneuroendocrinology 2005, 30, 815–825. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Reston, VA, USA, 2013; ISBN 089042554X. [Google Scholar]
- Leehr, E.J.; Krohmer, K.; Schag, K.; Dresler, T.; Zipfel, S.; Giel, K.E. Emotion regulation model in binge eating disorder and obesity-a systematic review. Neurosci. Biobehav. Rev. 2015, 49, 125–134. [Google Scholar] [CrossRef]
- Smith, D.G.; Robbins, T.W. The neurobiological underpinnings of obesity and binge eating: A rationale for adopting the food addiction model. Biol. Psychiatry 2013, 73, 804–810. [Google Scholar] [CrossRef]
- Turton, R.; Chami, R.; Treasure, J. Emotional eating, binge eating and animal models of binge-type eating disorders. Curr. Obes. Rep. 2017, 6, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Wilfley, D.E.; Wilson, G.T.; Agras, W.S. The clinical significance of binge eating disorder. Int. J. Eat. Disord. 2003, 34, S96–S106. [Google Scholar] [CrossRef]
- Paredes, M.F.; Greenwood, J.; Baraban, S.C. Neuropeptide Y modulates a G protein-coupled inwardly rectifying potassium current in the mouse hippocampus. Neurosci. Lett. 2003, 340, 9–12. [Google Scholar] [CrossRef]
- St-Pierre, J.A.; Nouel, D.; Dumont, Y.; Beaudet, A.; Quirion, R. Association of neuropeptide Y Y1 receptors with glutamate-positive and NPY-positive neurons in rat hippocampal cultures. Eur. J. Neurosci. 2000, 12, 1319–1330. [Google Scholar] [CrossRef]
- Geloso, M.C.; Corvino, V.; Di Maria, V.; Marchese, E.; Michetti, F. Cellular targets for neuropeptide Y-mediated control of adult neurogenesis. Front. Cell. Neurosci. 2015, 9, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Björntorp, P. Do stress reactions cause abdominal obesity and comorbidities? Obes. Rev. 2001, 2, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Bachman, E.S.; Dhillon, H.; Zhang, C.-Y.; Cinti, S.; Bianco, A.C.; Kobilka, B.K.; Lowell, B.B. βAR Signaling required for diet-induced thermogenesis and obesity resistance. Science 2002, 297, 843–845. [Google Scholar] [CrossRef] [Green Version]
- Nagy, A.; Rossant, J.; Nagy, R.; Abramow-Newerly, W.; Roder, J.C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 1993, 90, 8424–8428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dymecki, S.M. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc. Natl. Acad. Sci. USA 1996, 93, 6191–6196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayford, M.; Bach, M.E.; Huang, Y.Y.; Wang, L.; Hawkins, R.D.; Kandel, E.R. Control of memory formation through regulated expression of a CaMKII transgene. Science 1996. [Google Scholar] [CrossRef] [Green Version]
- Schönig, K.; Schwenk, F.; Rajewsky, K.; Bujard, H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res. 2002, 30, e134. [Google Scholar] [CrossRef] [Green Version]
- Crusio, W.E.; Goldowitz, D.; Holmes, A.; Wolfer, D. Standards for the publication of mouse mutant studies. Genes Brain Behav. 2009, 8, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.J.; Simpson, E.M.; Takahashi, J.S.; Lipp, H.P.; Nakanishi, S.; Wehner, J.M.; Giese, K.P.; Tully, T.; Abel, T.; Chapman, P.F.; et al. Mutant mice and neuroscience: Recommendations concerning genetic background. Neuron 1997, 19, 755–759. [Google Scholar] [CrossRef] [Green Version]
- Shimshek, D.R.; Jensen, V.; Celikel, T.; Geng, Y.; Schupp, B.; Bus, T.; Mack, V.; Marx, V.; Hvalby, Ø.; Seeburg, P.H.; et al. Forebrain-specific glutamate receptor B deletion impairs spatial memory but not hippocampal field long-term potentiation. J. Neurosci. 2006, 26, 8428–8440. [Google Scholar] [CrossRef] [PubMed]
- Palanza, P.; Howdeshell, K.L.; Parmigiani, S.; vom Saal, F.S. Exposure to a low dose of bisphenol A during fetal life or in adulthood alters maternal behavior in mice. Environ. Health Perspect. 2002, 110, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartolomucci, A.; Cabassi, A.; Govoni, P.; Ceresini, G.; Cero, C.; Berra, D.; Dadomo, H.; Franceschini, P.; Dell’Omo, G.; Parmigiani, S.; et al. Metabolic consequences and vulnerability to diet-induced obesity in male mice under chronic social stress. PLoS ONE 2009, 4, e4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Genotype | Cholesterol (mg/dL) | Cholesterol LDL (mg/dL) | Cholesterol HDL (mg/dL) | Triglycerides (mg/dL) | ||||
|---|---|---|---|---|---|---|---|---|
| SD | HFD | SD | HFD | SD | HFD | SD | HFD | |
| Npy1r2lox | 115.25 ± 6.06 | 157.45 ± 7.71 ## | 6.63 ± 1.05 | 10.89 ± 1.25 † | 62.75 ± 3.38 | 75.56 ± 3.31 † | 71.50 ± 3.76 | 83.34 ± 11.47 |
| Npy1rrfb | 100.43 ± 5.49 | 160.75 ± 10.04 §§§ | 5.43 ± 0.37 | 11.63 ± 1.52 § | 54.00 ± 3.16 | 76.75 ± 4.69 §§ | 76.86 ± 9.21 | 82.88 ± 8.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paterlini, S.; Panelli, R.; Gioiosa, L.; Parmigiani, S.; Franceschini, P.; Bertocchi, I.; Oberto, A.; Bartolomucci, A.; Eva, C.; Palanza, P. Conditional Inactivation of Limbic Neuropeptide Y-1 Receptors Increases Vulnerability to Diet-Induced Obesity in Male Mice. Int. J. Mol. Sci. 2021, 22, 8745. https://doi.org/10.3390/ijms22168745
Paterlini S, Panelli R, Gioiosa L, Parmigiani S, Franceschini P, Bertocchi I, Oberto A, Bartolomucci A, Eva C, Palanza P. Conditional Inactivation of Limbic Neuropeptide Y-1 Receptors Increases Vulnerability to Diet-Induced Obesity in Male Mice. International Journal of Molecular Sciences. 2021; 22(16):8745. https://doi.org/10.3390/ijms22168745
Chicago/Turabian StylePaterlini, Silvia, Riccardo Panelli, Laura Gioiosa, Stefano Parmigiani, Paolo Franceschini, Ilaria Bertocchi, Alessandra Oberto, Alessandro Bartolomucci, Carola Eva, and Paola Palanza. 2021. "Conditional Inactivation of Limbic Neuropeptide Y-1 Receptors Increases Vulnerability to Diet-Induced Obesity in Male Mice" International Journal of Molecular Sciences 22, no. 16: 8745. https://doi.org/10.3390/ijms22168745
APA StylePaterlini, S., Panelli, R., Gioiosa, L., Parmigiani, S., Franceschini, P., Bertocchi, I., Oberto, A., Bartolomucci, A., Eva, C., & Palanza, P. (2021). Conditional Inactivation of Limbic Neuropeptide Y-1 Receptors Increases Vulnerability to Diet-Induced Obesity in Male Mice. International Journal of Molecular Sciences, 22(16), 8745. https://doi.org/10.3390/ijms22168745