Anti-SARS-CoV-2 Strategies and the Potential Role of miRNA in the Assessment of COVID-19 Morbidity, Recurrence, and Therapy
Abstract
1. The Challenge
2. Knowing the Enemy
3. Potential Therapeutic Strategies towards COVID-19
3.1. Old Antimalarial Drugs
3.2. Investigation into the Role of Vitamins
3.3. Convalescent Plasma Transfusions
3.4. In Response to the Cytokine Storm Syndrome: Immunomodulation Effects in COVID-19 Management
4. Vaccination Strategy
4.1. The Vaccine Run
4.2. The Vaccine-Run Winners
4.3. Further Perspective and Another Challenge
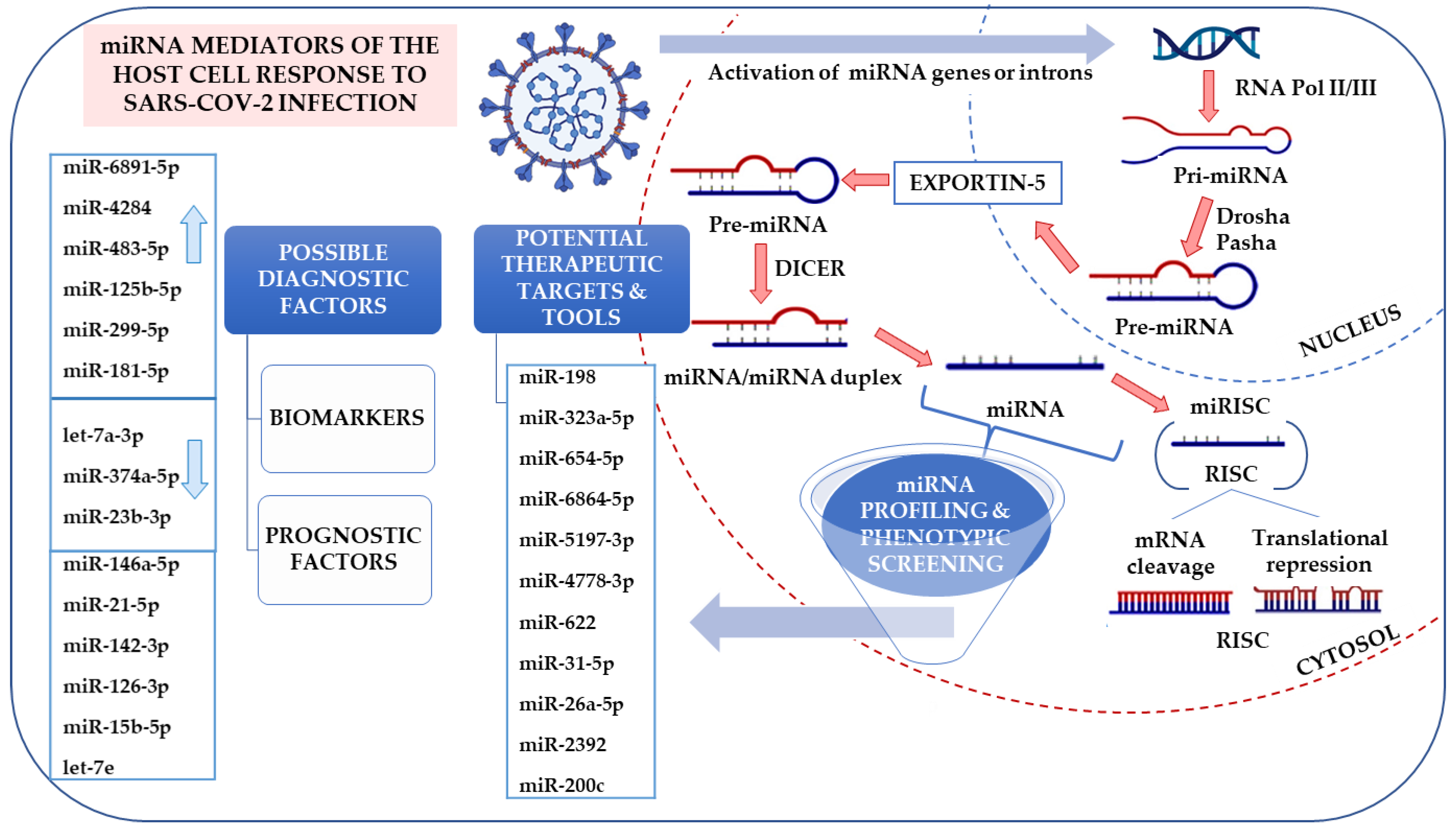
5. Alternative Approach—miRNA Profiling in Viral Infections
5.1. miRNA in Diagnostics and Therapy
5.2. The Contribution of miRNAs to the Virus Infection Course
5.3. Promising Fields
5.4. miRNAs as Diagnostic Biomarkers
5.5. The Role of miRNAs in Viral Infection and Therapy Perspective
5.6. miRNA-Based anti-COVID-19 Strategies
5.6.1. Bioinformatics—One More Challenge
Preliminary in Silico Studies
5.6.2. In Vitro and In Vivo Analyses
Recent In Vitro and In Vivo Studies
5.6.3. Molecular Targeted Therapy—The Progress and Future Strategy Perspective
5.6.4. Alternative Approach
5.7. Sensitivity and Specificity of microRNA Profiling in COVID-19 Patients
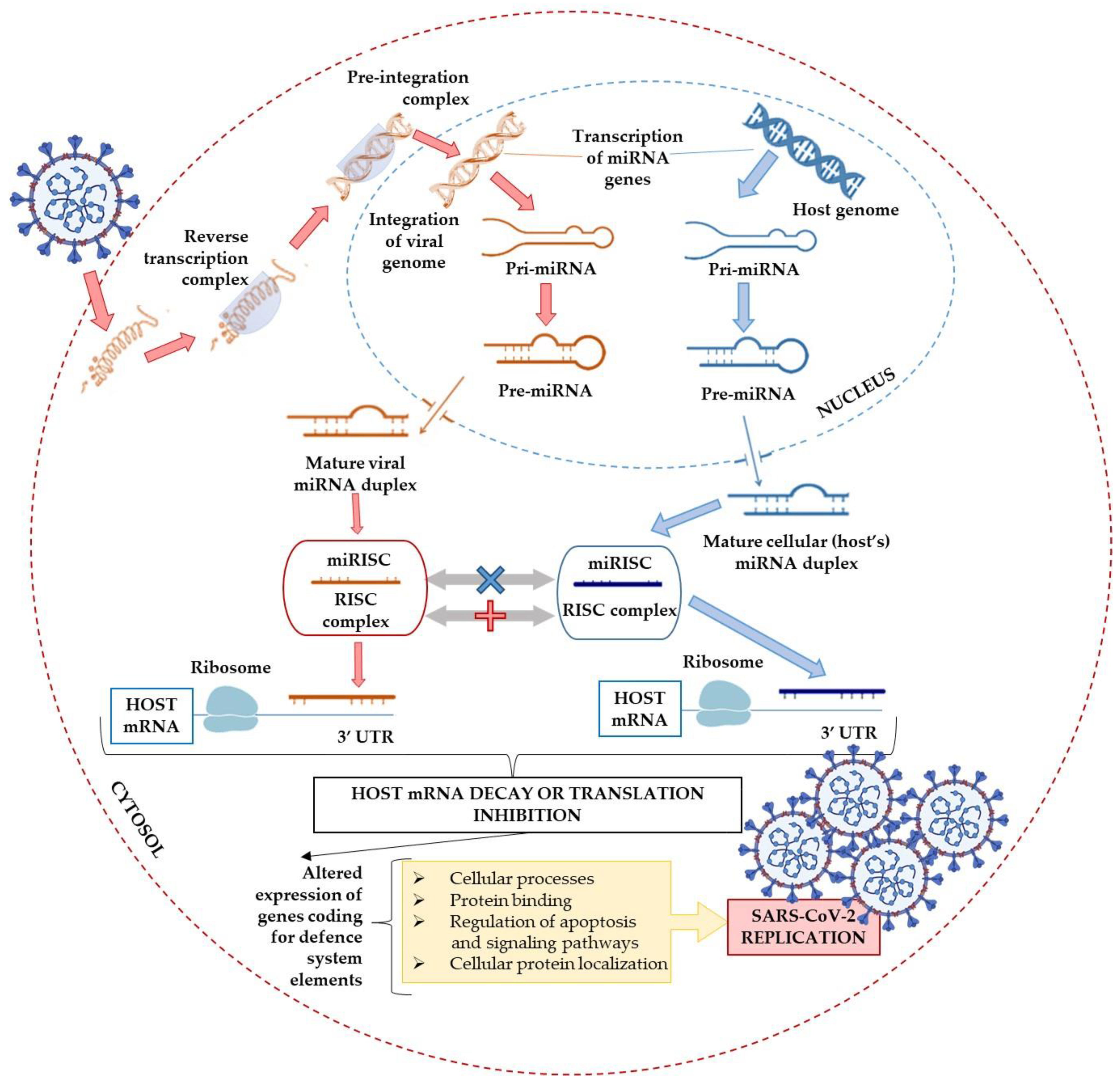
6. Alternative miRNAs-Associated Pathways
6.1. The DEAD-Box Helicases: The Double-Edged Sword in Viral Infections
6.2. Possible Interference with the miRNA Biogenesis Pathway
6.3. The miRNAs Delivery Systems
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. Available online: https://covid19.who.int/ (accessed on 7 August 2021).
- Callaway, E. The coronavirus is mutating—Does it matter? Nature 2020, 585, 174–177. [Google Scholar] [CrossRef]
- Shah, B.; Modi, P.; Sagar, S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020, 252, 117652. [Google Scholar] [CrossRef]
- Touret, F.; Gilles, M.; Barral, K.; Nougairède, A.; van Helden, J.; Decroly, E.; de Lamballerie, X.; Coutard, B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020, 10, 13093. [Google Scholar] [CrossRef] [PubMed]
- Seyedpour, S.; Khodaei, B.; Loghman, A.H.; Seyedpour, N.; Kisomi, M.F.; Balibegloo, M.; Nezamabadi, S.S.; Gholami, B.; Saghazadeh, A.; Rezaei, N. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: A systematic review of in vitro and in vivo studies. J. Cell. Physiol. 2021, 236, 2364–2392. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves First Treatment for COVID-19. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19 (accessed on 7 August 2021).
- Coronavirus (COVID-19) Drugs FDA. Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs (accessed on 7 August 2021).
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 2021, 113, 1221–1232. [Google Scholar] [CrossRef]
- Covid-19 Vaccine: First UK Citizens Receive Shot, a Landmark Moment in the Pandemic–CNN. Available online: https://edition.cnn.com/2020/12/08/europe/uk-pfizer-biontech-covid-vaccination-intl/index.html (accessed on 13 April 2021).
- Get the Facts about COVID-19 Vaccines—Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/coronavirus/in-depth/coronavirus-vaccine/art-20484859 (accessed on 13 April 2021).
- PANGO Lineages. Available online: https://cov-lineages.org/global_report.html (accessed on 13 April 2021).
- SARS-CoV-2 Variants of Concern as of 11 May 2021. Available online: https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed on 15 May 2021).
- Hudson B., S.; Kolte, V.; Khan, A.; Sharma, G. Dynamic tracking of variant frequencies depicts the evolution of mutation sites amongst SARS-CoV-2 genomes from India. J. Med. Virol. 2021, 93, 2534–2537. [Google Scholar] [CrossRef]
- Tang, J.W.; Toovey, O.T.R.; Harvey, K.N.; Hui, D.D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 2021, 82, e8–e10. [Google Scholar] [CrossRef] [PubMed]
- Conti, P.; Caraffa, A.; Gallenga, C.E.; Kritas, S.K.; Frydas, I.; Younes, A.; Di Emidio, P.; Tetè, G.; Pregliasco, F.; Ronconi, G. The British variant of the new coronavirus-19 (SARS-CoV-2) should not create a vaccine problem. J. Biol. Regul. Homeost. Agents 2021, 35, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Beyerstedt, S.; Casaro, E.B.; Rangel, É.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Verdecchia, P.; Cavallini, C.; Spanevello, A.; Angeli, F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020, 76, 14–20. [Google Scholar] [CrossRef]
- Clark, I.A. The advent of the cytokine storm. Immunol. Cell Biol. 2007, 85, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Carfì, A.; Bernabei, R.; Landi, F. Persistent symptoms in patients after acute COVID-19. JAMA J. Am. Med. Assoc. 2020, 324, 603–605. [Google Scholar] [CrossRef]
- Fotuhi, M.; Mian, A.; Meysami, S.; Raji, C.A. Neurobiology of COVID-19. J. Alzheimer’s Dis. 2020, 76, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, N.; Nadjafi, S.; Ashtary, B. Overview of COVID-19 and neurological complications. Rev. Neurosci. 2021. [Google Scholar] [CrossRef]
- Perrin, R.; Riste, L.; Hann, M.; Walther, A.; Mukherjee, A.; Heald, A. Into the looking glass: Post-viral syndrome post COVID-19. Med. Hypotheses 2020, 144, 110055. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, Y.; Sun, C.; Wang, X.; Guo, Y.; Qiu, S.; Ma, K. A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 2021, 54, 12–16. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Khullar, N.; Reddy, A.P.; Reddy, P.H. Therapeutic Strategies in the Development of Anti-viral Drugs and Vaccines Against SARS-CoV-2 Infection. Mol. Neurobiol. 2020, 57, 4856–4877. [Google Scholar] [CrossRef]
- Amini Pouya, M.; Afshani, S.M.; Maghsoudi, A.S.; Hassani, S.; Mirnia, K. Classification of the present pharmaceutical agents based on the possible effective mechanism on the COVID-19 infection. DARU J. Pharm. Sci. 2020, 28, 745–764. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Yang, N.; Shen, H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Guan, Z.; Li, H.; Ye, M.; Chen, X.; Shen, J.; Zhou, Y.; Shi, Z.L.; Zhou, P.; et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Kumari, P.; Singh, A.; Ngasainao, M.R.; Shakeel, I.; Kumar, S.; Lal, S.; Singhal, A.; Sohal, S.S.; Singh, I.K.; Hassan, M.I. Potential diagnostics and therapeutic approaches in COVID-19. Clin. Chim. Acta 2020, 510, 488–497. [Google Scholar] [CrossRef]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef] [PubMed]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pohlmann, S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2014, 88, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, Y.X.; Jiang, L.J.; Chen, Q.; Wang, T.; Ye, D.W. Targeting JAK-STAT Signaling to Control Cytokine Release Syndrome in COVID-19. Trends Pharmacol. Sci. 2020, 41, 531–543. [Google Scholar] [CrossRef]
- Hu, T.Y.; Frieman, M.; Wolfram, J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 2020, 15, 247–249. [Google Scholar] [CrossRef]
- Huchting, J. Targeting viral genome synthesis as broad-spectrum approach against RNA virus infections. Antivir. Chem. Chemother. 2020, 28. [Google Scholar] [CrossRef]
- Muhammad, Y.; Kani, Y.A.; Iliya, S.; Muhammad, J.B.; Binji, A.; El-Fulaty Ahmad, A.; Kabir, M.B.; Umar Bindawa, K.; Ahmed, A. Deficiency of antioxidants and increased oxidative stress in COVID-19 patients: A cross-sectional comparative study in Jigawa, Northwestern Nigeria. SAGE Open Med. 2021, 9. [Google Scholar] [CrossRef]
- Brenner, H.; Schöttker, B. Vitamin D Insufficiency May Account for Almost Nine of Ten COVID-19 Deaths: Time to Act. Comment on: “Vitamin D Deficiency and Outcome of COVID-19 Patients”. Nutrients 2020, 12, 3642. [Google Scholar] [CrossRef]
- Ali, R.M.; Al-Shorbagy, M.Y.; Helmy, M.W.; El-Abhar, H.S. Role of Wnt4/β-catenin, Ang II/TGFβ, ACE2, NF-κB, and IL-18 in attenuating renal ischemia/reperfusion-induced injury in rats treated with Vit D and pioglitazone. Eur. J. Pharmacol. 2018, 831, 68–76. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, R.; Kim, S.H.; Shah, H.; Zhang, S.; Liang, J.H.; Fang, Y.; Gentili, M.; Leary, C.N.O.; Elledge, S.J.; et al. SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Han, N.; Hwang, W.; Tzelepis, K.; Schmerer, P.; Yankova, E.; MacMahon, M.; Lei, W.; Katritsis, M.N.; Liu, A.; Felgenhauer, U.; et al. Identification of SARS-CoV-2-induced pathways reveals drug repurposing strategies. Sci. Adv. 2021, 7, eabh3032. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods in Mol. Biol. 2020, 2203, 1–29. [Google Scholar] [CrossRef]
- Pujari, R.; Thommana, M.V.; Ruiz Mercedes, B.; Serwat, A. Therapeutic Options for COVID-19: A Review. Cureus 2020, 12, e10480. [Google Scholar] [CrossRef]
- Horby, P.W.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Emberson, J.; Palfreeman, A.; Raw, J.; Elmahi, E.; Prudon, B.; et al. Lopinavir–ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2020, 396, 1345–1352. [Google Scholar] [CrossRef]
- Pruijssers, A.J.; George, A.S.; Schäfer, A.; Leist, S.R.; Gralinksi, L.E.; Dinnon, K.H.; Yount, B.L.; Agostini, M.L.; Stevens, L.J.; Chappell, J.D.; et al. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 32, 107940. [Google Scholar] [CrossRef]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.-X.; et al. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef]
- Hegerova, L.; Gooley, T.A.; Sweerus, K.A.; Maree, C.; Bailey, N.; Bailey, M.; Dunleavy, V.; Patel, K.; Alcorn, K.; Haley, R.; et al. Use of convalescent plasma in hospitalized patients with COVID-19: Case series. Blood 2020, 136, 759–762. [Google Scholar] [CrossRef]
- Davies, M.; Osborne, V.; Lane, S.; Roy, D.; Dhanda, S.; Evans, A.; Shakir, S. Remdesivir in Treatment of COVID-19: A Systematic Benefit–Risk Assessment. Drug Saf. 2020, 43, 645–656. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.P.; Forleo-Neto, E.; Musser, B.J.; Isa, F.; Chan, K.-C.; Sarkar, N.; Bar, K.J.; Barnabas, R.V.; Barouch, D.H.; Cohen, M.S.; et al. Subcutaneous REGEN-COV Antibody Combination to Prevent Covid-19. N. Eng. J. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Meo, S.A.; Klonoff, D.C.; Akram, J. Efficacy of chloroquine and hydroxychloroquine in the treatment of COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4539–4547. [Google Scholar] [CrossRef] [PubMed]
- Keyaerts, E.; Vijgen, L.; Maes, P.; Neyts, J.; Ranst, M. Van In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004, 323, 264–268. [Google Scholar] [CrossRef]
- Gasmi, A.; Peana, M.; Noor, S.; Lysiuk, R.; Menzel, A.; Gasmi Benahmed, A.; Bjørklund, G. Chloroquine and hydroxychloroquine in the treatment of COVID-19: The never-ending story. Appl. Microbiol. Biotechnol. 2021, 105, 1333–1343. [Google Scholar] [CrossRef]
- Firth, A.; Prathapan, P. Azithromycin: The First Broad-spectrum Therapeutic. Eur. J. Med. Chem. 2020, 207, 112739. [Google Scholar] [CrossRef]
- Abaleke, E.; Abbas, M.; Abbasi, S.; Abbott, A.; Abdelaziz, A.; Abdelbadiee, S.; Abdelfattah, M.; Abdul, B.; Rasheed, A.A.; Abdul-Kadir, R.; et al. Azithromycin in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2021, 397, 605–612. [Google Scholar] [CrossRef]
- Hydroxychloroquine or Chloroquine for COVID-19: Drug Safety Communication - FDA Cautions Against Use Outside of the Hospital Setting or a Clinical Trial Due to Risk of Heart Rhythm Problems. Available online: https://www.fda.gov/safety/medical-product-safety-information/hydroxychloroquine-or-chloroquine-covid-19-drug-safety-communication-fda-cautions-against-use (accessed on 13 April 2021).
- Horby, P.; Mafham, M.; Linsell, L.; Bell, J.L.; Staplin, N.; Emberson, J.R.; Wiselka, M.; Ustianowski, A.; Elmahi, E.; Prudon, B.; et al. Effect of Hydroxychloroquine in Hospitalized Patients with COVID-19: Preliminary results from a multi-centre, randomized, controlled trial. medRxiv 2020. [Google Scholar] [CrossRef]
- Hemilä, H.; Louhiala, P. Vitamin C for preventing and treating pneumonia. Cochrane Database Syst. Rev. 2013, 8, CD005532. [Google Scholar] [CrossRef]
- Cerullo, G.; Negro, M.; Parimbelli, M.; Pecoraro, M.; Perna, S.; Liguori, G.; Rondanelli, M.; Cena, H.; D’Antona, G. The Long History of Vitamin C: From Prevention of the Common Cold to Potential Aid in the Treatment of COVID-19. Front. Immunol. 2020, 11, 1. [Google Scholar] [CrossRef]
- Carr, A.C.; Rowe, S. The emerging role of vitamin c in the prevention and treatment of covid-19. Nutrients 2020, 12, 3286. [Google Scholar] [CrossRef]
- Tomasa-Irriguible, T.M. COVID-19: Up to 87% Critically Ill Patients Had Low Vitamin C Values. Nutr. J. 2020. [Google Scholar] [CrossRef]
- Hoang, X.; Shaw, G.; Fang, W.; Han, B. Possible application of high-dose vitamin C in the prevention and therapy of coronavirus infection. J. Glob. Antimicrob. Resist. 2020, 23, 256–262. [Google Scholar] [CrossRef]
- Hiedra, R.; Lo, K.B.; Elbashabsheh, M.; Gul, F.; Wright, R.M.; Albano, J.; Azmaiprashvili, Z.; Patarroyo Aponte, G. The Use of IV vitamin C for patients with COVID-19: A single center observational study. Expert Rev. Anti. Infect. Ther. 2020, 18, 1259–1261. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.C. A new clinical trial to test high-dose vitamin C in patients with COVID-19. Crit. Care 2020, 24, 133. [Google Scholar] [CrossRef]
- REMAP-CAP Investigators; Gordon, A.C.; Mouncey, P.R.; Al-Beidh, F.; Rowan, K.M.; Nichol, A.D.; Arabi, Y.M.; Annane, D.; Beane, A.; van Bentum-Puijk, W.; et al. Interleukin-6 Receptor Antagonists in Critically Ill Patients with Covid-19. N. Eng. J. Med. 2021, 384, 1491–1502. [Google Scholar] [CrossRef]
- Abani, O.; Abbas, A.; Abbas, F.; Abbas, M.; Abbasi, S.; Abbass, H.; Abbott, A.; Abdallah, N.; Abdelaziz, A.; Abdelfattah, M.; et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): A randomised, controlled, open-label, platform trial. Lancet 2021, 397, 1637–1645. [Google Scholar] [CrossRef]
- Biesalski, H.K. Vitamin D deficiency and co-morbidities in COVID-19 patients—A fatal relationship? NFS J. 2020, 20, 10–21. [Google Scholar] [CrossRef]
- Razdan, K.; Singh, K.; Singh, D. Vitamin D Levels and COVID-19 Susceptibility: Is there any Correlation? Med. Drug Discov. 2020, 7, 100051. [Google Scholar] [CrossRef] [PubMed]
- Hansdottir, S.; Monick, M.M.; Lovan, N.; Powers, L.; Gerke, A.; Hunninghake, G.W. Vitamin D Decreases Respiratory Syncytial Virus Induction of NF-κB–Linked Chemokines and Cytokines in Airway Epithelium While Maintaining the Antiviral State. J. Immunol. 2010, 184, 965–974. [Google Scholar] [CrossRef]
- Campbell, P.A.; Young, M.W.; Lee, R.C. Vitamin D Clinical Pharmacology: Relevance to COVID-19 Pathogenesis. J. Natl. Med. Assoc. 2020, 113, 208. [Google Scholar] [CrossRef]
- COVID-19 and Vitamin D Supplementation: A Multicenter Randomized Controlled Trial of High Dose Versus Standard Dose Vitamin D3 in High-Risk COVID-19 Patients. Available online: https://clinicaltrials.gov/ct2/show/NCT04344041 (accessed on 13 April 2021).
- Benskin, L.L. A Basic Review of the Preliminary Evidence That COVID-19 Risk and Severity Is Increased in Vitamin D Deficiency. Front. Public Health 2020, 8, 513. [Google Scholar] [CrossRef]
- Sulli, A.; Gotelli, E.; Casabella, A.; Paolino, S.; Pizzorni, C.; Alessandri, E.; Grosso, M.; Ferone, D.; Smith, V.; Cutolo, M. Vitamin d and lung outcomes in elderly covid-19 patients. Nutrients 2021, 13, 717. [Google Scholar] [CrossRef]
- Brenner, H. Vitamin D supplementation to prevent COVID-19 infections and deaths—accumulating evidence from epidemiological and intervention studies calls for immediate action. Nutrients 2021, 13, 411. [Google Scholar] [CrossRef]
- Ripoll, J.G.; van Helmond, N.; Senefeld, J.W.; Wiggins, C.C.; Klassen, S.A.; Baker, S.E.; Larson, K.F.; Murphy, B.M.; Andersen, K.J.; Ford, S.K.; et al. Convalescent Plasma for Infectious Diseases: Historical Framework and Use in COVID-19. Clin. Microbiol. Newsl. 2021, 43, 23–32. [Google Scholar] [CrossRef]
- Simonovich, V.A.; Burgos Pratx, L.D.; Scibona, P.; Beruto, M.V.; Vallone, M.G.; Vázquez, C.; Savoy, N.; Giunta, D.H.; Pérez, L.G.; Sánchez, M.d.L.; et al. A Randomized Trial of Convalescent Plasma in Covid-19 Severe Pneumonia. N. Engl. J. Med. 2021, 384, 619–629. [Google Scholar] [CrossRef]
- Abani, O.; Abbas, A.; Abbas, F.; Abbas, M.; Abbasi, S.; Abbass, H.; Abbott, A.; Abdallah, N.; Abdelaziz, A.; Abdelfattah, M.; et al. Convalescent plasma in patients admitted to hospital with COVID-19 (RECOVERY): A randomised controlled, open-label, platform trial. Lancet 2021, 397, 2049–2059. [Google Scholar] [CrossRef]
- Rowaiye, A.B.; Okpalefe, O.A.; Adejoke, O.O.; Ogidigo, J.O.; Oladipo, O.H.; Ogu, A.C.; Oli, A.N.; Olofinase, S.; Onyekwere, O.; Abubakar, A.R.; et al. Attenuating the Effects of Novel COVID-19 (SARS-CoV-2) Infection-Induced Cytokine Storm and the Implications. J. Inflamm. Res. 2021, 14, 1487–1510. [Google Scholar] [CrossRef]
- Mahase, E. Covid-19: Low dose steroid cuts death in ventilated patients by one third, trial finds. BMJ 2020, 369, m2422. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, X.; Huang, Y.; Shan, H.; Huang, J. Successful use of methylprednisolone for treating severe COVID-19. J. Allergy Clin. Immunol. 2020, 146, 325–327. [Google Scholar] [CrossRef] [PubMed]
- RECOVERY Collaborative Group; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with Covid-19. N. Eng. J. Med. 2020, 384, 693–704. [Google Scholar] [CrossRef]
- Mehta, P.; Ciurtin, C.; Scully, M.; Levi, M.; Chambers, R.C. JAK inhibitors in COVID-19: The need for vigilance regarding increased inherent thrombotic risk. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef] [PubMed]
- Cantini, F.; Niccoli, L.; Matarrese, D.; Nicastri, E.; Stobbione, P.; Goletti, D. Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J. Infect. 2020, 81, 318–356. [Google Scholar] [CrossRef]
- Kalil, A.C.; Patterson, T.F.; Mehta, A.K.; Tomashek, K.M.; Wolfe, C.R.; Ghazaryan, V.; Marconi, V.C.; Ruiz-Palacios, G.M.; Hsieh, L.; Kline, S.; et al. Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19. N. Engl. J. Med. 2020, 384, 795–807. [Google Scholar] [CrossRef]
- The Different Types of COVID-19 Vaccines. Available online: https://www.who.int/news-room/feature-stories/detail/the-race-for-a-covid-19-vaccine-explained (accessed on 13 April 2021).
- COVID-19 Vaccine Tracker | RAPS. Available online: https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker (accessed on 29 May 2021).
- WHO Says India Covid Variant of “Global Concern”. Available online: https://www.bbc.com/news/world-asia-india-57067190 (accessed on 15 May 2021).
- Coronavirus Variants are Spreading in India—What Scientists Know so Far. Available online: https://www.nature.com/articles/d41586-021-01274-7 (accessed on 15 May 2021).
- Yadav, P.D.; Mohandas, S.; Shete, A.M.; Nyayanit, D.A.; Gupta, N.; Patil, D.Y.; Sapkal, G.N.; Potdar, V.; Kadam, M.; Kumar, A.; et al. SARS CoV-2 variant B.1.617.1 is highly pathogenic in hamsters than B.1 variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ferreira, I.; Datir, R.; Papa, G.; Kemp, S.; Meng, B.; Singh, S.; Pandey, R.; Ponnusamy, K.; Radhakrishnan, V.; Sato, K.; et al. SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Krüger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Sidarovich, A.; Moldenhauer, A.-S.; Winkler, M.S.; Schulz, S.; et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades 1 antibodies induced by infection and vaccinatio. bioRxiv 2021. [Google Scholar] [CrossRef]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Das, S.; Agarwal, A.; Singh, S.; Abraham, P.; et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gupta, R.K. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021, 1–2. [Google Scholar] [CrossRef]
- Mellet, J.; Pepper, M.S. A COVID-19 Vaccine: Big Strides Come with Big Challenges. Vaccines 2021, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Thanh Le, T.; Andreadakis, Z.; Kumar, A.; Gómez Román, R.; Tollefsen, S.; Saville, M.; Mayhew, S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020, 19, 305–306. [Google Scholar] [CrossRef]
- BCG Vaccination to Protect Healthcare Workers against COVID-19. Available online: https://clinicaltrials.gov/ct2/show/NCT04327206 (accessed on 13 April 2021).
- Rivas, M.N.; Ebinger, J.E.; Wu, M.; Sun, N.; Braun, J.; Sobhani, K.; van Eyk, J.E.; Cheng, S.; Arditi, M. BCG vaccination history associates with decreased SARS-CoV-2 seroprevalence across a diverse cohort of health care workers. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Vaccines. Available online: https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-vaccines (accessed on 13 April 2021).
- Loo, K.Y.; Letchumanan, V.; Ser, H.L.; Teoh, S.L.; Law, J.W.F.; Tan, L.T.H.; Mutalib, N.S.A.; Chan, K.G.; Lee, L.H. COVID-19: Insights into potential vaccines. Microorganisms 2021, 9, 605. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. What Pfizer’s landmark COVID vaccine results mean for the pandemic. Nature 2020. [Google Scholar] [CrossRef]
- Moderna COVID-19 Vaccine Retains Neutralizing Activity Against Emerging Variants First Identified in the U.K. and the Republic of South Africa. Available online: https://investors.modernatx.com/news-releases/news-release-details/moderna-covid-19-vaccine-retains-neutralizing-activity-against/ (accessed on 1 May 2021).
- Wu, K.; Werner, A.P.; Moliva, J.I.; Koch, M.; Choi, A.; Stewart-Jones, G.B.E.; Bennett, H.; Boyoglu-Barnum, S.; Shi, W.; Graham, B.S.; et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021. [Google Scholar] [CrossRef]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Jones, I.; Roy, P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet 2021, 397, 642–643. [Google Scholar] [CrossRef]
- Tanne, J.H. Covid-19: All Johnson and Johnson vaccine in use is safe, says US regulator. BMJ 2021, 373, 897. [Google Scholar] [CrossRef]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.H.; Michailidis, E.; et al. Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants. Elife 2020, 9, 1. [Google Scholar] [CrossRef]
- Xie, X.; Zou, J.; Fontes-Garfias, C.R.; Xia, H.; Swanson, K.A.; Cutler, M.; Cooper, D.; Menachery, V.D.; Weaver, S.; Dormitzer, P.R.; et al. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv 2021. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Monego, S.D.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020. [Google Scholar] [CrossRef]
- Challen, R.; Brooks-Pollock, E.; Read, J.M.; Dyson, L.; Tsaneva-Atanasova, K.; Danon, L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: Matched cohort study. BMJ 2021, 372, 579. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef]
- Fay, E.J.; Langlois, R.A. MicroRNA-Attenuated virus vaccines. Non-Coding RNA 2018, 4, 25. [Google Scholar] [CrossRef]
- Yee, P.; Poh, C. Development of Novel miRNA-based Vaccines and Antivirals against Enterovirus 71. Curr. Pharm. Des. 2016, 22, 6694–6700. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef]
- Ding, S.W.; Voinnet, O. Antiviral Immunity Directed by Small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- El-Nabi, S.H.; Elhiti, M.; El-Sheekh, M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses 2020, 143, 110203. [Google Scholar] [CrossRef]
- Solleti, S.K.; Bhattacharya, S.; Ahmad, A.; Wang, Q.; Mereness, J.; Rangasamy, T.; Mariani, T.J. MicroRNA expression profiling defines the impact of electronic cigarettes on human airway epithelial cells. Sci. Rep. 2017, 7, 1081. [Google Scholar] [CrossRef]
- Głobińska, A.; Pawełczyk, M.; Kowalski, M.L. MicroRNAs and the immune response to respiratory virus infections. Expert Rev. Clin. Immunol. 2014, 10, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Mahdavi, F.; Badrzadeh, F.; Hosseini-Fard, S.R.; Heidary, M.; Jeda, A.S.; Mohammadi, T.; Roshani, M.; Yousefimashouf, R.; Keyvani, H.; et al. The emerging role of microRNAs in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Int. Immunopharmacol. 2021, 90, 107204. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Leon-Icaza, S.A.; Zeng, M.; Rosas-Taraco, A.G. microRNAs in viral acute respiratory infections: Immune regulation, biomarkers, therapy, and vaccines. ExRNA 2019, 1, 1. [Google Scholar] [CrossRef]
- Guterres, A.; de Azeredo Lima, C.H.; Miranda, R.L.; Gadelha, M.R. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect. Genet. Evol. 2020, 85, 104417. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.M.; Park, C.; Choi, M.Y. Update on non-canonical microRNAs. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewski, R.; Dabrowski, M.; Jakiela, B.; Matalon, S.; Harrod, K.S.; Sanak, M.; Collawn, J.F. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L444–L455. [Google Scholar] [CrossRef]
- Mukhopadhyay, D.; Mussa, B.M. Identification of novel hypothalamic micrornas as promising therapeutics for sars-cov-2 by regulating ace2 and tmprss2 expression: An in silico analysis. Brain Sci. 2020, 10, 666. [Google Scholar] [CrossRef]
- Pierce, J.B.; Simion, V.; Icli, B.; Pérez-Cremades, D.; Cheng, H.S.; Feinberg, M.W. Computational analysis of targeting SARS-CoV-2, viral entry proteins ace2 and tmprss2, and interferon genes by host micrornas. Genes (Basel) 2020, 11, 1354. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.T.-S.; Salmena, L. Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium. Genes (Basel) 2020, 11, 1002. [Google Scholar] [CrossRef]
- Girardi, E.; López, P.; Pfeffer, S. On the importance of host MicroRNAs during viral infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Abu-Izneid, T.; AlHajri, N.; Ibrahim, A.M.; Javed, M.N.; Salem, K.M.; Pottoo, F.H.; Kamal, M.A. Micro-RNAs in the regulation of immune response against SARS CoV-2 and other viral infections. J. Adv. Res. 2020, 30, 133–145. [Google Scholar] [CrossRef]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef]
- Khan, M.A.A.K.; Sany, M.R.U.; Islam, M.S.; Islam, A.B.M.M.K. Epigenetic Regulator miRNA Pattern Differences Among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 World-Wide Isolates Delineated the Mystery Behind the Epic Pathogenicity and Distinct Clinical Characteristics of Pandemic COVID-19. Front. Genet. 2020, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Amahong, K.; Sun, X.; Lian, X.; Liu, J.; Sun, H.; Lou, Y.; Zhu, F.; Qiu, Y. The miRNA: A small but powerful RNA for COVID-19. Brief Bioinform. 2021, 22, 1137–1149. [Google Scholar] [CrossRef]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Oran, D.P.; Topol, E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection: A Narrative Review. Ann. Intern. Med. 2020, 173, 362–367. [Google Scholar] [CrossRef]
- GISAID—Initiative. Available online: https://www.gisaid.org/ (accessed on 17 April 2021).
- Sevgin, O.; Sevgin, K. Systematic Review of microRNAs in the SARS-CoV-2 Infection: Are microRNAs Potential Therapy for COVID-19? J. Genet. Genome Res. 2021, 8, 053. [Google Scholar] [CrossRef]
- miRBase. Available online: http://www.mirbase.org/ (accessed on 17 April 2021).
- Juzenas, S.; Venkatesh, G.; Hübenthal, M.; Hoeppner, M.P.; Du, Z.G.; Paulsen, M.; Rosenstiel, P.; Senger, P.; Hofmann-Apitius, M.; Keller, A.; et al. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017, 45, 9290–9301. [Google Scholar] [CrossRef]
- Yeung, M.L.; Bennasser, Y.; Le, S.Y.; Jeang, K.T. siRNA, miRNA and HIV: Promises and challenges. Cell Res. 2005, 15, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1. [Google Scholar] [CrossRef]
- Li, Y.; Chan, E.Y.; Li, J.; Ni, C.; Peng, X.; Rosenzweig, E.; Tumpey, T.M.; Katze, M.G. MicroRNA Expression and Virulence in Pandemic Influenza Virus-Infected Mice. J. Virol. 2010, 84, 3023–3032. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Jaggi, N.; Chauhan, M.; Yallapu, S.C. COVID-19: Fighting the invisible enemy with microRNA. Expert Rev. Anti. Infect. Ther. 2021, 19, 137–145. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef] [PubMed]
- McLean, H.Q.; Belongia, E.A.; Kieke, B.A.; Meece, J.K.; Fry, A.M. Impact of Late Oseltamivir Treatment on Influenza Symptoms in the Outpatient Setting: Results of a Randomized Trial. Open Forum Infect. Dis. 2015, 2, 100. [Google Scholar] [CrossRef]
- Chugh, P.; Dittmer, D.P. Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA 2012, 3, 601–616. [Google Scholar] [CrossRef]
- Huang, J.; Lai, J.; Liang, B.; Jiang, J.; Ning, C.; Liao, Y.; Zang, N.; Wang, M.; Qin, F.; Yu, J.; et al. mircoRNA-3162-3p is a potential biomarker to identify new infections in HIV-1-infected patients. Gene 2018, 662, 21–27. [Google Scholar] [CrossRef]
- Yahyaei, S.; Biasin, M.; Saulle, I.; Gnudi, F.; De Luca, M.; Tasca, K.I.; Trabattoni, D.; Lo Caputo, S.; Mazzotta, F.; Clerici, M. Identification of a Specific miRNA Profile in HIV-Exposed Seronegative Individuals. JAIDS J. Acquir. Immune Defic. Syndr. 2016, 73, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 2019, 43, 307–316. [Google Scholar] [CrossRef]
- Cui, L.; Qi, Y.; Li, H.; Ge, Y.; Zhao, K.; Qi, X.; Guo, X.; Shi, Z.; Zhou, M.; Zhu, B.; et al. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PLoS ONE 2011, 6, 27071. [Google Scholar] [CrossRef]
- El-Radhi, A.S. Fever in Common Infectious Diseases. Clin. Man. Fever Child. 2018, 85–140. [Google Scholar] [CrossRef]
- Cui, C.; Griffiths, A.; Li, G.; Silva, L.M.; Kramer, M.F.; Gaasterland, T.; Wang, X.-J.; Coen, D.M. Prediction and Identification of Herpes Simplex Virus 1-Encoded MicroRNAs. J. Virol. 2006, 80, 5499–5508. [Google Scholar] [CrossRef]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef]
- Liu, M.; John, C.M.; Jarvis, G.A. Induction of Endotoxin Tolerance by Pathogenic Neisseria Is Correlated with the Inflammatory Potential of Lipooligosaccharides and Regulated by MicroRNA-146a. J. Immunol. 2014, 192, 1768–1777. [Google Scholar] [CrossRef]
- Kocerha, J.; Kouri, N.; Baker, M.; Finch, N.C.; DeJesus-Hernandez, M.; Gonzalez, J.; Chidamparam, K.; Josephs, K.A.; Boeve, B.F.; Graff-Radford, N.R.; et al. Altered microRNA expression in frontotemporal lobar degeneration with TDP-43 pathology caused by progranulin mutations. BMC Genom. 2011, 12, 527. [Google Scholar] [CrossRef]
- Piscopo, P.; Albani, D.; Castellano, A.E.; Forloni, G.; Confaloni, A. Frontotemporal lobar degeneration and microRNAs. Front. Aging Neurosci. 2016, 8, 17. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Bala, S.; Petrasek, J.; Mundkur, S.; Catalano, D.; Levin, I.; Ward, J.; Alao, H.; Kodys, K.; Szabo, G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012, 56, 1946–1957. [Google Scholar] [CrossRef]
- Turchinovich, A.; Samatov, T.R.; Tonevitsky, A.G.; Burwinkel, B. Circulating miRNAs: Cell-cell communication function? Front. Genet. 2013, 4, 119. [Google Scholar] [CrossRef]
- Scheel, T.K.H.; Luna, J.M.; Liniger, M.; Nishiuchi, E.; Rozen-Gagnon, K.; Shlomai, A.; Auray, G.; Gerber, M.; Fak, J.; Keller, I.; et al. A Broad RNA Virus Survey Reveals Both miRNA Dependence and Functional Sequestration. Cell Host Microbe 2016, 19, 409–423. [Google Scholar] [CrossRef]
- Masaki, T.; Arend, K.C.; Li, Y.; Yamane, D.; McGivern, D.R.; Kato, T.; Wakita, T.; Moorman, N.J.; Lemon, S.M. MiR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe 2015, 17, 217–228. [Google Scholar] [CrossRef]
- Sharma, N.; Verma, R.; Kumawat, K.L.; Basu, A.; Singh, S.K. miR-146a suppresses cellular immune response during Japanese encephalitis virus JaOArS982 strain infection in human microglial cells. J. Neuroinflammation 2015, 12, 30. [Google Scholar] [CrossRef]
- Ho, B.C.; Yang, P.C.; Yu, S.L. MicroRNA and pathogenesis of enterovirus infection. Viruses 2016, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Slonchak, A.; Shannon, R.P.; Pali, G.; Khromykh, A.A. Human MicroRNA miR-532-5p Exhibits Antiviral Activity against West Nile Virus via Suppression of Host Genes SESTD1 and TAB3 Required for Virus Replication. J. Virol. 2016, 90, 2388–2402. [Google Scholar] [CrossRef]
- Wu, N.; Gao, N.; Fan, D.; Wei, J.; Zhang, J.; An, J. MiR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926cells. Microbes Infect. 2014, 16, 911–922. [Google Scholar] [CrossRef]
- Bruscella, P.; Bottini, S.; Baudesson, C.; Pawlotsky, J.M.; Feray, C.; Trabucchi, M. Viruses and miRNAs: More friends than foes. Front. Microbiol. 2017, 8, 824. [Google Scholar] [CrossRef]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef] [PubMed]
- Witkos, T.; Koscianska, E.; Krzyzosiak, W. Practical Aspects of microRNA Target Prediction. Curr. Mol. Med. 2011, 11, 93–109. [Google Scholar] [CrossRef]
- TargetScanHuman 7.2. Available online: http://www.targetscan.org/vert_72/ (accessed on 17 April 2021).
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for sequence-based miRNA target prediction: What to choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef]
- Lekprasert, P.; Mayhew, M.; Ohler, U. Assessing the utility of thermodynamic features for microRNA target prediction under relaxed seed and no conservation requirements. PLoS ONE 2011, 6, e20622. [Google Scholar] [CrossRef][Green Version]
- Sardar, R.; Satish, D.; Gupta, D. Identification of Novel SARS-CoV-2 Drug Targets by Host MicroRNAs and Transcription Factors Co-regulatory Interaction Network Analysis. Front. Genet. 2020, 11, 571274. [Google Scholar] [CrossRef]
- Mariappan, V.; Manoharan, P.S.; Shanmugam, L.; Rao, S.R.; Pillai, A.B. Potential biomarkers for the early prediction of SARS-COV-2 disease outcome. Microb. Pathog. 2021, 158, 105057. [Google Scholar] [CrossRef]
- Schäfer, A.; Baric, R.S. Epigenetic landscape during coronavirus infection. Pathogens 2017, 6, 8. [Google Scholar] [CrossRef]
- Espinoza, J.A.; González, P.A.; Kalergis, A.M. Modulation of Antiviral Immunity by Heme Oxygenase-1. Am. J. Pathol. 2017, 187, 487–493. [Google Scholar] [CrossRef]
- Danesh, A.; Cameron, C.M.; León, A.J.; Ran, L.; Xu, L.; Fang, Y.; Kelvin, A.A.; Rowe, T.; Chen, H.; Guan, Y.; et al. Early gene expression events in ferrets in response to SARS coronavirus infection versus direct interferon-alpha2b stimulation. Virology 2011, 409, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, H.; Gao, S.; Jiang, W.; Huang, W. Cellular MicroRNAs Inhibit Replication of the H1N1 Influenza A Virus in Infected Cells. J. Virol. 2010, 84, 8849–8860. [Google Scholar] [CrossRef]
- Zhu, Z.; Qi, Y.; Ge, A.; Zhu, Y.; Xu, K.; Ji, H.; Shi, Z.; Cui, L.; Zhou, M. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses 2014, 6, 1525–1539. [Google Scholar] [CrossRef] [PubMed]
- Balmeh, N.; Mahmoudi, S.; Mohammadi, N.; Karabedianhajiabadi, A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Informatics Med. Unlocked 2020, 20, 100407. [Google Scholar] [CrossRef]
- Fulzele, S.; Sahay, B.; Yusufu, I.; Lee, T.J.; Sharma, A.; Kolhe, R.; Isales, C.M. COVID-19 virulence in aged patients might be impacted by the host cellular MicroRNAs abundance/profile. Aging Dis. 2020, 11, 509–522. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Rakhmetullina, A.; Aisina, D. How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV, and MERS-CoV. Res. Sq. 2020, rs.3.rs-16264. [Google Scholar] [CrossRef]
- Hosseini Rad SM, A.; McLellan, A.D. Implications of SARS-CoV-2 Mutations for Genomic RNA Structure and Host microRNA Targeting. Int. J. Mol. Sci. 2020, 21, 4807. [Google Scholar] [CrossRef] [PubMed]
- Mockly, S.; Seitz, H. Inconsistencies and Limitations of Current MicroRNA Target Identification Methods. Met. Mol. Biol. 2019, 1970, 291–314. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20. [Google Scholar] [CrossRef]
- Pinzón, N.; Li, B.; Martinez, L.; Sergeeva, A.; Presumey, J.; Apparailly, F.; Seitz, H. MicroRNA target prediction programs predict many false positives. Genome Res. 2017, 27, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, O.; Sugai, K.; Yamaguchi, R.; Tashiro, S.; Nagoshi, N.; Kohyama, J.; Iida, T.; Ohkubo, T.; Itakura, G.; Isoda, M.; et al. Laying the Groundwork for a First-In-Human Study of an Induced Pluripotent Stem Cell-Based Intervention for Spinal Cord Injury. Stem Cells 2019, 37, 6–13. [Google Scholar] [CrossRef]
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced pluripotent stem cells and their use in human models of disease and development. Physiol. Rev. 2019, 99, 79–114. [Google Scholar] [CrossRef]
- Lee, S.H.; Cunha, D.; Piermarocchi, C.; Paternostro, G.; Pinkerton, A.; Ladriere, L.; Marchetti, P.; Eizirik, D.L.; Cnop, M.; Levine, F. High-throughput screening and bioinformatic analysis to ascertain compounds that prevent saturated fatty acid-induced β-cell apoptosis. Biochem. Pharmacol. 2017, 138, 140–149. [Google Scholar] [CrossRef]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Chen, Y.; Luo, D.; Zhuang, Z.; Jin, H.; Zhou, H.; Li, X.; Lin, H.; Zheng, X.; Zhang, J.; et al. Therapeutic potential of C1632 by inhibition of SARS-CoV-2 replication and viral-induced inflammation through upregulating let-7. Signal Transduct. Target. Ther. 2021, 6, 1–3. [Google Scholar] [CrossRef]
- Tan, J.; Liu, S.; Zhuang, L.; Chen, L.; Dong, M.; Zhang, J.; Xin, Y. Transmission and clinical characteristics of asymptomatic patients with SARS-CoV-2 infection. Future Virol. 2020, 15, 10. [Google Scholar] [CrossRef]
- Lessler, J.; Reich, N.G.; Brookmeyer, R.; Perl, T.M.; Nelson, K.E.; Cummings, D.A. Incubation periods of acute respiratory viral infections: A systematic review. Lancet Infect. Dis. 2009, 9, 291–300. [Google Scholar] [CrossRef]
- Tarhini, H.; Recoing, A.; Bridier-nahmias, A.; Rahi, M.; Lambert, C.; Martres, P.; Lucet, J.-C.; Rioux, C.; Bouzid, D.; Lebourgeois, S.; et al. Long-Term Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infectiousness Among Three Immunocompromised Patients: From Prolonged Viral Shedding to SARS-CoV-2 Superinfection. J. Infect. Dis. 2021, 223, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.-H.; Boucau, J.; Bowman, K.; et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef] [PubMed]
- Baang, J.H.; Smith, C.; Mirabelli, C.; Valesano, A.L.; Manthei, D.M.; Bachman, M.A.; Wobus, C.E.; Adams, M.; Washer, L.; Martin, E.T.; et al. Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 Replication in an Immunocompromised Patient. J. Infect. Dis. 2021, 223, 23–27. [Google Scholar] [CrossRef]
- Avanzato, V.A.; Matson, M.J.; Seifert, S.N.; Pryce, R.; Williamson, B.N.; Anzick, S.L.; Barbian, K.; Judson, S.D.; Fischer, E.R.; Martens, C.; et al. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 2020, 183, 1901–1912. [Google Scholar] [CrossRef]
- Aydillo, T.; Gonzalez-Reiche, A.S.; Aslam, S.; van de Guchte, A.; Khan, Z.; Obla, A.; Dutta, J.; van Bakel, H.; Aberg, J.; García-Sastre, A.; et al. Shedding of Viable SARS-CoV-2 after Immunosuppressive Therapy for Cancer. N. Engl. J. Med. 2020, 383, 2586–2588. [Google Scholar] [CrossRef]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef]
- Wenzel, P.; Kopp, S.; Gobel, S.; Jansen, T.; Geyer, M.; Hahn, F.; Kreitner, K.F.; Escher, F.; Schultheiss, H.P.; Münzel, T. Evidence of SARS-CoV-2 mRNA in endomyocardial biopsies of patients with clinically suspected myocarditis tested negative for COVID-19 in nasopharyngeal swab. Cardiovasc. Res. 2020, 116, 1661–1663. [Google Scholar] [CrossRef]
- Liu, Q.; Du, J.; Yu, X.; Xu, J.; Huang, F.; Li, X.; Zhang, C.; Li, X.; Chang, J.; Shang, D.; et al. MiRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Chatterjee, S.; Xiao, K.; Riedel, I.; Wang, Y.; Foo, R.; Bär, C.; Thum, T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020, 148, 46–49. [Google Scholar] [CrossRef]
- Shukla, A.K.; Banerjee, M. Angiotensin-Converting-Enzyme 2 and Renin-Angiotensin System Inhibitors in COVID-19: An Update. High Blood Press. Cardiovasc. Prev. 2021, 28, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hemida, M.G.; Ye, X.; Thair, S.; Yang, D. Exploiting the therapeutic potential of microRNAs in viral diseases: Expectations and limitations. Mol. Diagn. Ther. 2010, 14, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Underwood, P.C.; Adler, G.K. The renin angiotensin aldosterone system and insulin resistance in humans. Curr. Hypertens. Rep. 2013, 15, 59–70. [Google Scholar] [CrossRef]
- Cao, X.; Song, L.N.; Yang, J.K. ACE2 and energy metabolism: The connection between COVID-19 and chronic metabolic disorders. Clin. Sci. 2021, 135, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Thunders, M.; Delahunt, B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J. Clin. Pathol. 2020, 73, 773–776. [Google Scholar] [CrossRef]
- Choudhary, S.; Sreenivasulu, K.; Mitra, P.; Misra, S.; Sharma, P. Role of genetic variants and gene expression in the susceptibility and severity of COVID-19. Ann. Lab. Med. 2020, 41, 129–138. [Google Scholar] [CrossRef]
- Nersisyan, S.; Shkurnikov, M.; Turchinovich, A.; Knyazev, E.; Tonevitsky, A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS ONE 2020, 15, e0235987. [Google Scholar] [CrossRef] [PubMed]
- Kuba, K.; Imai, Y.; Penninger, J.M. Angiotensin-converting enzyme 2 in lung diseases. Curr. Opin. Pharmacol. 2006, 6, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Matarese, A.; Gambardella, J.; Sardu, C.; Santulli, G. MiR-98 regulates tmprss2 expression in human endothelial cells: Key implications for covid-19. Biomedicines 2020, 8, 462. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.L.; Zuluaga-Ramirez, V.; Gajghate, S.; Reichenbach, N.L.; Polyak, B.; Persidsky, Y.; Rom, S. miR-98 reduces endothelial dysfunction by protecting blood–brain barrier (BBB) and improves neurological outcomes in mouse ischemia/reperfusion stroke model. J. Cereb. Blood Flow Metab. 2020, 40, 1953–1965. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, M.; He, Q.; Li, Z.; Zhao, Y.; Wang, W.; Ma, J.; Li, Y.; Chang, G. Microrna-98 rescues proliferation and alleviates ox-LDL-induced apoptosis in HUVECs by targeting LOX-1. Exp. Ther. Med. 2017, 13, 1702–1710. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.W.; Yang, B.; Wong, W.; Blakeley, P.; Seah, I.; Tan, Q.S.W.; Wang, H.; Bhargava, M.; Lin, H.A.; Chai, C.H.; et al. A Pilot Study on MicroRNA Profile in Tear Fluid to Predict Response to Anti-VEGF Treatments for Diabetic Macular Edema. J. Clin. Med. 2020, 9, 2920. [Google Scholar] [CrossRef]
- Li, H.W.; Meng, Y.; Xie, Q.; Yi, W.J.; Lai, X.L.; Bian, Q.; Wang, J.; Wang, J.F.; Yu, G. miR-98 protects endothelial cells against hypoxia/reoxygenation induced-apoptosis by targeting caspase-3. Biochem. Biophys. Res. Commun. 2015, 467, 595–601. [Google Scholar] [CrossRef]
- Petrilli, C.M.; Jones, S.A.; Yang, J.; Rajagopalan, H.; O’Donnell, L.; Chernyak, Y.; Tobin, K.A.; Cerfolio, R.J.; Francois, F.; Horwitz, L.I. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: Prospective cohort study. BMJ 2020, 369, 1966. [Google Scholar] [CrossRef]
- Shi, Y.; Yu, X.; Zhao, H.; Wang, H.; Zhao, R.; Sheng, J. Host susceptibility to severe COVID-19 and establishment of a host risk score: Findings of 487 cases outside Wuhan. Crit. Care 2020, 24, 108. [Google Scholar] [CrossRef]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 2020, 12, 10087–10098. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nagata, N.; Shirato, K.; Kawase, M.; Takeda, M.; Taguchi, F. Efficient Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by the Transmembrane Protease TMPRSS2. J. Virol. 2010, 84, 12658–12664. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Muller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [PubMed]
- Baughn, L.B.; Sharma, N.; Elhaik, E.; Sekulic, A.; Bryce, A.H.; Fonseca, R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin. Proc. 2020, 95, 1989–1999. [Google Scholar] [CrossRef]
- Jin, J.M.; Bai, P.; He, W.; Wu, F.; Liu, X.F.; Han, D.M.; Liu, S.; Yang, J.K. Gender differences in patients with COVID-19: Focus on severity and mortality. medRxiv 2020, 8, 152. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; Nakaya, H.I. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Ganai, S.A.; Malli Kalladi, S.; Mahadevan, V. HDAC inhibition through valproic acid modulates the methylation profiles in human embryonic kidney cells. J. Biomol. Struct. Dyn. 2015, 33, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Beacon, T.H.; Delcuve, G.P.; Davie, J.R. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus 1. Genome 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Kapila, S.; Kapila, R.; Kumar, S.; Upadhyay, D.; Kaur, M.; Sharma, C. Tmprss2 specific miRNAs as promising regulators for SARS-CoV-2 entry checkpoint. Virus Res. 2021, 294, 198275. [Google Scholar] [CrossRef] [PubMed]
- Donyavi, T.; Bokharaei-Salim, F.; Baghi, H.B.; Khanaliha, K.; Alaei Janat-Makan, M.; Karimi, B.; Sadri Nahand, J.; Mirzaei, H.; Khatami, A.; Garshasbi, S.; et al. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021, 97, 107641. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 21, 122–125. [Google Scholar] [CrossRef]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 105020. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Svegliati Baroni, S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with non-clinical response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021, 3, 468–475. [Google Scholar] [CrossRef]
- Farr, R.; Cheng, A.; Kedzierska, K. Altered microRNA expression in COVID-19 patients enables identication of SARS-CoV-2 infection. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Centa, A.; Fonseca, A.S.; Da Silva Ferreira, S.G.; Azevedo, M.L.V.; De Paula, C.B.V.; Nagashima, S.; MacHado-Souz, C.; Dos Santos Miggiolaro, A.F.R.; Baena, C.P.; De Noronha, L.; et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021, 320, 405–412. [Google Scholar] [CrossRef]
- McDonald, J.T.; Javier Enguita, F.; Taylor, D.; Griffin, R.J.; Priebe, W.; Emmett, M.R.; McGrath, M.; Sajadi, M.M.; Clement, J.; Dybas, J.M.; et al. The Great Deceiver: miR-2392’s Hidden Role in Driving SARS-CoV-2 Infection. bioRxiv 2021. [Google Scholar] [CrossRef]
- Papannarao, J.B.; Schwenke, D.O.; Manning, P.; Katare, R. Upregulated miR-200c may increase the risk of obese individuals to severe COVID-19. medRxiv 2021. [Google Scholar] [CrossRef]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene 2006, 367, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.K.; Linder, P. DEAD box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell 2001, 8, 251–262. [Google Scholar] [CrossRef]
- Taschuk, F.; Cherry, S. DEAD-box helicases: Sensors, regulators, and effectors for antiviral defense. Viruses 2020, 12, 181. [Google Scholar] [CrossRef]
- Wang, X.; Wang, R.; Luo, M.; Li, C.; Wang, H.X.; Huan, C.C.; Qu, Y.R.; Liao, Y.; Mao, X. (DEAD)-box RNA helicase 3 modulates NF-κB signal pathway by controlling the phosphorylation of PP2A-C subunit. Oncotarget 2017, 8, 33197–33213. [Google Scholar] [CrossRef]
- Lorgeoux, R.P.; Pan, Q.; Le Duff, Y.; Liang, C. DDX17 promotes the production of infectious HIV-1 particles through modulating viral RNA packaging and translation frameshift. Virology 2013, 443, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Chen, G.; Jia, H.; He, X.; Jing, Z. DDX5 RNA helicases: Emerging roles in viral infection. Int. J. Mol. Sci. 2018, 19, 1122. [Google Scholar] [CrossRef]
- Squeglia, F.; Romano, M.; Ruggiero, A.; Maga, G.; Berisio, R. Host DDX Helicases as Possible SARS-CoV-2 Proviral Factors: A Structural Overview of Their Hijacking Through Multiple Viral Proteins. Front. Chem. 2020, 8, 602162. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352. [Google Scholar] [CrossRef]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Davis-Dusenbery, B.N.; Hata, A. Mechanisms of control of microRNA biogenesis. J. Biochem. 2010, 148, 381–392. [Google Scholar] [CrossRef]
- Otsuka, M.; Jing, Q.; Georgel, P.; New, L.; Chen, J.; Mols, J.; Kang, Y.J.; Jiang, Z.; Du, X.; Cook, R.; et al. Hypersusceptibility to Vesicular Stomatitis Virus Infection in Dicer1-Deficient Mice Is Due to Impaired miR24 and miR93 Expression. Immunity 2007, 27, 123–134. [Google Scholar] [CrossRef]
- Ostermann, E.; Tuddenham, L.; Macquin, C.; Alsaleh, G.; Schreiber-Becker, J.; Tanguy, M.; Bahram, S.; Pfeffer, S.; Georgel, P. Deregulation of type I IFN-dependent genes correlates with increased susceptibility to cytomegalovirus acute infection of dicer mutant mice. PLoS ONE 2012, 7, 43744. [Google Scholar] [CrossRef]
- Wang, M.; Gu, B.; Chen, X.; Wang, Y.; Li, P.; Wang, K. The Function and Therapeutic Potential of Epstein-Barr Virus-Encoded MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 17, 657–668. [Google Scholar] [CrossRef]
- Cai, L.; Li, J.; Zhang, X.; Lu, Y.; Wang, J.; Lyu, X.; Chen, Y.; Liu, J.; Cai, H.; Wang, Y.; et al. Gold nano-particles (AuNPs) carrying anti-EBV-miR-BART7-3p inhibit growth of EBV-positive nasopharyngeal carcinoma. Oncotarget 2015, 6, 7838–7850. [Google Scholar] [CrossRef] [PubMed]
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143. [Google Scholar] [CrossRef]
- Gangemi, S.; Tonacci, A. AntagomiRs: A novel therapeutic strategy for challenging COVID-19 cytokine storm. Cytokine Growth Factor Rev. 2020, 58, 111. [Google Scholar] [CrossRef]
- Desjarlais, M.; Wirth, M.; Lahaie, I.; Ruknudin, P.; Hardy, P.; Rivard, A.; Chemtob, S. Nutraceutical Targeting of Inflammation-Modulating microRNAs in Severe Forms of COVID-19: A Novel Approach to Prevent the Cytokine Storm. Front. Pharmacol. 2020, 11, 2108. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Kwon, T.H.; Kim, A. Delivery of therapeutic miRNA using polymer-based formulation. Drug Deliv. Transl. Res. 2019, 9, 1043–1056. [Google Scholar] [CrossRef]
- Yang, N. An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig. 2015, 5, 179–181. [Google Scholar] [CrossRef]
- Lee, S.W.L.; Paoletti, C.; Campisi, M.; Osaki, T.; Adriani, G.; Kamm, R.D.; Mattu, C.; Chiono, V. MicroRNA delivery through nanoparticles. J. Control. Release 2019, 313, 80–95. [Google Scholar] [CrossRef]
- O’Neill, C.P.; Dwyer, R.M. Nanoparticle-Based Delivery of Tumor Suppressor microRNA for Cancer Therapy. Cells 2020, 9, 521. [Google Scholar] [CrossRef]
- Zhu, L.; Mahato, R.I. Lipid and polymeric carrier-mediated nucleic acid delivery. Expert Opin. Drug Deliv. 2010, 7, 1209–1226. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.X.M.; Caballero, R.; Ali, H.; Roy, D.; Cassol, E.; Kumar, A. Transfection of hard-to-transfect primary human macrophages with Bax siRNA to reverse Resveratrol-induced apoptosis. RNA Biol. 2020, 17, 755–764. [Google Scholar] [CrossRef]
- Diener, Y.; Jurk, M.; Kandil, B.; Choi, Y.H.; Wild, S.; Bissels, U.; Bosio, A. RNA-based, transient modulation of gene expression in human haematopoietic stem and progenitor cells. Sci. Rep. 2015, 5, 17184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Viennois, E.; Xu, C.; Merlin, D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 2016, 4, e1134415. [Google Scholar] [CrossRef] [PubMed]
- Kalarikkal, S.P.; Sundaram, G.M. Edible plant-derived exosomal microRNAs: Exploiting a cross-kingdom regulatory mechanism for targeting SARS-CoV-2. Toxicol. Appl. Pharmacol. 2021, 414, 115425. [Google Scholar] [CrossRef] [PubMed]
- Bernier, A.; Sagan, S.M. The diverse roles of microRNAs at the host–virus interface. Viruses 2018, 10, 440. [Google Scholar] [CrossRef] [PubMed]




| Downregulated miRNAs | Upregulated miRNAs |
|---|---|
| miR-183-5p | miR-16-2-3p |
| miR-941 | miR-5695 |
| miR-627-5p | miR-618 |
| miR-144-3p | miR-10399-3p |
| miR-21-5p | miR-6501-5p |
| miR-20a-5p | miR-361-3p |
| miR-146b-5p | miR-4659a-3p |
| miR-454-3p | miR-142-5p |
| miR-18a-5p | miR-4685-3p |
| miR-340-5p | miR-454-5p |
| miR-17-5p | miR-30c-5p |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narożna, M.; Rubiś, B. Anti-SARS-CoV-2 Strategies and the Potential Role of miRNA in the Assessment of COVID-19 Morbidity, Recurrence, and Therapy. Int. J. Mol. Sci. 2021, 22, 8663. https://doi.org/10.3390/ijms22168663
Narożna M, Rubiś B. Anti-SARS-CoV-2 Strategies and the Potential Role of miRNA in the Assessment of COVID-19 Morbidity, Recurrence, and Therapy. International Journal of Molecular Sciences. 2021; 22(16):8663. https://doi.org/10.3390/ijms22168663
Chicago/Turabian StyleNarożna, Maria, and Błażej Rubiś. 2021. "Anti-SARS-CoV-2 Strategies and the Potential Role of miRNA in the Assessment of COVID-19 Morbidity, Recurrence, and Therapy" International Journal of Molecular Sciences 22, no. 16: 8663. https://doi.org/10.3390/ijms22168663
APA StyleNarożna, M., & Rubiś, B. (2021). Anti-SARS-CoV-2 Strategies and the Potential Role of miRNA in the Assessment of COVID-19 Morbidity, Recurrence, and Therapy. International Journal of Molecular Sciences, 22(16), 8663. https://doi.org/10.3390/ijms22168663







