Analysis of the Applicability of microRNAs in Peripheral Blood Leukocytes as Biomarkers of Sensitivity and Exposure to Fractionated Radiotherapy towards Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. Sample Description
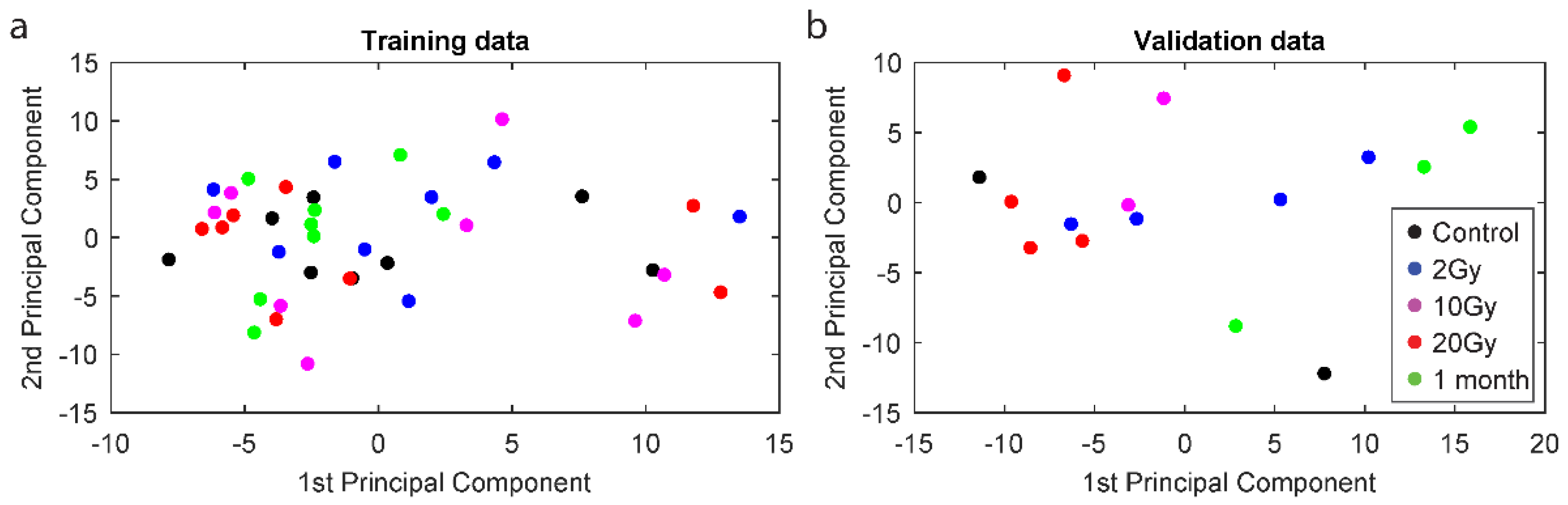
2.2. Principal Component Analysis
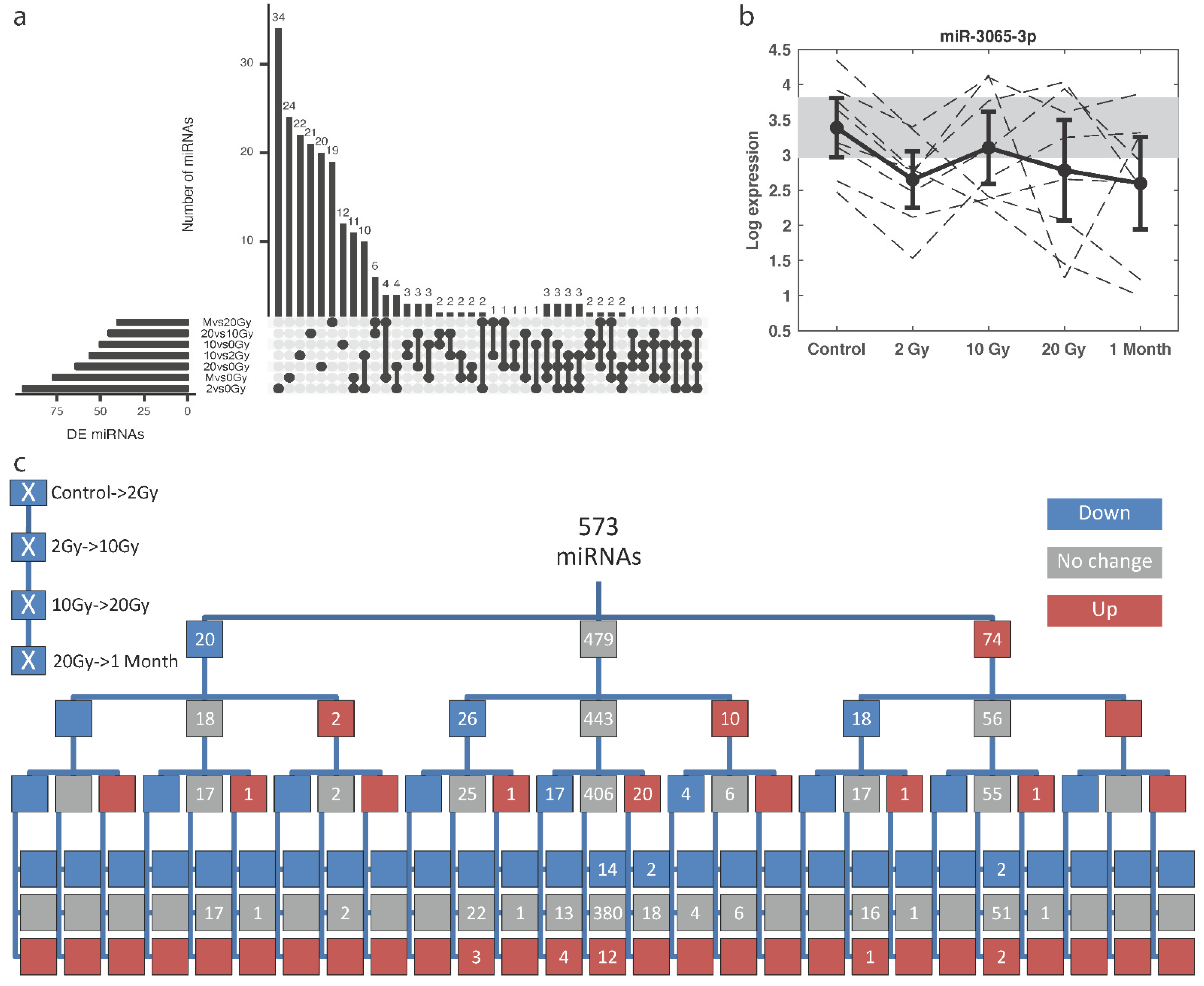
2.3. Radiation-Responsive miRNAs
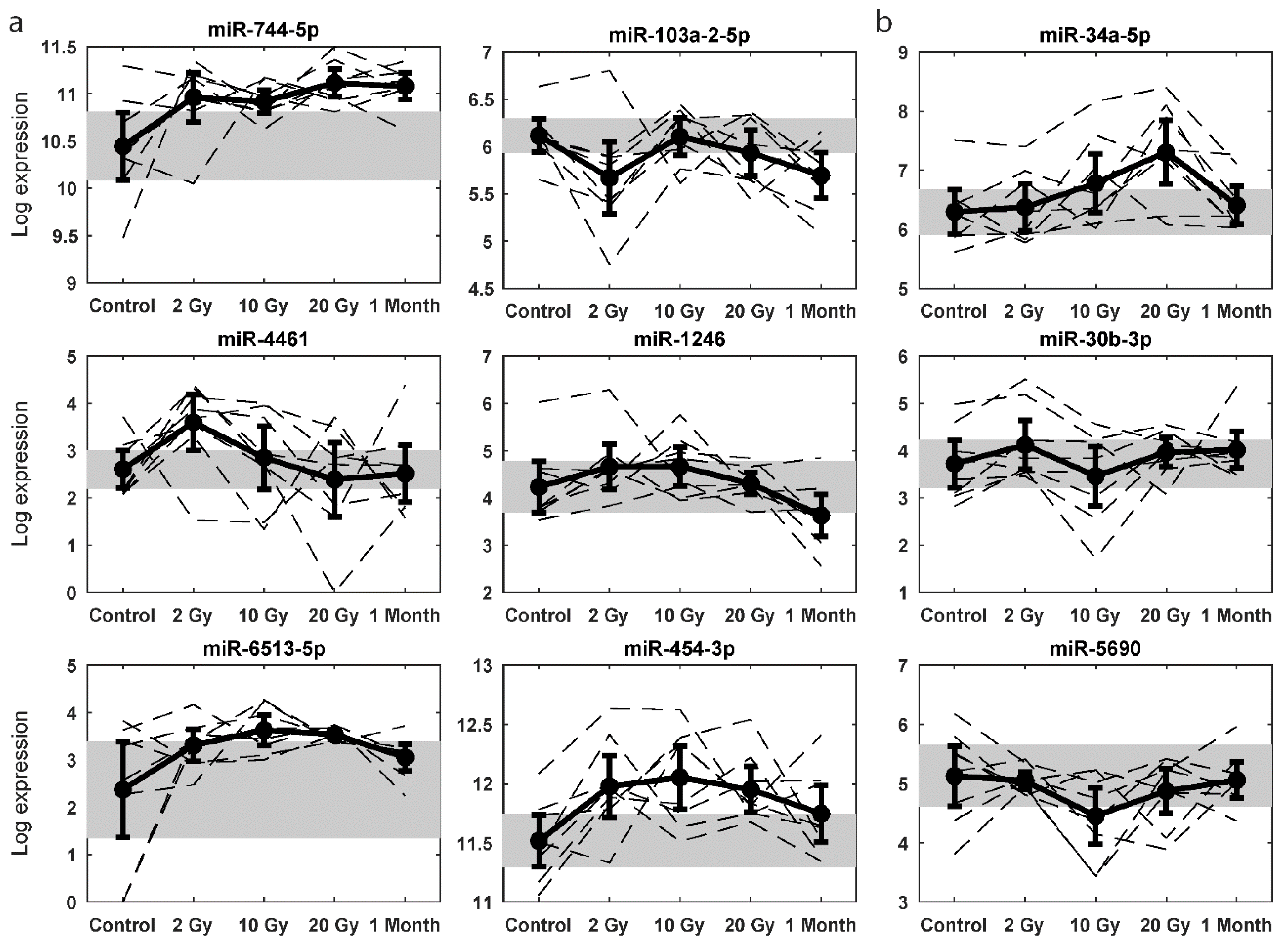
2.4. Dose Predictive miRNA Signatures
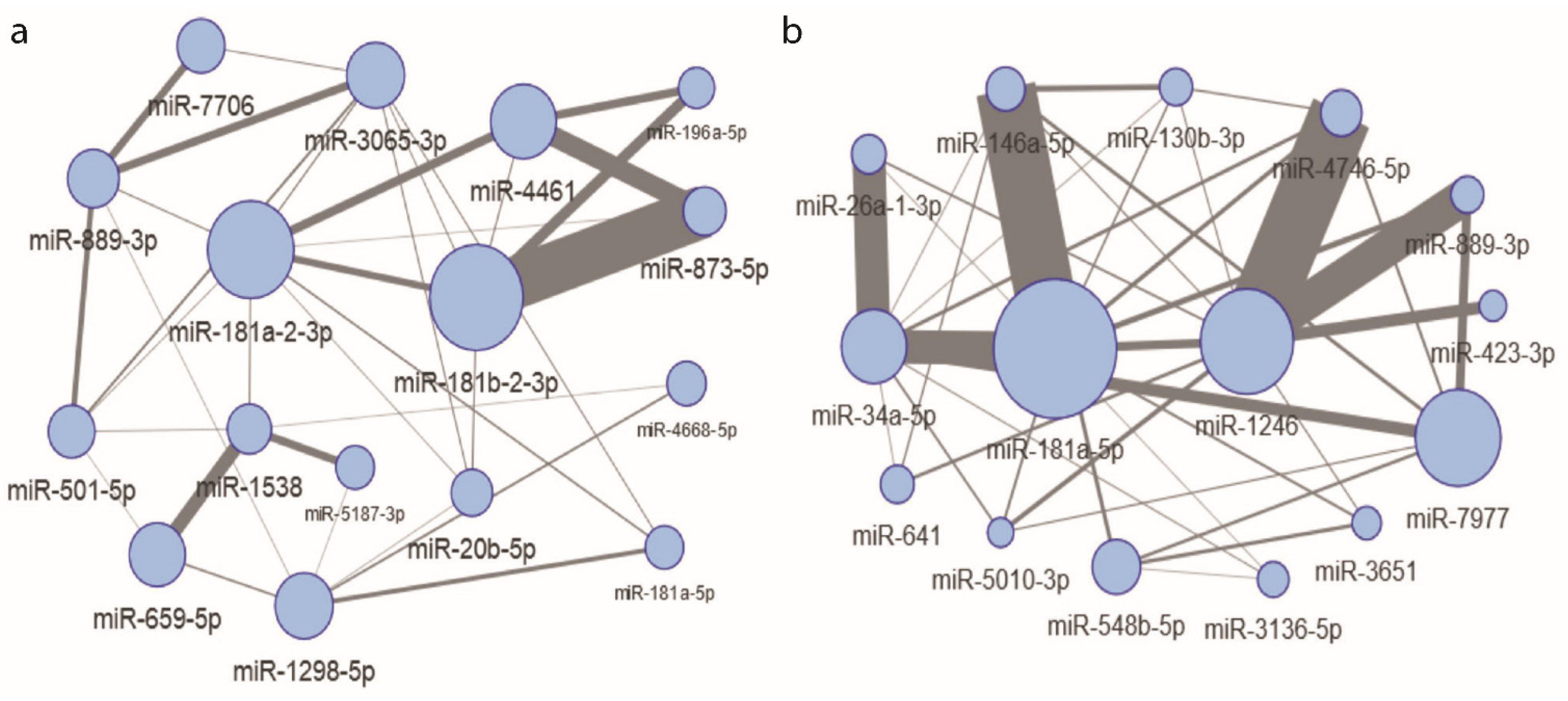
2.5. Interacting, Dose Informative miRNAs
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Sample Preparation
4.3. RNA Sequencing and Reads Pre-Processing
4.4. Data Cleaning and Imputation
4.5. Statistics of Sequencing Data
4.6. Filtering and Normalisation
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- ICRP. Prevention of Accidental Exposures to Patients Undergoing Radiation Therapy; International Commission on Radiological Protection: Ottawa, ON, Canada, 2001. [Google Scholar]
- Dorr, W. Radiobiology of tissue reactions. Ann. ICRP 2015, 44, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Newhauser, W.D.; Berrington de Gonzalez, A.; Schulte, R.; Lee, C. A Review of Radiotherapy-Induced Late Effects Research after Advanced Technology Treatments. Front. Oncol 2016, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Bentzen, S.M. Potential clinical impact of normal-tissue intrinsic radiosensitivity testing. Radiother. Oncol 1997, 43, 121–131. [Google Scholar] [CrossRef]
- Averbeck, D.; Candeias, S.; Chandna, S.; Foray, N.; Friedl, A.A.; Haghdoost, S.; Jeggo, P.A.; Lumniczky, K.; Paris, F.; Quintens, R.; et al. Establishing mechanisms affecting the individual response to ionizing radiation. Int. J. Radiat. Biol. 2020, 96, 297–323. [Google Scholar] [CrossRef] [Green Version]
- Bergom, C.; West, C.M.; Higginson, D.S.; Abazeed, M.E.; Arun, B.; Bentzen, S.M.; Bernstein, J.L.; Evans, J.D.; Gerber, N.K.; Kerns, S.L.; et al. The Implications of Genetic Testing on Radiation Therapy Decisions: A Guide for Radiation Oncologists. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, 698–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasanna, P.G.S.; Narayanan, D.; Zhang, K.; Rahbar, A.; Coleman, C.N.; Vikram, B. Radiation Biomarkers: Can Small Businesses Drive Accurate Radiation Precision Medicine? Radiat. Res. 2020, 193, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, P.; Hauptmann, M.; Bouffler, S.; Wojcik, A. Human individual radiation sensitivity and prospects for prediction. Ann. ICRP 2018, 47, 126–141. [Google Scholar] [CrossRef]
- Malicki, J. The importance of accurate treatment planning, delivery, and dose verification. Rep. Pract. Oncol. Radiother. J. Greatpoland Cancer Cent. Pozn. Pol. Soc. Radiat. Oncol. 2012, 17, 63–65. [Google Scholar] [CrossRef] [Green Version]
- Mijnheer, B.; Beddar, S.; Izewska, J.; Reft, C. In vivo dosimetry in external beam radiotherapy. Med. Phys. 2013, 40, 070903. [Google Scholar] [CrossRef]
- Tanderup, K.; Beddar, S.; Andersen, C.E.; Kertzscher, G.; Cygler, J.E. In vivo dosimetry in brachytherapy. Med. Phys. 2013, 40, 070902. [Google Scholar] [CrossRef] [Green Version]
- Deperas-Kaminska, M.; Bajinskis, A.; Marczyk, M.; Polanska, J.; Wersäll, P.; Lidbrink, E.; Ainsbury, E.A.; Guipaud, O.; Bender, M.A.; Haghdoost, S.; et al. Radiation induced changes in levels of selected proteins in peripheral blood serum of breast cancer patients as a potential triage biodosimeter for large-scale radiological emergencies. Health Phys. 2014, 107, 555–563. [Google Scholar] [CrossRef]
- Jacob, N.K.; Cooley, J.V.; Yee, T.N.; Jacob, J.; Alder, H.; Wickramasinghe, P.; Maclean, K.H.; Chakravarti, A. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS ONE 2013, 8, e57603. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; He, J.; Wang, J.; Ding, N.; Wang, B.; Lin, S.; Zhang, X.; Hua, J.; Li, H.; Hu, B. Serum microRNAs as Early Indicators for Estimation of Exposure Degree in Response to Ionizing Irradiation. Radiat. Res. 2017, 188, 342–354. [Google Scholar] [CrossRef]
- Hunten, S.; Kaller, M.; Drepper, F.; Oeljeklaus, S.; Bonfert, T.; Erhard, F.; Dueck, A.; Eichner, N.; Friedel, C.C.; Meister, G.; et al. p53-Regulated Networks of Protein, mRNA, miRNA, and lncRNA Expression Revealed by Integrated Pulsed Stable Isotope Labeling With Amino Acids in Cell Culture (pSILAC) and Next Generation Sequencing (NGS) Analyses. Mol. Cell. Proteom. MCP 2015, 14, 2609–2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Hu, Y.; Fang, J.Y.; Xu, J. Gain-of-function miRNA signature by mutant p53 associates with poor cancer outcome. Oncotarget 2016, 7, 11056–11066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. TIG 2015, 31, 613–626. [Google Scholar] [CrossRef] [Green Version]
- Miyamae, M.; Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; Shiozaki, A.; et al. Plasma microRNA profiles: Identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br. J. Cancer 2015, 113, 1467–1476. [Google Scholar] [CrossRef]
- Chaudhry, M.A.; Omaruddin, R.A.; Brumbaugh, C.D.; Tariq, M.A.; Pourmand, N. Identification of radiation-induced microRNA transcriptome by next-generation massively parallel sequencing. J. Radiat. Res. 2013, 54, 808–822. [Google Scholar] [CrossRef] [Green Version]
- Dluzen, D.F.; Kim, Y.; Bastian, P.; Zhang, Y.; Lehrmann, E.; Becker, K.G.; Noren Hooten, N.; Evans, M.K. MicroRNAs Modulate Oxidative Stress in Hypertension through PARP-1 Regulation. Oxid. Med. Cell. Longev. 2017, 2017, 3984280. [Google Scholar] [CrossRef] [Green Version]
- Yuan, D.; Xu, J.; Wang, J.; Pan, Y.; Fu, J.; Bai, Y.; Zhang, J.; Shao, C. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 2016, 7, 32707–32722. [Google Scholar] [CrossRef] [Green Version]
- Sakha, S.; Muramatsu, T.; Ueda, K.; Inazawa, J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38750. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Ding, N.; Hu, W.; He, J.; Xu, S.; Pei, H.; Hua, J.; Zhou, G.; Wang, J. Down-regulation of BTG1 by miR-454-3p enhances cellular radiosensitivity in renal carcinoma cells. Radiat. Oncol. 2014, 9, 179. [Google Scholar] [CrossRef] [Green Version]
- Kabacik, S.; Manning, G.; Raffy, C.; Bouffler, S.; Badie, C. Time, dose and ataxia telangiectasia mutated (ATM) status dependency of coding and noncoding RNA expression after ionizing radiation exposure. Radiat. Res. 2015, 183, 325–337. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Xiao, H.; Li, Y.; Dong, J.; Luo, D.; Li, H.; Feng, G.; Wang, H.; Fan, S. Total abdominal irradiation exposure impairs cognitive function involving miR-34a-5p/BDNF axis. Biochim. Biophys. Acta 2017, 1863, 2333–2341. [Google Scholar] [CrossRef]
- Cortez, M.A.; Valdecanas, D.; Niknam, S.; Peltier, H.J.; Diao, L.; Giri, U.; Komaki, R.; Calin, G.A.; Gomez, D.R.; Chang, J.Y.; et al. In Vivo Delivery of miR-34a Sensitizes Lung Tumors to Radiation Through RAD51 Regulation. Mol. Ther. Nucleic Acids 2015, 4, e270. [Google Scholar] [CrossRef] [PubMed]
- IAEA. Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies; International Energy Agency: Vienna, Austria, 2011. [Google Scholar]
- Wojcik, A.; Stephan, G.; Sommer, S.; Buraczewska, I.; Kuszewski, T.; Wieczorek, A.; Gozdz, S. Chromosomal aberrations and micronuclei in lymphocytes of breast cancer patients after an accident during radiotherapy with 8 MeV electrons. Radiat. Res. 2003, 160, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Sak, A.; Grehl, S.; Erichsen, P.; Engelhard, M.; Grannass, A.; Levegrun, S.; Pottgen, C.; Groneberg, M.; Stuschke, M. gamma-H2AX foci formation in peripheral blood lymphocytes of tumor patients after local radiotherapy to different sites of the body: Dependence on the dose-distribution, irradiated site and time from start of treatment. Int. J. Radiat. Biol. 2007, 83, 639–652. [Google Scholar] [CrossRef] [PubMed]
- D’Alesio, V.; Pacelli, R.; Durante, M.; Canale Cama, G.; Cella, L.; Gialanella, G.; Grossi, G.; Pugliese, M.; Punzo, G.; Sardi, I.; et al. Lymph nodes in the irradiated field influence the yield of radiation-induced chromosomal aberrations in lymphocytes from breast cancer patients. Int. J. Radiat. Oncol. Biol. Phys. 2003, 57, 732–738. [Google Scholar] [CrossRef]
- O’Brien, G.; Cruz-Garcia, L.; Majewski, M.; Grepl, J.; Abend, M.; Port, M.; Tichy, A.; Sirak, I.; Malkova, A.; Donovan, E.; et al. FDXR is a biomarker of radiation exposure in vivo. Sci. Rep. 2018, 8, 684. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Tichy, A.; Sirak, I.; Badie, C. Radiotherapy-Associated Long-term Modification of Expression of the Inflammatory Biomarker Genes ARG1, BCL2L1, and MYC. Front. Immunol. 2017, 8, 412. [Google Scholar] [CrossRef] [Green Version]
- Amundson, S.A.; Grace, M.B.; McLeland, C.B.; Epperly, M.W.; Yeager, A.; Zhan, Q.; Greenberger, J.S.; Fornace, A.J., Jr. Human in vivo radiation-induced biomarkers: Gene expression changes in radiotherapy patients. Cancer Res. 2004, 64, 6368–6371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trim_Galore. Core Script: Trim_Galore--Small_Rna $RAW.Fastq.gz--Length 18--Gzip -o $TRIMMED.Fastq.gz. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 12 October 2016).
- Zhao, S.; Gordon, W.; Du, S.; Zhang, C.; He, W.; Xi, L.; Mathur, S.; Agostino, M.; Paradis, T.; von Schack, D.; et al. QuickMIRSeq: A pipeline for quick and accurate quantification of both known miRNAs and isomiRs by jointly processing multiple samples from microRNA sequencing. BMC Bioinform. 2017, 18, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Tam, S.; Tsao, M.S.; McPherson, J.D. Optimization of miRNA-seq data preprocessing. Brief. Bioinform. 2015, 16, 950–963. [Google Scholar] [CrossRef] [Green Version]
- Krol, L.; Polanska, J. Multidimensional Feature Selection and Interaction Mining with Decision Tree Based Ensemble Methods; Springer: Cham, Switzerlnad, 2017; pp. 118–125. [Google Scholar]
| Patient ID | Age | Smoker | Group | Total Dose [Gy] | ||||
|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 10 | 20 | 46–50/1 Month | ||||
| 1 | 61 | No | Validation | |||||
| 2 | 65 | No | Validation | |||||
| 3 | 56 | No | Validation | |||||
| 4 | 72 | No | Validation | |||||
| 5 | 74 | No | Validation | |||||
| 6 | 66 | NK | Validation | |||||
| 7 | 57 | Yes | Training | |||||
| 8 | 67 | No | Training | |||||
| 9 | 64 | No | Training | |||||
| 10 | 75 | No | Validation | |||||
| 11 | 81 | No | Validation | |||||
| 12 | 65 | No | Training | |||||
| 13 | 58 | Yes | Training | |||||
| 14 | 65 | No | Training | |||||
| 15 | 68 | Yes | Training | |||||
| 16 | 60 | No | Training | |||||
| Dose | 2 vs. 0 Gy | 10 vs. 2 Gy | 20 vs. 10 Gy | M vs. 20 Gy | 10 vs. 0 Gy | 20 vs. 0 Gy | M vs. 0 Gy |
|---|---|---|---|---|---|---|---|
| Up-regulated miRNAs | 74 | 12 | 24 | 22 | 36 | 46 | 61 |
| Down-regulated miRNAs | 20 | 44 | 21 | 18 | 14 | 18 | 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marczyk, M.; Polańska, J.; Wojcik, A.; Lundholm, L. Analysis of the Applicability of microRNAs in Peripheral Blood Leukocytes as Biomarkers of Sensitivity and Exposure to Fractionated Radiotherapy towards Breast Cancer. Int. J. Mol. Sci. 2021, 22, 8705. https://doi.org/10.3390/ijms22168705
Marczyk M, Polańska J, Wojcik A, Lundholm L. Analysis of the Applicability of microRNAs in Peripheral Blood Leukocytes as Biomarkers of Sensitivity and Exposure to Fractionated Radiotherapy towards Breast Cancer. International Journal of Molecular Sciences. 2021; 22(16):8705. https://doi.org/10.3390/ijms22168705
Chicago/Turabian StyleMarczyk, Michal, Joanna Polańska, Andrzej Wojcik, and Lovisa Lundholm. 2021. "Analysis of the Applicability of microRNAs in Peripheral Blood Leukocytes as Biomarkers of Sensitivity and Exposure to Fractionated Radiotherapy towards Breast Cancer" International Journal of Molecular Sciences 22, no. 16: 8705. https://doi.org/10.3390/ijms22168705
APA StyleMarczyk, M., Polańska, J., Wojcik, A., & Lundholm, L. (2021). Analysis of the Applicability of microRNAs in Peripheral Blood Leukocytes as Biomarkers of Sensitivity and Exposure to Fractionated Radiotherapy towards Breast Cancer. International Journal of Molecular Sciences, 22(16), 8705. https://doi.org/10.3390/ijms22168705







