The Oxoglutarate Binding Site and Regulatory Mechanism Are Conserved in Ammonium Transporter Inhibitors GlnKs from Methanococcales
Abstract
1. Introduction
2. Results
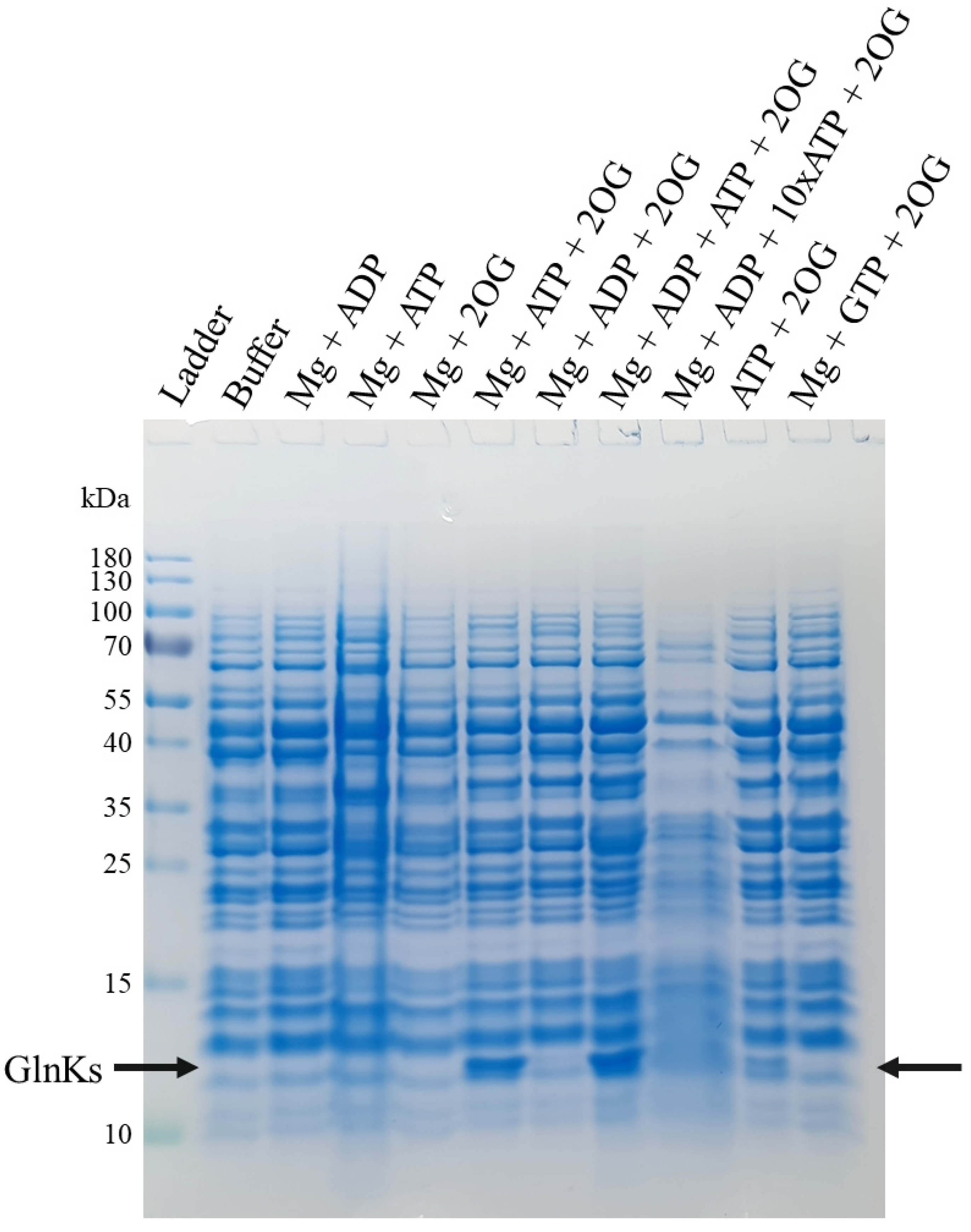
2.1. 2OG, Mg and ATP Are Required to Disrupt MtGlnKs from the Membrane Fraction
2.2. MtGlnK1 and MtGlnK2 Show Remarkable Purification and Crystallization Behaviors
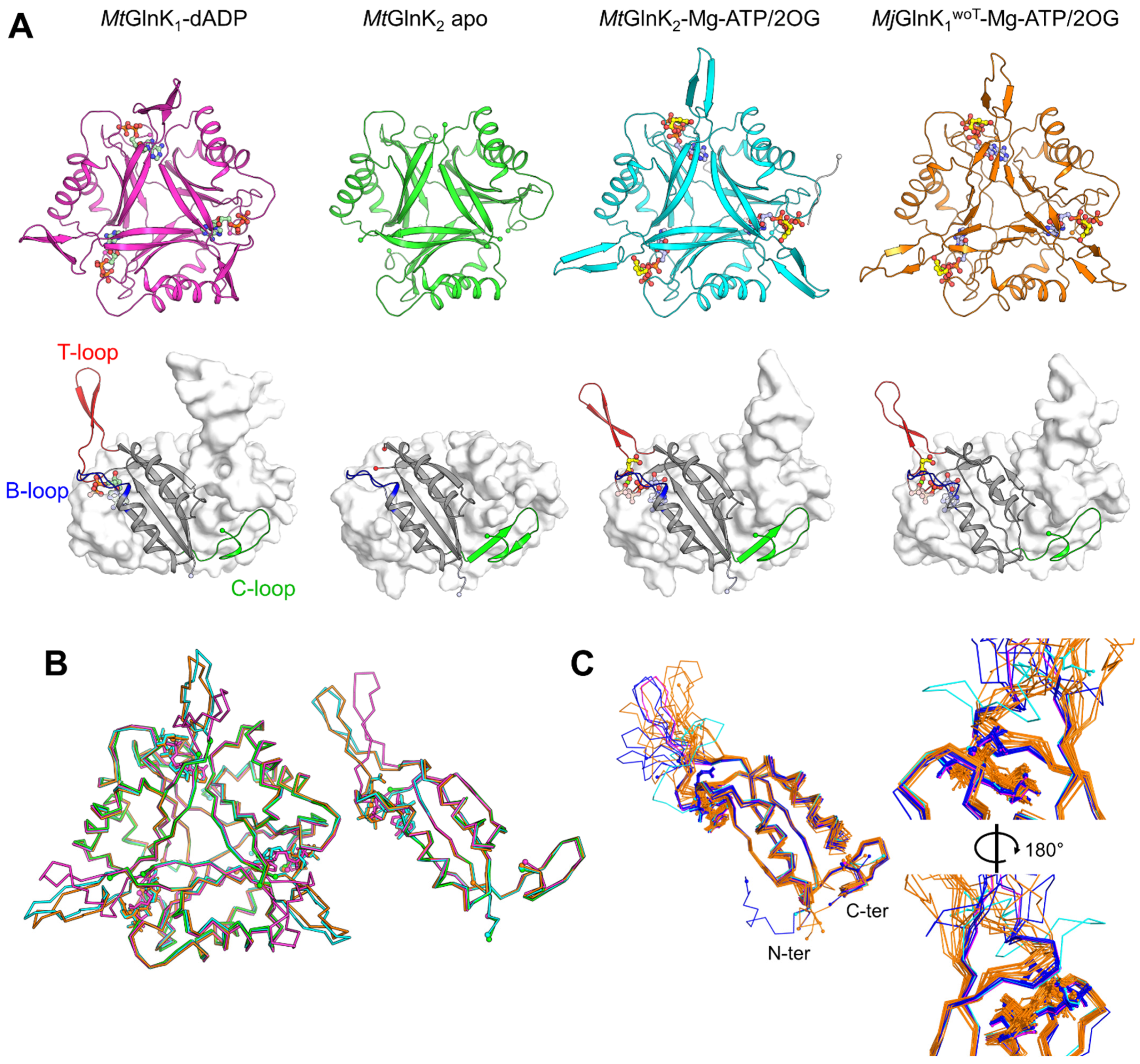
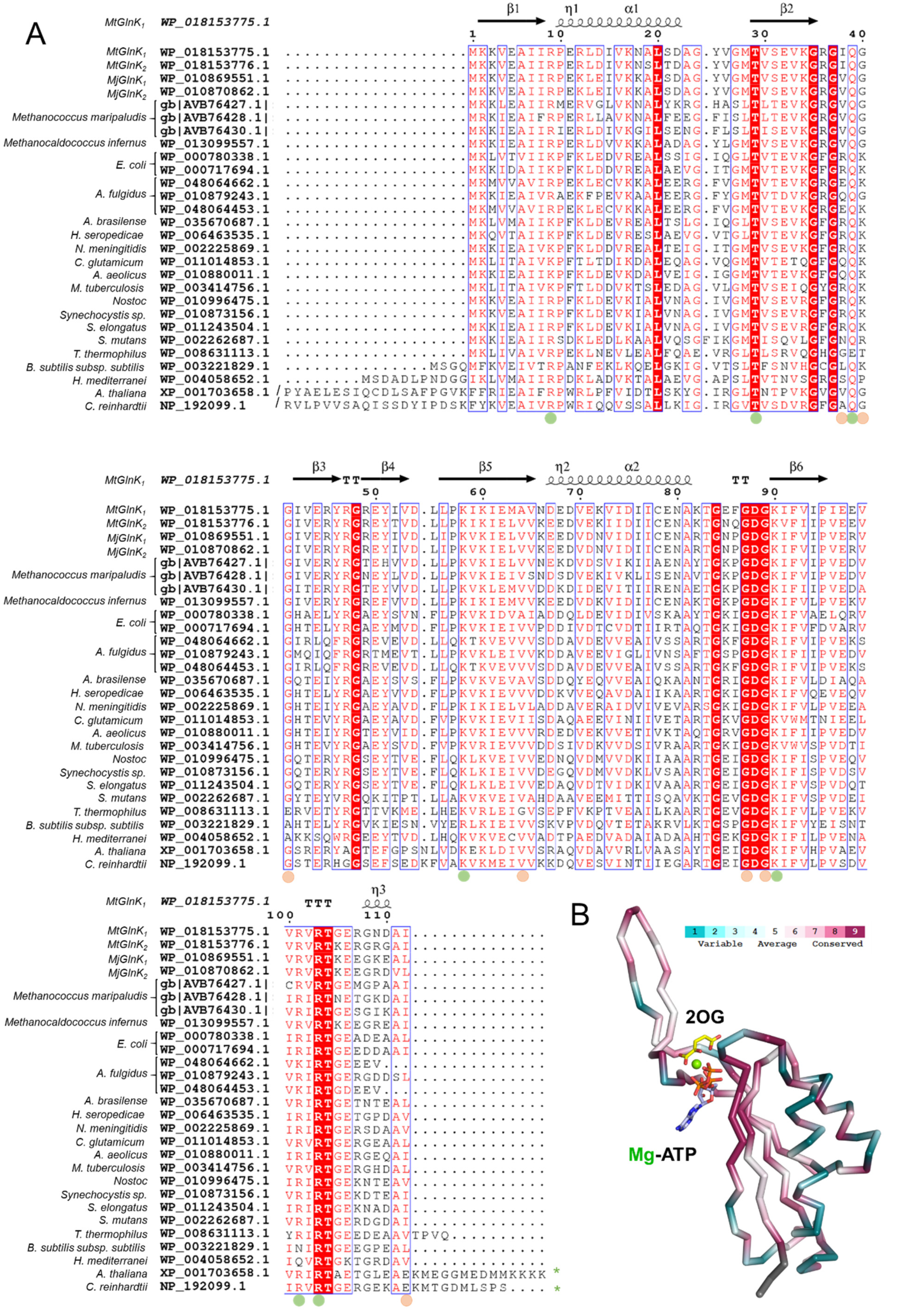
2.3. Conformational Rearrangement upon Ligand Binding
2.4. The Free Carboxy-Terminal Is Indirectly Stabilizing 2OG Binding
3. Discussion
4. Materials and Methods
4.1. Cultivation of M. thermolithotrophicus
4.2. Membrane Experiment, Binding Assay
4.3. Protein Identification by Mass Spectrometry
4.4. GlnKs Expression and Purification
4.5. Crystallization
4.6. X-Ray Crystallography Data Collection, Refinement and Analyses
4.7. Structural Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huergo, L.F.; Chandra, G.; Merrick, M. PII signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013, 37, 251–283. [Google Scholar] [CrossRef]
- Conroy, M.J.; Durand, A.; Lupo, D.; Li, X.D.; Bullough, P.A.; Winkler, F.K.; Merrick, M. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc. Natl. Acad. Sci. USA 2007, 104, 1213–1218. [Google Scholar] [CrossRef]
- Gruswitz, F.; O’Connell, J., 3rd; Stroud, R.M. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc. Natl. Acad. Sci. USA 2007, 104, 42–47. [Google Scholar] [CrossRef]
- Shapiro, B.M. The Glutamine Synthetase Deadenylylating Enzyme System from Escherichia coli. Resolution into Two Components, Specific Nucleotide Stimulation, and Cofactor Requirements. Biochemistry 1969, 8, 659–670. [Google Scholar] [CrossRef]
- Ehlers, C.; Weidenbach, K.; Veit, K.; Forchhammer, K.; Schmitz, R.A. Unique mechanistic features of post-translational regulation of glutamine synthetase activity in Methanosarcina mazei strain Gö1 in response to nitrogen availability. Mol. Microbiol. 2005, 55, 1841–1854. [Google Scholar] [CrossRef]
- Kessler, P.S.; Daniel, C.; Leigh, J.A. Ammonia Switch-Off of Nitrogen Fixation in the Methanogenic Archaeon Methanococcus maripaludis: Mechanistic Features and Requirement for the Novel GlnB Homologues, NifI1 and NifI2. J. Bacteriol. 2001, 183, 882–889. [Google Scholar] [CrossRef]
- Leigh, J.A.; Dodsworth, J.A. Nitrogen Regulation in Bacteria and Archaea. Annu. Rev. Microbiol. 2007, 61, 349–377. [Google Scholar] [CrossRef]
- Little, R.; Colombo, V.; Leech, A.; Dixon, R. Direct Interaction of the NifL Regulatory Protein with the GlnK Signal Transducer Enables the Azotobacter vinelandii NifL-NifA Regulatory System to Respond to Conditions Replete for Nitrogen. J. Biol. Chem. 2002, 277, 15472–15481. [Google Scholar] [CrossRef]
- He, L.; Soupene, E.; Ninfa, A.; Kustu, S. Physiological role for the GlnK protein of enteric bacteria: Relief of NifL inhibition under nitrogen-limiting conditions. J. Bacteriol. 1998, 180, 6661–6667. [Google Scholar] [CrossRef]
- Scholl, J.; Dengler, L.; Bader, L.; Forchhammer, K. Phosphoenolpyruvate carboxylase from the cyanobacterium Synechocystis sp. PCC 6803 is under global metabolic control by PII signaling. Mol. Microbiol. 2020, 114, 292–307. [Google Scholar] [CrossRef]
- Selim, K.A.; Haase, F.; Hartmann, M.D.; Hagemann, M.; Forchhammer, K. PII-like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response. Proc. Natl. Acad. Sci. USA 2018, 115, E4861–E4869. [Google Scholar] [CrossRef] [PubMed]
- Ninfa, A.J.; Jiang, P. PII signal transduction proteins: Sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr. Opin. Microbiol. 2005, 8, 168–173. [Google Scholar] [CrossRef]
- Jiang, P.; Ninfa, A.J. Escherichia coli PII Signal Transduction Protein Controlling Nitrogen Assimilation Acts As a Sensor of Adenylate Energy Charge in Vitro. Biochemistry 2007, 46, 12979–12996. [Google Scholar] [CrossRef]
- Forchhammer, K.; Luddecke, J. Sensory properties of the PII signalling protein family. FEBS J. 2016, 283, 425–437. [Google Scholar] [CrossRef]
- Sant’Anna, F.H.; Trentini, D.B.; de Souto Weber, S.; Cecagno, R.; da Silva, S.C.; Schrank, I.S. The PII Superfamily Revised: A Novel Group and Evolutionary Insights. J. Mol. Evol. 2009, 68, 322–336. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Segal, A.; Stadtman, E.R. Modulation of Glutamine Synthetase Adenylylation and Deadenylylation Is Mediated by Metabolic Transformation of the PII-Regulatory Protein. Proc. Natl. Acad. Sci. USA 1971, 68, 2949–2953. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.; Schleberger, P.; Lu, W.; Wacker, T.; Pfluger, T.; Litz, C.; Andrade, S.L. Mechanism of Disruption of the Amt-GlnK Complex by PII-Mediated Sensing of 2-Oxoglutarate. PLoS ONE 2011, 6, e26327. [Google Scholar] [CrossRef] [PubMed]
- Pedro-Roig, L.; Lange, C.; Bonete, M.J.; Soppa, J.; Maupin-Furlow, J. Nitrogen regulation of protein-protein interactions and transcript levels of GlnK PII regulator and AmtB ammonium transporter homologs in Archaea. Microbiologyopen 2013, 2, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, Ö.; Kalthoff, C.; Raunser, S.; Kuhlbrandt, W. Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake. EMBO J. 2007, 26, 589–599. [Google Scholar] [CrossRef]
- Leigh, J.A. Nitrogen Fixation In Methanogens: The Archaeal Perspective. Curr. Issues Mol. Biol. 2000, 2, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Radchenko, M.V.; Thornton, J.; Merrick, M. Control of AmtB-GlnK Complex Formation by Intracellular Levels of ATP, ADP, and 2-Oxoglutarate. J. Biol. Chem. 2010, 285, 31037–31045. [Google Scholar] [CrossRef]
- Truan, D.; Huergo, L.F.; Chubatsu, L.S.; Merrick, M.; Li, X.D.; Winkler, F.K. A New PII Protein Structure Identifies the 2-Oxoglutarate Binding Site. J. Mol. Biol. 2010, 400, 531–539. [Google Scholar] [CrossRef]
- Moure, V.R.; Razzera, G.; Araújo, L.M.; Oliveira, M.A.S.; Gerhardt, E.C.M.; Müller-Santos, M.; Almeida, F.; Pedrosa, F.O.; Valente, A.P.; Souza, E.M.; et al. Heat stability of Proteobacterial PII protein facilitate purification using a single chromatography step. Protein Expr. Purif. 2012, 81, 83–88. [Google Scholar] [CrossRef]
- Cheah, E.; Carr, P.D.; Suffolk, P.M.; Vasudevan, S.G.; Dixon, N.E.; Ollis, D.L. Structure of the Escherichia coli signal transducing protein PII. Structure 1994, 2, 981–990. [Google Scholar] [CrossRef]
- Landau, M.; Mayrose, I.; Rosenberg, Y.; Glaser, F.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005, 33, W299–W302. [Google Scholar] [CrossRef]
- Glaser, F.; Pupko, T.; Paz, I.; Bell, R.E.; Bechor-Shental, D.; Martz, E.; Ben-Tal, N. ConSurf: Identification of Functional Regions in Proteins by Surface-Mapping of Phylogenetic Information. Bioinformatics 2003, 19, 163–164. [Google Scholar] [CrossRef]
- Arcondéguy, T.; Jack, R.; Merrick, M. PII Signal Transduction Proteins, Pivotal Players in Microbial Nitrogen Control. Microbiol. Mol. Biol. Rev. 2001, 65, 80–105. [Google Scholar] [CrossRef]
- Helfmann, S.; Lü, W.; Litz, C.; Andrade, S.L. Cooperative Binding of MgATP and MgADP in the Trimeric PII Protein GlnK2 from Archaeoglobus fulgidus. J. Mol. Biol. 2010, 402, 165–177. [Google Scholar] [CrossRef]
- Chellamuthu, V.R.; Ermilova, E.; Lapina, T.; Luddecke, J.; Minaeva, E.; Herrmann, C.; Hartmann, M.D.; Forchhammer, K. A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom. Cell 2014, 159, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Wacker, T.; Garcia-Celma, J.J.; Lewe, P.; Andrade, S.L. Direct observation of electrogenic NH4+ transport in ammonium transport (Amt) proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 9995–10000. [Google Scholar] [CrossRef]
- Pedro-Roig, L.; Camacho, M.; Bonete, M.J. Regulation of ammonium assimilation in Haloferax mediterranei: Interaction between glutamine synthetase and two GlnK proteins. Biochim. Biophys. Acta 2013, 1834, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]

| Compound | Amount | Final Molarity |
|---|---|---|
| K2HPO4 | 55.7 mg | 0.32 mM |
| KH2PO4 | 55.8 mg | 0.41 mM |
| KCl | 1 g | 13.4 mM |
| NaCl | 25.13 g | 430 mM |
| NaHCO3 | 0.81 g | 10 mM |
| CaCl2·2H2O | 367.5 mg | 2.5 mM |
| MgCl2·6H2O | 7.725 g | 38 mM |
| NH4Cl | 1.18 g | 22.06 mM |
| Fe(NH4)2(SO4)2·12H2O | 29.92 mg | 0.031 mM |
| Nitrilotriacetic acid | 61.16 mg | 0.32 mM |
| 2 mM Na2SeO3·5H2O stock | 10 µL | 0.02 µM |
| Na2WO4·2H2O | 3.3 mg | 0.01 mM |
| Na2MoO4·2H2O | 2.42 mg | 0.01 mM |
| MES | 9.76 g | 50 mM |
| Na2SO4 | 1.42 g | 10 mM |
| 1.5 mM resazurin | 1 mL | 0.0015 mM |
| Trace element solution | 10 mL |
| Compound | Amount | Final Molarity |
|---|---|---|
| MnCl2·6H2O | 91.4 mg | 0.45 mM |
| FeCl3·6H2O | 183.3 mg | 0.68 mM |
| CaCl2·2H2O | 60.26 mg | 0.76 mM |
| CoCl2·6H2O | 180.8 mg | 0.76 mM |
| ZnCl2 | 90 mg | 0.66 mM |
| CuSO4·5H2O | 35.21 mg | 0.14 mM |
| Na2MoO4·2H2O | 46 mg | 0.19 mM |
| NiCl2·6H2O | 90 mg | 0.38 mM |
| VCl3 | 30 mg | 0.19 mM |
| MtGlnK1 with dADP | MtGlnK2 apo | MtGlnK2 with ATP, Mg and 2OG | MjGlnK1 with ATP, Mg and 2OG | |
|---|---|---|---|---|
| Data collection | ||||
| Synchrotron source | SLS, PXIII | SLS, PXIII | SLS, PXIII | SLS, PXIII |
| Wavelength (Å) | 1.00003 | 0.99187 | 0.99985 | 0.99187 |
| Space group | P321 | P21212 | P43212 | H32 |
| Resolution (Å) | 75.60–1.94 (2.15–1.94) | 73.16–2.30 (2.40–2.30) | 57.33–1.16 (1.26–1.16) | 44.53–1.20 (1.24–1.20) |
| Cell dimensions | ||||
| a, b, c (Å) | 87.296, 87.296, 46.010 | 103.461, 103.460, 178.491 | 58.238, 58.238, 229.304 | 89.053, 89.053, 98.059 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Rmerge(%) a | 5.3 (171.1) | 13.3 (115.1) | 4.9 (64.3) | 5.7 (120.9) |
| Rpim (%) a | 1.3 (40.3) | 5.5 (50.8) | 1.0 (28.6) | 2.0 (44.3) |
| CC1/2 a | 0.999 (0.773) | 0.998 (0.671) | 0.999 (0.774) | 0.999 (0.642) |
| I/σI a | 27.3 (1.8) | 10.2 (1.5) | 29.1 (1.7) | 17.8 (1.7) |
| Spherical completeness a | 66.3 (12.7) | 82.1 (35) | 78.1 (17.7) | 67.8 (34.6) |
| Ellipsoidal completeness a | 89.1 (60.2) | 96.2 (81.3) | 93.2 (51.7) | 89.8 (99.6) |
| Redundancy a | 19.3 (18.6) | 6.7 (6.0) | 21.6 (5.2) | 9.4 (8.3) |
| Nr. unique reflections a | 10,108 (502) | 70,360 (3519) | 108,153 (5406) | 31,673 (1585) |
| Refinement | ||||
| Resolution (Å) | 39.30–1.94 | 73.16–2.30 | 57.33–1.16 | 44.53–1.20 |
| Number of reflections | 9995 | 70,326 | 108,153 | 31,668 |
| Rwork/Rfree b (%) | 19.66/22.57 | 22.15/24.45 | 11.68/13.66 | 13.11/15.54 |
| Number of atoms | ||||
| Protein | 882 | 9066 | 2788 | 953 |
| Ligands/ions | 28 | 13 | 145 | 52 |
| Solvent | 23 | 104 | 516 | 178 |
| Mean B-value (Å2) | 62.0 | 37.0 | 20.0 | 21.0 |
| Molprobity clash score, all atoms | 1.63 | 3.89 | 0.67 | 0 |
| Ramachandran plot | ||||
| Favored regions (%) | 100 | 98.75 | 100 | 100 |
| Outlier regions (%) | 0 | 0 | 0 | 0 |
| rmsd c bond lengths (Å) | 0.013 | 0.006 | 0.014 | 0.012 |
| rmsd c bond angles (°) | 1.454 | 0.888 | 1.592 | 1.565 |
| PDB ID code | 7P4V | 7P4Y | 7P50 | 7P52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, M.-C.; Wagner, T. The Oxoglutarate Binding Site and Regulatory Mechanism Are Conserved in Ammonium Transporter Inhibitors GlnKs from Methanococcales. Int. J. Mol. Sci. 2021, 22, 8631. https://doi.org/10.3390/ijms22168631
Müller M-C, Wagner T. The Oxoglutarate Binding Site and Regulatory Mechanism Are Conserved in Ammonium Transporter Inhibitors GlnKs from Methanococcales. International Journal of Molecular Sciences. 2021; 22(16):8631. https://doi.org/10.3390/ijms22168631
Chicago/Turabian StyleMüller, Marie-Caroline, and Tristan Wagner. 2021. "The Oxoglutarate Binding Site and Regulatory Mechanism Are Conserved in Ammonium Transporter Inhibitors GlnKs from Methanococcales" International Journal of Molecular Sciences 22, no. 16: 8631. https://doi.org/10.3390/ijms22168631
APA StyleMüller, M.-C., & Wagner, T. (2021). The Oxoglutarate Binding Site and Regulatory Mechanism Are Conserved in Ammonium Transporter Inhibitors GlnKs from Methanococcales. International Journal of Molecular Sciences, 22(16), 8631. https://doi.org/10.3390/ijms22168631






