The Role of miRNA in the Pathophysiology of Neuroendocrine Tumors
Abstract
:1. Introduction
2. Biogenesis and Function of miRNAs
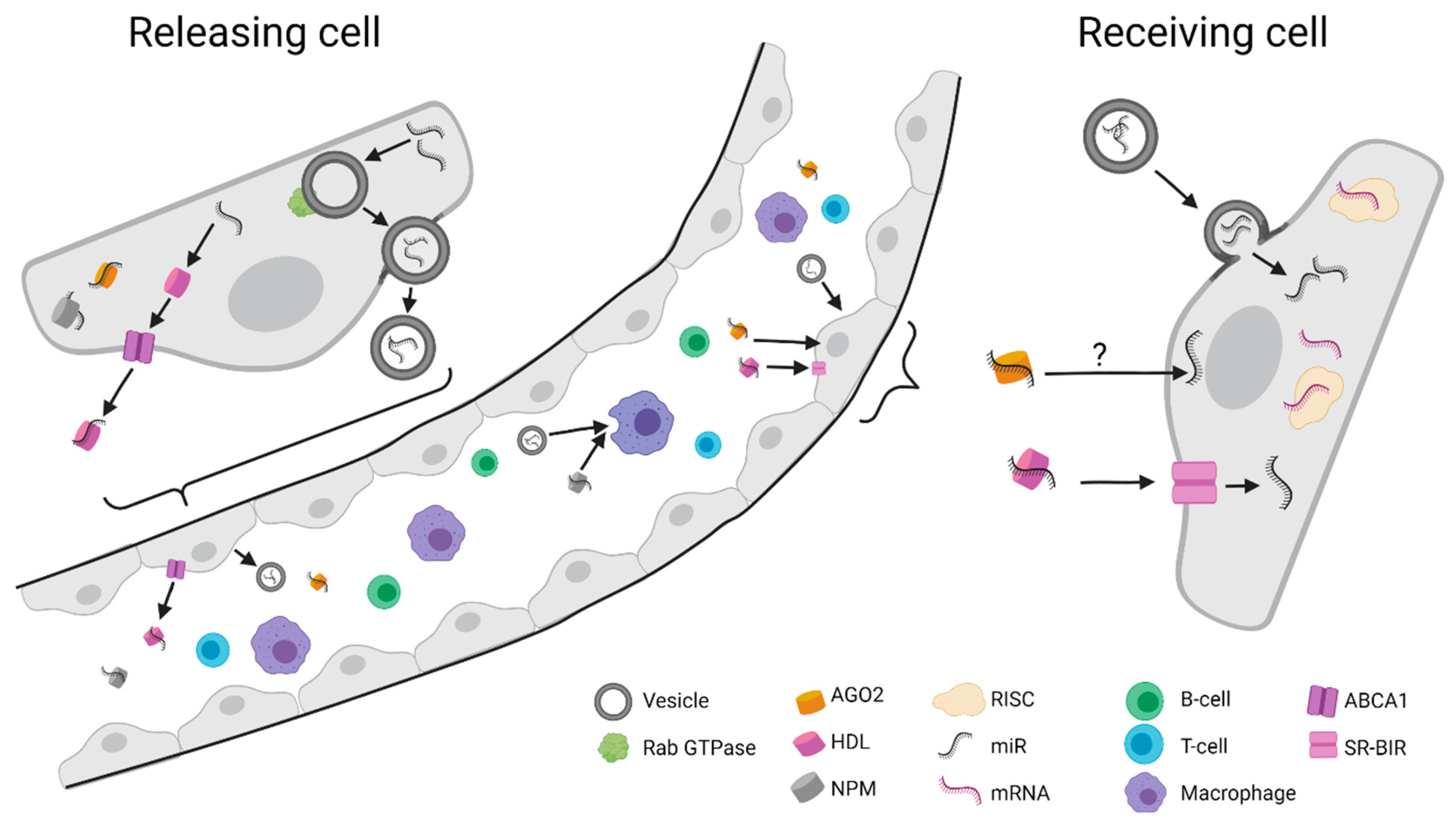
3. Circulating miRNAs
4. miRNA in the Pathophysiology of NET
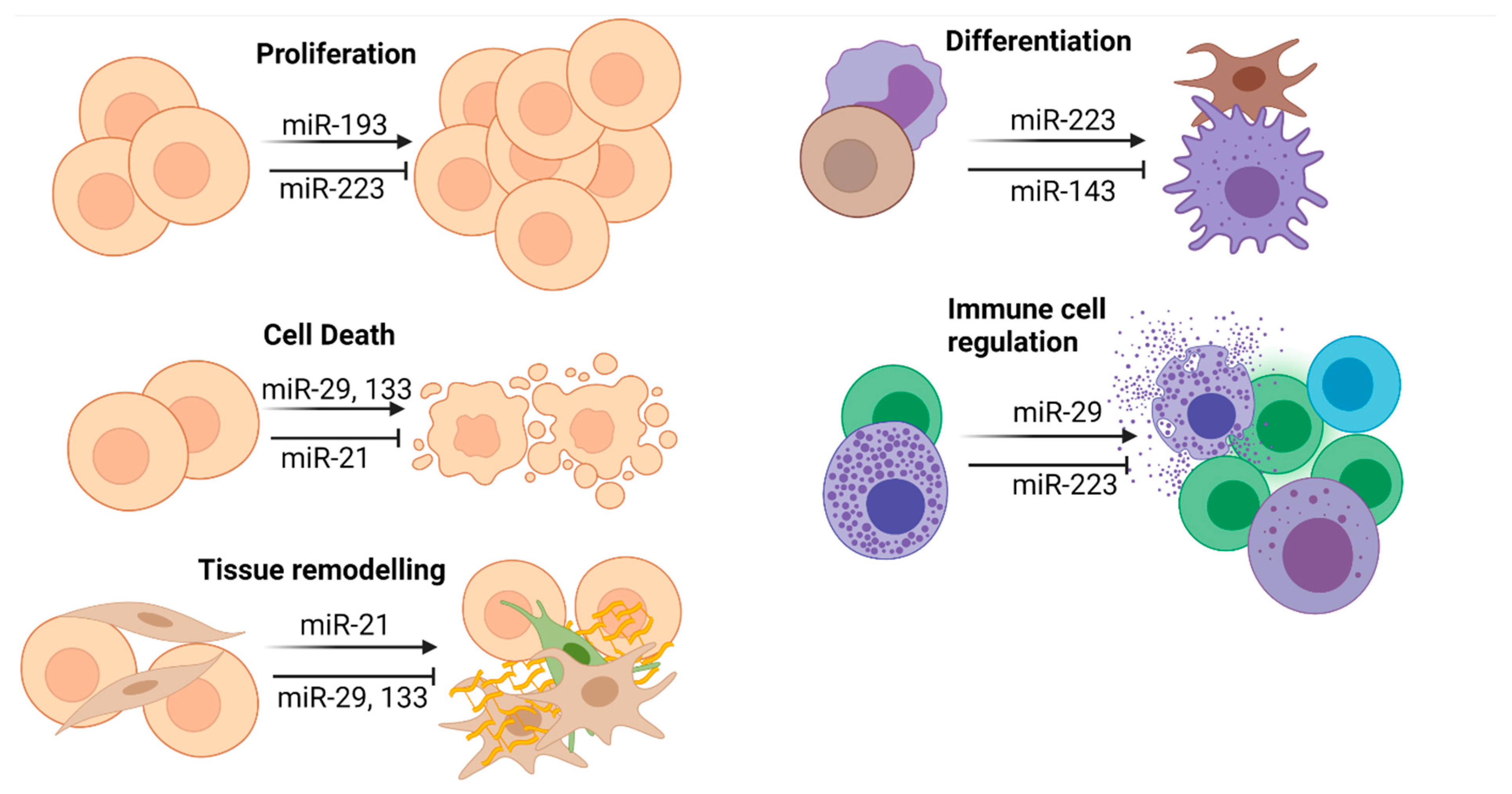
4.1. General Considerations
4.2. miR-21
4.3. miR-29a
4.4. miR-133
4.5. miR-223
5. Circulating miRNAs for the Diagnosis and Prognosis of NET
6. Diagnostic Circulating miRNAs
7. Prognostic Circulating miRNAs
8. Therapeutic Use of miRNAs
9. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BCL6 | B-cell lymphoma 6 |
| BACH1 | BTB and CNC homology 1 |
| CgA | Chromogranin A |
| FBXO11 | F-box only protein 11 |
| FGF2 | Fibroblast growth factor |
| HSP70 | Heat shock protein 70 |
| HDL | High-density lipoprotein |
| HIF-1α | Hypoxia-inducible factor 1α |
| INoS | Inducible nitric oxide synthases |
| INF | Interferon |
| LOXL | Lysyl oxidase-like protein |
| HMGA2 | Mammalian high-mobility-group protein AT-hook 2 |
| MMP | Matrix metalloproteinase |
| mRNA | Messenger RNA |
| miRNA | MicroRNA |
| NET | Neuroendocrine tumor |
| NF-kb | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| OS | Overall survival |
| pNET | Pancreatic NET |
| PTEN | Phosphatase and tensin homolog |
| PI3K | Phosphoinositide 3-kinases |
| AKT | Protein kinase B |
| RISC | RNA-induced silencing complex |
| STAT3 | Signal transducer and activator of transcription 3 |
| siNET | Small intestinal NET |
| SSAs | Somatostatin analogues |
| YAP | Yes-associated protein |
References
- Sackstein, P.E.; O’Neil, D.S.; Neugut, A.I.; Chabot, J.; Fojo, T. Epidemiologic trends in neuroendocrine tumors: An examination of incidence rates and survival of specific patient subgroups over the past 20 years. Semin. Oncol. 2018, 45, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Dasari, A. Epidemiology, Incidence, and Prevalence of Neuroendocrine Neoplasms: Are There Global Differences? Curr. Oncol. Rep. 2021, 23, 43. [Google Scholar] [CrossRef] [PubMed]
- Leoncini, E.; Boffetta, P.; Shafir, M.; Aleksovska, K.; Boccia, S.; Rindi, G. Increased incidence trend of low-grade and high-grade neuroendocrine neoplasms. Endocrine 2017, 58, 368–379. [Google Scholar] [CrossRef] [Green Version]
- Jensen, R.T.; Bodei, L.; Capdevila, J.; Couvelard, A.; Falconi, M.; Grozinsky-Glasberg, S.; Kloppel, G.; Lamberts, S.W.J.; Peeters, M.; Rindi, G.; et al. Unmet Needs in Functional and Nonfunctional Pancreatic Neuroendocrine Neoplasms. Neuroendocrinology 2019, 108, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Marotta, V.; Zatelli, M.C.; Sciammarella, C.; Ambrosio, M.R.; Bondanelli, M.; Colao, A.; Faggiano, A. Chromogranin A as circulating marker for diagnosis and management of neuroendocrine neoplasms: More flaws than fame. Endocr. Related Cancer 2018, 25, R11–R29. [Google Scholar] [CrossRef] [PubMed]
- Lyubimova, N.V.; Churikova, T.K.; Kushlinskii, N.E. Chromogranin As a Biochemical Marker of Neuroendocrine Tumors. Bull. Exp. Biol. Med. 2016, 160, 702–704. [Google Scholar] [CrossRef] [PubMed]
- Al-Toubah, T.; Cives, M.; Valone, T.; Blue, K.; Strosberg, J.R. Sensitivity and Specificity of the NETest: A Validation Study. Neuroendocrinology 2020, 111, 580–585. [Google Scholar] [CrossRef]
- Modlin, I.M.; Kidd, M.; Malczewska, A.; Drozdov, I.; Bodei, L.; Matar, S.; Chung, K.-S. The NETest: The Clinical Utility of Multigene Blood Analysis in the Diagnosis and Management of Neuroendocrine Tumors. Endocrinol. Metab. Clin. N. Am. 2018, 47, 485–504. [Google Scholar] [CrossRef]
- Roderburg, C.; Urban, G.-W.; Bettermann, K.; Vucur, M.; Zimmermann, H.W.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.A.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2010, 53, 209–218. [Google Scholar] [CrossRef]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendell, J.T.; Olson, E.N. MicroRNAs in Stress Signaling and Human Disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Rindi, G.; Wiedenmann, B. Neuroendocrine neoplasia goes molecular—Time for a change. Nat. Rev. Clin. Oncol. 2019, 16, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Benz, F.; Roy, S.; Trautwein, C.; Roderburg, C.; Luedde, T. Circulating MicroRNAs as Biomarkers for Sepsis. Int. J. Mol. Sci. 2016, 17, 78. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Broughton, J.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruebel, K.; Leontovich, A.A.; Stilling, G.A.; Zhang, S.; Righi, A.; Jin, L.; Lloyd, R.V. MicroRNA expression in ileal carcinoid tumors: Downregulation of microRNA-133a with tumor progression. Mod. Pathol. 2010, 23, 367–375. [Google Scholar] [CrossRef]
- Miller, H.C.; Frampton, A.E.; Malczewska, A.; Ottaviani, S.; Stronach, E.A.; Flora, R.; Kaemmerer, D.; Schwach, G.; Pfragner, R.; Faiz, O.; et al. MicroRNAs associated with small bowel neuroendocrine tumours and their metastases. Endocr. Relat. Cancer 2016, 23, 711–726. [Google Scholar] [CrossRef] [Green Version]
- Mandal, R.; Hardin, H.; Baus, R.; Rehrauer, W.; Lloyd, R.V. Analysis of miR-96 and miR-133a Expression in Gastrointestinal Neuroendocrine Neoplasms. Endocr. Pathol. 2017, 28, 345–350. [Google Scholar] [CrossRef]
- Heverhagen, A.E.; Legrand, N.; Wagner, V.; Fendrich, V.; Bartsch, D.K.; Slater, E.P. Overexpression of MicroRNA miR-7-5p Is a Potential Biomarker in Neuroendocrine Neoplasms of the Small Intestine. Neuroendocrinology 2018, 106, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Hellberg, T.; Mohr, R.; Geisler, L.; Knorr, J.; Wree, A.; Demir, M.; Benz, F.; Lambrecht, J.; Loosen, S.H.; Tacke, F.; et al. Serum levels of miR-223 but not miR-21 are decreased in patients with neuroendocrine tumors. PLoS ONE 2020, 15, e0244504. [Google Scholar] [CrossRef] [PubMed]
- Roldo, C.; Missiaglia, E.; Hagan, J.; Falconi, M.; Capelli, P.; Bersani, S.; Calin, G.; Volinia, S.; Liu, C.-G.; Scarpa, A.; et al. MicroRNA Expression Abnormalities in Pancreatic Endocrine and Acinar Tumors Are Associated with Distinctive Pathologic Features and Clinical Behavior. J. Clin. Oncol. 2006, 24, 4677–4684. [Google Scholar] [CrossRef]
- Thorns, C.; Schurmann, C.; Gebauer, N.; Wallaschofski, H.; Kümpers, C.; Bernard, V.; Feller, A.C.; Keck, T.; Habermann, J.K.; Begum, N.; et al. Global microRNA profiling of pancreatic neuroendocrine neoplasias. Anticancer. Res. 2014, 34, 2249–2254. [Google Scholar]
- Yang, X.; Nanayakkara, J.; Claypool, D.; Saghafinia, S.; Wong, J.J.M.; Xu, M.; Wang, X.; Nicol, C.J.B.; Michael, I.P.; Hafner, M.; et al. A miR-375/YAP axis regulates neuroendocrine differentiation and tumorigenesis in lung carcinoid cells. Sci. Rep. 2021, 11, 10455. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, H.; Yu, S.; Li, C.; Bai, J.; Gui, S.; Zhang, Y.; Zhao, P. Study on miRNAs’ expression for the invasion of pituitary adenomas. Turk. Neurosurg. 2017. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.-H.; Lee, M.-J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related MicroRNAs are Abundant in Breast Milk Exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Cogswell, J.P.; Ward, J.; Taylor, I.A.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C.; et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimer’s Dis. 2008, 14, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Quintero, B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016, 49, 281–303. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawa, Y.; Kawahara, K.-I.; Tancharoen, S.; Meng, X.; Sameshima, H.; Ito, T.; Masuda, Y.; Imaizumi, H.; Hashiguchi, T.; Maruyama, I. Nucleophosmin may act as an alarmin: Implications for severe sepsis. J. Leukoc. Biol. 2009, 86, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Raposo, G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2, 2. [Google Scholar] [CrossRef]
- Henderson, M.C.; Azorsa, D.O. The Genomic and Proteomic Content of Cancer Cell-Derived Exosomes. Front. Oncol. 2012, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Hsu, C.; Morohashi, Y.; Yoshimura, S.-I.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M.A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. J. Cell Biol. 2010, 189, 223–232. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.-Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Barros, F.M.; Carneiro, F.; Machado, J.C.; Melo, S.A. Exosomes and Immune Response in Cancer: Friends or Foes? Front. Immunol. 2018, 9, 730. [Google Scholar] [CrossRef]
- Ohshima, K.; Inoue, K.; Fujiwara, A.; Hatakeyama, K.; Kanto, K.; Watanabe, Y.; Muramatsu, K.; Fukuda, Y.; Ogura, S.-I.; Yamaguchi, K.; et al. Let-7 MicroRNA Family Is Selectively Secreted into the Extracellular Environment via Exosomes in a Metastatic Gastric Cancer Cell Line. PLoS ONE 2010, 5, e13247. [Google Scholar] [CrossRef] [Green Version]
- Cavalcanti, E.; Galleggiante, V.; Coletta, S.; Stasi, E.; Chieppa, M.; Armentano, R.; Serino, G. Altered miRNAs Expression Correlates With Gastroenteropancreatic Neuroendocrine Tumors Grades. Front. Oncol. 2020, 10, 1187. [Google Scholar] [CrossRef]
- Mairinger, F.; Ting, S.; Werner, R.; Walter, R.; Hager, T.; Vollbrecht, C.; Christoph, D.C.; Worm, K.; Mairinger, T.; Sheu-Grabellus, S.-Y.; et al. Different micro-RNA expression profiles distinguish subtypes of neuroendocrine tumors of the lung: Results of a profiling study. Mod. Pathol. 2014, 27, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Liu, L.; Qiu, Y.; Liu, L. MicroRNA-29a Inhibits Growth, Migration and Invasion of Melanoma A375 Cells In Vitro by Directly Targeting BMI1. Cell. Physiol. Biochem. 2018, 50, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Bondgaard, A.-L.R.Ø.; Poulsen, T.T.; Poulsen, H.S.; Skov, B.G. Different expression of EZH2, BMI1 and Ki67 in low and high grade neuroendocrine tumors of the lung. Cancer Biomark. 2012, 11, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Bommer, G.; Gerin, I.; Feng, Y.; Kaczorowski, A.J.; Kuick, R.; Love, R.E.; Zhai, Y.; Giordano, T.; Qin, Z.; Moore, B.; et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr. Biol. 2007, 17, 1298–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoffersen, N.R.; Shalgi, R.; Frankel, L.; Leucci, E.; Lees, M.; Klausen, M.; Pilpel, Y.; Nielsen, F.C.; Oren, M.; Lund, A. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010, 17, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Kress, T.R.; Cannell, I.; Brenkman, A.B.; Samans, B.; Gaestel, M.; Roepman, P.; Burgering, B.M.; Bushell, M.; Rosenwald, A.; Eilers, M. The MK5/PRAK Kinase and Myc Form a Negative Feedback Loop that Is Disrupted during Colorectal Tumorigenesis. Mol. Cell 2011, 41, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Craig, V.J.; Cogliatti, S.B.; Imig, J.; Renner, C.; Neuenschwander, S.; Rehrauer, H.; Schlapbach, R.; Dirnhofer, S.; Tzankov, A.; Müller, A. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood 2011, 117, 6227–6236. [Google Scholar] [CrossRef] [PubMed]
- Cole, K.A.; Attiyeh, E.F.; Mosse, Y.P.; Laquaglia, M.J.; Diskin, S.J.; Brodeur, G.M.; Maris, J.M. A Functional Screen Identifies miR-34a as a Candidate Neuroblastoma Tumor Suppressor Gene. Mol. Cancer Res. 2008, 6, 735–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Z.; Zhan, G.; Ye, D.; Ren, Y.; Cheng, L.; Wu, Z.; Guo, J. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med. Oncol. 2012, 29, 2473–2480. [Google Scholar] [CrossRef]
- Sun, F.; Fu, H.; Liu, Q.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008, 582, 1564–1568. [Google Scholar] [CrossRef] [Green Version]
- He, L.; He, X.; Lim, L.P.; De Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef] [Green Version]
- Tivnan, A.; Tracey, L.; Buckley, P.G.; Alcock, L.C.; Davidoff, A.M.; Stallings, R.L. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer 2011, 11, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, A.; Ruike, Y.; Terasawa, K.; Shimizu, K.; Tsujimoto, G. MicroRNA-34a Inhibits Cell Proliferation by Repressing Mitogen-Activated Protein Kinase Kinase 1 during Megakaryocytic Differentiation of K562 Cells. Mol. Pharmacol. 2010, 77, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.; Chen, Y.; Stallings, R.L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 2007, 26, 5017–5022. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.H.; Kim, H.S.; Kim, N.G.; Lee, I.; Choi, H.-S.; Li, X.-Y.; Kang, S.E.; Cha, S.; Ryu, J.K.; Na, J.M.; et al. p53 and MicroRNA-34 Are Suppressors of Canonical Wnt Signaling. Sci. Signal. 2011, 4, ra71. [Google Scholar] [CrossRef] [Green Version]
- Vacante, F.; Denby, L.; Sluimer, J.; Baker, A.H. The function of miR-143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vasc. Pharmacol. 2019, 112, 24–30. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Guo, H.; Qian, G.; Ge, S.; Ji, H.; Hu, X.; Chen, W. MiR-145, a new regulator of the DNA Fragmentation Factor-45 (DFF45)-mediated apoptotic network. Mol. Cancer 2010, 9, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregersen, L.H.; Jacobsen, A.B.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-145 Targets YES and STAT1 in Colon Cancer Cells. PLoS ONE 2010, 5, e8836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Li, M.; Wang, L.; Zhou, Y.; Zhou, J.; Pan, H.; Qu, P. microRNA-145 inhibits cell proliferation, migration and invasion by targeting matrix metallopeptidase-11 in renal cell carcinoma. Mol. Med. Rep. 2014, 10, 393–398. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Wang, Z.; Sun, G.; Wan, Y.; Guo, J.; Fu, X. miR-145 Inhibits Migration and Invasion of Glioma Stem Cells by Targeting ABCG2. NeuroMolecular Med. 2014, 16, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Borralho, P.; Kren, B.T.; Castro, R.; da Silva, I.M.; Steer, C.J.; Rodrigues, C.M.P. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009, 276, 6689–6700. [Google Scholar] [CrossRef]
- Guo, H.; Chen, Y.; Hu, X.; Qian, G.; Ge, S.; Zhang, J. The regulation of toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol. Cancer 2013, 12, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osaki, M.; Takeshita, F.; Sugimoto, Y.; Kosaka, N.; Yamamoto, Y.; Yoshioka, Y.; Kobayashi, E.; Yamada, T.; Kawai, A.; Inoue, T.; et al. MicroRNA-143 Regulates Human Osteosarcoma Metastasis by Regulating Matrix Metalloprotease-13 Expression. Mol. Ther. 2011, 19, 1123–1130. [Google Scholar] [CrossRef]
- Chen, X.; Guo, X.; Zhang, H.; Xiang, Y.; Chen, J.; Yin, Y.; Cai, X.; Wang, K.; Wang, G.; Ba, Y.; et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009, 28, 1385–1392. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhou, K.; Wu, L.; Gu, H.; Huang, Z.; Xu, J. Downregulation of microRNA-143 promotes osteogenic differentiation of human adipose-derived mesenchymal stem cells through the k-Ras/MEK/ERK signaling pathway. Int. J. Mol. Med. 2020, 46, 965–976. [Google Scholar] [CrossRef]
- Kent, O.A.; Fox-Talbot, K.; Halushka, M.K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 2013, 32, 2576–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Shan, A.; Su, Y.; Cheng, Y.; Gu, W.; Wang, W.; Ning, G.; Cao, Y. miR-144/451 Promote Cell Proliferation via Targeting PTEN/AKT Pathway in Insulinomas. Endocrinology 2015, 156, 2429–2439. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.H.; Zhao, J.X.; Jiang, M.Y.; Yang, L.P.; Sun, M.L.; Wang, H.W. MiR-193 promotes cell proliferation and invasion by ING5/PI3K/AKT pathway of triple-negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3122–3129. [Google Scholar] [PubMed]
- Munoz, J.L.; Walker, N.D.; Mareedu, S.; Pamarthi, S.H.; Sinha, G.; Greco, S.J.; Rameshwar, P. Cycling Quiescence in Temozolomide Resistant Glioblastoma Cells Is Partly Explained by microRNA-93 and -193-Mediated Decrease of Cyclin D. Front. Pharmacol. 2019, 10, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozaki, K.-I.; Imoto, I.; Mogi, S.; Omura, K.; Inazawa, J. Exploration of Tumor-Suppressive MicroRNAs Silenced by DNA Hypermethylation in Oral Cancer. Cancer Res. 2008, 68, 2094–2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, K.; He, Y.; Zhang, H.; Fei, Q.; Niu, D.; Wang, D.; Ding, X.; Xu, H.; Chen, X.; Zhu, J. DNA Methylation-regulated miR-193a-3p Dictates Resistance of Hepatocellular Carcinoma to 5-Fluorouracil via Repression of SRSF2 Expression. J. Biol. Chem. 2012, 287, 5639–5649. [Google Scholar] [CrossRef] [Green Version]
- Mood, A.D.; Premachandra, I.D.U.A.; Hiew, S.; Wang, F.; Scott, K.A.; Oldenhuis, N.J.; Liu, H.; Van Vranken, D.L. Potent Antifungal Synergy of Phthalazinone and Isoquinolones with Azoles Against Candida albicans. ACS Med. Chem. Lett. 2017, 8, 168–173. [Google Scholar] [CrossRef] [Green Version]
- Kietzmann, L.; Guhr, S.S.; Meyer, T.N.; Ni, L.; Sachs, M.; Panzer, U.; Stahl, R.A.; Saleem, M.A.; Kerjaschki, N.; Gebeshuber, C.A.; et al. MicroRNA-193a Regulates the Transdifferentiation of Human Parietal Epithelial Cells toward a Podocyte Phenotype. J. Am. Soc. Nephrol. 2015, 26, 1389–1401. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Qian, L.; Lv, L.; Ding, B.; Zhou, G.; Cheng, X.; Niu, S.; Liang, Y. miR-193a-3p regulation of chemoradiation resistance in oesophageal cancer cells via the PSEN1 gene. Gene 2016, 579, 139–145. [Google Scholar] [CrossRef]
- Alberici, A.; Moratto, D.; Benussi, L.; Gasparini, L.; Ghidoni, R.; Gatta, L.B.; Finazzi, D.; Frisoni, G.B.; Trabucchi, M.; Growdon, J.H.; et al. Presenilin 1 Protein Directly Interacts with Bcl-2. J. Biol. Chem. 1999, 274, 30764–30769. [Google Scholar] [CrossRef] [Green Version]
- Seviour, E.G.; Sehgal, V.; Mishra, D.; Rupaimoole, R.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Lee, J.-S.; Sood, A.K.; Kim, M.P.; Mills, G.; et al. Targeting KRas-dependent tumour growth, circulating tumour cells and metastasis in vivo by clinically significant miR-193a-3p. Oncogene 2017, 36, 1339–1350. [Google Scholar] [CrossRef] [Green Version]
- Iliopoulos, D.; Rotem, A.; Struhl, K. Inhibition of miR-193a expression by Max and RXRalpha activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011, 71, 5144–5153. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Li, J.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Sun, L.; Zhang, Y.; Cui, Y.; Zhang, F.; et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 2015, 34, 413–423. [Google Scholar] [CrossRef]
- Kirschmann, D.A.; Seftor, E.A.; Fong, S.F.T.; Nieva, D.R.C.; Sullivan, C.M.; Edwards, E.M.; Sommer, P.; Csiszar, K.; Hendrix, M.J.C. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002, 62, 4478–4483. [Google Scholar] [PubMed]
- Deng, H.; Lv, L.; Li, Y.; Zhang, C.; Meng, F.; Pu, Y.; Xiao, J.; Qian, L.; Zhao, W.; Liu, Q.; et al. miR-193a-3p regulates the multi-drug resistance of bladder cancer by targeting the LOXL4 gene and the Oxidative Stress pathway. Mol. Cancer 2014, 13, 234. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Gao, L.; Luo, X.; Wang, L.; Gao, X.; Wang, W.; Sun, J.; Dou, L.; Li, J.; Xu, C.; et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood 2013, 121, 499–509. [Google Scholar] [CrossRef]
- Pu, Y.; Zhao, F.; Cai, W.; Meng, X.; Li, Y.; Cai, S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin. Exp. Metastasis 2016, 33, 359–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Q.; Wang, T.; Lu, P.; Zhang, R.; Zou, J.; Yuan, S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma. J. Neurosci. Res. 2014, 92, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, L. The small RNA miR-375—A pancreatic islet abundant miRNA with multiple roles in endocrine beta cell function. Mol. Cell. Endocrinol. 2017, 456, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Avnit-Sagi, T.; Kantorovich, L.; Kredo-Russo, S.; Hornstein, E.; Walker, M.D. The promoter of the pri-miR-375 gene directs expression selectively to the endocrine pancreas. PLoS ONE 2009, 4, e5033. [Google Scholar] [CrossRef]
- Gao, S.; Wang, K.; Wang, X. miR-375 targeting autophagy-related 2B (ATG2B) suppresses autophagy and tumorigenesis in cisplatin-resistant osteosarcoma cells. Neoplasma 2020, 67, 724–734. [Google Scholar] [CrossRef]
- Zhao, S.-P.; Yu, C.; Xiang, K.-M.; Yang, M.-S.; Liu, Z.-L.; Yang, B.-C. miR-375 Inhibits Autophagy and Further Promotes Inflammation and Apoptosis of Acinar Cells by Targeting ATG7. Pancreas 2020, 49, 543–551. [Google Scholar] [CrossRef]
- Li, L.; Jia, L.; Ding, Y. Upregulation of miR-375 inhibits human liver cancer cell growth by modulating cell proliferation and apoptosis via targeting ErbB2. Oncol. Lett. 2018, 16, 3319–3326. [Google Scholar] [CrossRef]
- Kang, W.; Huang, T.; Zhou, Y.; Zhang, J.; Lung, R.W.M.; Tong, J.H.M.; Chan, A.; Zhang, B.; Wong, C.C.; Wu, F.; et al. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, L.; Lim, J.; Burd, B.; Harris, T.M.; Ow, T.J.; Kawachi, N.; Belbin, T.J.; Angeletti, R.; Prystowsky, M.B.; Childs, G.; et al. miR-375 Regulates Invasion-Related Proteins Vimentin and L-Plastin. Am. J. Pathol. 2017, 187, 1523–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Huang, K.; Jie, Z.; Wu, Y.; Chen, J.; Chen, Z.; Fang, X.; Shen, S. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol. Cancer 2018, 17, 1–16. [Google Scholar] [CrossRef]
- Li, G.F.; Cheng, Y.Y.; Li, B.J.; Zhang, C.; Zhang, X.X.; Su, J.; Wang, C.; Chang, L.; Zhang, D.Z.; Tan, C.L.; et al. miR-375 inhibits the proliferation and invasion of glioblastoma by regulating Wnt5a. Neoplasma 2019, 66, 350–356. [Google Scholar] [CrossRef]
- Wu, C.P.; Bi, Y.J.; Liu, D.M.; Wang, L.Y. Hsa-miR-375 promotes the progression of inflammatory bowel disease by upregulating TLR4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7543–7549. [Google Scholar]
- Bosch, F.; Bazhin, A.V.; Heublein, S.; Bruwer, K.; Knosel, T.; Reiter, F.P.; Auernhammer, C.J.; Guba, M.O.; Spitzweg, C.; Werner, J.; et al. Treatment with somatostatin analogs induces differentially expressed let-7c-5p and mir-3137 in small intestine neuroendocrine tumors. BMC Cancer 2019, 19, 575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, N.; Knief, J.; Kacprowski, T.; Lazar-Karsten, P.; Keck, T.; Billmann, F.; Schmid, S.; Luley, K.; Lehnert, H.; Brabant, G.; et al. MicroRNA analysis of gastroenteropancreatic neuroendocrine tumors and metastases. Oncotarget 2018, 9, 28379–28390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Døssing, K.B.V.; Binderup, T.; Kaczkowski, B.; Jacobsen, A.; Rossing, M.; Winther, O.; Federspiel, B.; Knigge, U.; Kjær, A.; Friis-Hansen, L. Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes 2014, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Thornton, J.E.; Gregory, R.I. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012, 22, 474–482. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Ng, S.C.; Segrè, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.; Kharas, M.G.; et al. The Lin28/let-7 Axis Regulates Glucose Metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Kim, H.; Kim, H.W.; Lee, J.-C.; Paik, K.-H.; Kang, J.; Kim, J.; Yoon, Y.-S.; Han, H.-S.; Sohn, I.; et al. High Expression of MicroRNA-196a Indicates Poor Prognosis in Resected Pancreatic Neuroendocrine Tumor. Medicine 2015, 94, e2224. [Google Scholar] [CrossRef]
- Liu, Y.; Nie, H.; Zhang, K.; Ma, D.; Yang, G.; Zheng, Z.; Liu, K.; Yu, B.; Zhai, C.; Yang, S. A feedback regulatory loop between HIF-1α and miR-21 in response to hypoxia in cardiomyocytes. FEBS Lett. 2014, 588, 3137–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Multhoff, G.; Vaupel, P. Hypoxia Compromises Anti-Cancer Immune Responses. Adv. Exp. Med. Biol. 2020, 1232, 131–143. [Google Scholar] [CrossRef]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Fan, M.; Pfeffer, L.M. IFN Induces miR-21 through a Signal Transducer and Activator of Transcription 3–Dependent Pathway as a Suppressive Negative Feedback on IFN-Induced Apoptosis. Cancer Res. 2010, 70, 8108–8116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Duan, S.; Cermak, L.; Pagan, J.; Rossi, M.; Martinengo, C.; Di Celle, P.F.; Chapuy, B.; Shipp, M.; Chiarle, R.; Pagano, M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 2012, 481, 90–93. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.H.; Pfeffer, S.R.; Sims, M.; Yue, J.; Wang, Y.; Linga, V.G.; Paulus, E.; Davidoff, A.M.; Pfeffer, L.M. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J. Biol. Chem. 2015, 290, 6037–6046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Chen, X.; Du, Y.; Yao, W.; Shen, L.; Wang, C.; Hu, Z.; Zhuang, R.; Ning, G.; Zhang, C.; et al. Serum MicroRNA Expression Profile as a Biomarker in the Diagnosis and Prognosis of Pancreatic Cancer. Clin. Chem. 2012, 58, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, P.; Li, A.; Jiang, W.; Wang, H.; Wang, J.; Xie, K. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J. Exp. Clin. Cancer Res. 2012, 31, 97. [Google Scholar] [CrossRef] [Green Version]
- Abd-El-Fattah, A.A.; Sadik, N.A.H.; Shaker, O.; Aboulftouh, M.L. Differential MicroRNAs Expression in Serum of Patients with Lung Cancer, Pulmonary Tuberculosis, and Pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884. [Google Scholar] [CrossRef]
- Liu, G.-H.; Zhou, Z.-G.; Chen, R.; Wang, M.-J.; Zhou, B.; Li, Y.; Sun, X.-F. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumor Biol. 2013, 34, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhuijze, A.; Dooley, J.; Humblet-Baron, S.; Sreenivasan, J.; Koenders, M.; Schlenner, S.M.; Linterman, M.; Liston, A. Defective germinal center B-cell response and reduced arthritic pathology in microRNA-29a-deficient mice. Cell. Mol. Life Sci. 2017, 74, 2095–2106. [Google Scholar] [CrossRef]
- Steiner, D.F.; Thomas, M.F.; Hu, J.K.; Yang, Z.; Babiarz, J.E.; Allen, C.D.; Matloubian, M.; Blelloch, R.; Ansel, K.M. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity 2011, 35, 169–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, N.L.; Gilchrist, D.S.; Akbar, M.; Reilly, J.H.; Kerr, S.C.; Campbell, A.L.; Murrell, G.A.C.; Liew, F.Y.; Kurowska-Stolarska, M.; McInnes, I. MicroRNA29a regulates IL-33-mediated tissue remodelling in tendon disease. Nat. Commun. 2015, 6, 6774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brain, O.; Owens, B.M.; Pichulik, T.; Allan, P.; Khatamzas, E.; Leslie, A.; Steevels, T.; Sharma, S.; Mayer, A.; Catuneanu, A.M.; et al. The Intracellular Sensor NOD2 Induces MicroRNA-29 Expression in Human Dendritic Cells to Limit IL-23 Release. Immunity 2013, 39, 521–536. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; Tong, B.D.; Wu, Y.J.; Xiong, W. MicroRNA-29 mediates TGFbeta1-induced extracellular matrix synthesis by targeting wnt/beta-catenin pathway in human orbital fibroblasts. Int. J. Clin. Exp. Pathol. 2014, 7, 7571–7577. [Google Scholar]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Meng, X.-M.; Huang, X.R.; Chung, A.C.; Feng, Y.-L.; Hui, D.; Yu, C.-M.; Sung, J.J.Y.; Lan, H.Y. miR-29 Inhibits Bleomycin-induced Pulmonary Fibrosis in Mice. Mol. Ther. 2012, 20, 1251–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzon, R.; Heaphy, C.E.A.; Havelange, V.; Fabbri, M.; Volinia, S.; Tsao, T.; Zanesi, N.; Kornblau, S.M.; Marcucci, G.; Calin, G.A.; et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009, 114, 5331–5341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Fang, J.-H.; Yun, J.-P.; Yang, J.; Zhang, Y.; Jia, W.-H.; Zhuang, S.-M. Effects of MicroRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010, 51, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-S.; Hur, K.; Xu, G.; Choi, B.; Okugawa, Y.; Toiyama, Y.; Oshima, H.; Oshima, M.; Lee, H.-J.; Kim, V.N.; et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015, 64, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Arechaga-Ocampo, E.; López-Camarillo, C.; Villegas-Sepulveda, N.; la Rosa, C.H.G.-D.; Perez-Añorve, I.X.; Roldan-Perez, R.; Flores-Perez, A.; Peña-Curiel, O.; Angeles-Zaragoza, O.; Corona, R.R.; et al. Tumor suppressor miR-29c regulates radioresistance in lung cancer cells. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Shin-Hyuk, K.; Shim, H.G.; Hwang, T.; Kim, H.; Kang, S.-H.; Dho, Y.-S.; Park, S.-H.; Kim, S.J.; Park, C.-K. Restoration of miR-29b exerts anti-cancer effects on glioblastoma. Cancer Cell Int. 2017, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Kønig, S.M.; Rissler, V.; Terkelsen, T.; Lambrughi, M.; Papaleo, E. Alterations of the interactome of Bcl-2 proteins in breast cancer at the transcriptional, mutational and structural level. PLoS Comput. Biol. 2019, 15, e1007485. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Lee, J.H.; Ha, M.; Nam, J.-W.; Kim, V.N. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat. Struct. Mol. Biol. 2009, 16, 23–29. [Google Scholar] [CrossRef]
- Kuo, T.-Y.; Hsi, E.; Yang, I.-P.; Tsai, P.-C.; Wang, J.-Y.; Juo, S.-H.H. Computational Analysis of mRNA Expression Profiles Identifies MicroRNA-29a/c as Predictor of Colorectal Cancer Early Recurrence. PLoS ONE 2012, 7, e31587. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, K.; Seki, N.; Mataki, H.; Matsushita, R.; Kamikawaji, K.; Kumamoto, T.; Takagi, K.; Goto, Y.; Nishikawa, R.; Kato, M.; et al. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int. J. Oncol. 2016, 48, 450–460. [Google Scholar] [CrossRef]
- Gebeshuber, C.A.; Zatloukal, K.; Martinez, J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009, 10, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-J.; Liu, B.-Y.; Yan, S.; Jiang, T.-H.; Cheng, H.-Q.; Jiang, H.-S.; Cao, Y.; Mao, A.-W. MicroRNA-29a Promotes Pancreatic Cancer Growth by Inhibiting Tristetraprolin. Cell. Physiol. Biochem. 2015, 37, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jiang, K.; Zhao, F. Oxymatrine suppresses proliferation and facilitates apoptosis of human ovarian cancer cells through upregulating microRNA-29b and downregulating matrix metalloproteinase-2 expression. Mol. Med. Rep. 2015, 12, 5369–5374. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Li, J.; Wang, Y.; Liu, C.; Jia, H.; Jiang, C.; Wang, Y.; Luo, M.; Zhao, H.; Dong, L.; et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis 2014, 35, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.; Li, J.; Chen, Z.; Li, X.; Zheng, S.; Yi, D.; Zhong, A.; Chen, J. miR-29c suppresses pancreatic cancer liver metastasis in an orthotopic implantation model in nude mice and affects survival in pancreatic cancer patients. Carcinogenesis 2015, 36, 676–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Crampton, S.P.; Hughes, C.C. Wnt Signaling Induces Matrix Metalloproteinase Expression and Regulates T Cell Transmigration. Immunity 2007, 26, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Zhu, Y.; Gao, J.; Fu, J.; Liu, C.; Liu, Y.; Song, C.; Zhu, S.; Leng, Y.; Wang, G.; et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br. J. Cancer 2014, 110, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.-G.; Gu, J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012, 36, e61–e67. [Google Scholar] [CrossRef]
- Peng, Q.; Feng, Z.; Shen, Y.; Zhu, J.; Zou, L.; Shen, Y.; Zhu, Y. Integrated analyses of microRNA-29 family and the related combination biomarkers demonstrate their widespread influence on risk, recurrence, metastasis and survival outcome in colorectal cancer. Cancer Cell Int. 2019, 19, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.; Zhang, Y.; Qu, K.; Yang, X.; Fu, J.; Chen, W.; Li, X. MicroRNA-29B (mir-29b) regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3. Oncotarget 2015, 6, 40799–40814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.A.; Wappett, M.; Delpuech, O.; Brown, H.; Chresta, C.M. Enhanced MAPK signaling drives ETS1-mediated induction of miR-29b leading to downregulation of TET1 and changes in epigenetic modifications in a subset of lung SCC. Oncogene 2016, 35, 4345–4357. [Google Scholar] [CrossRef] [Green Version]
- Özdirik, B.; Stueven, A.K.; Mohr, R.; Geisler, L.; Wree, A.; Knorr, J.; Demir, M.; Vucur, M.; Loosen, S.H.; Benz, F.; et al. Analysis of miR-29 Serum Levels in Patients with Neuroendocrine Tumors—Results from an Exploratory Study. J. Clin. Med. 2020, 9, 2881. [Google Scholar] [CrossRef] [PubMed]
- Sjogren, R.J.; Egan, B.; Katayama, M.; Zierath, J.R.; Krook, A. Temporal analysis of reciprocal miRNA-mRNA expression patterns predicts regulatory networks during differentiation in human skeletal muscle cells. Physiol. Genom. 2015, 47, 45–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumazaki, M.; Shinohara, H.; Taniguchi, K.; Ueda, H.; Nishi, M.; Ryo, A.; Akao, Y. Understanding of tolerance in TRAIL-induced apoptosis and cancelation of its machinery by α-mangostin, a xanthone derivative. Oncotarget 2015, 6, 25828–25842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, X.; Zhang, Y.; Hu, Z.; Yang, D.; Wang, C.; Guo, M.; Cai, Q. Identification of miRNomes in human stomach and gastric carcinoma reveals miR-133b/a-3p as therapeutic target for gastric cancer. Cancer Lett. 2015, 369, 58–66. [Google Scholar] [CrossRef]
- Crawford, M.; Batte, K.; Yu, L.; Wu, X.; Nuovo, G.J.; Marsh, C.B.; Otterson, G.A.; Nana-Sinkam, S.P. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem. Biophys. Res. Commun. 2009, 388, 483–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, T.; Zhou, X.; Wang, J.; Du, Y.; Xu, J.; Huang, Z.; Zhu, W.; Shu, Y.; Liu, P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014, 588, 1168–1177. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Li, M.; Li, L.; Yang, X.; Lan, G.; Zhang, Y. MiR-133b Is Down-Regulated in Human Osteosarcoma and Inhibits Osteosarcoma Cells Proliferation, Migration and Invasion, and Promotes Apoptosis. PLoS ONE 2013, 8, e83571. [Google Scholar] [CrossRef]
- Hu, G.; Chen, D.; Li, X.; Yang, K.; Wang, H.; Wu, W. miR-133b regulates the MET proto-oncogene and inhibits the growth of colorectal cancer cells in vitro and in vivo. Cancer Biol. Ther. 2010, 10, 190–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novello, C.; Pazzaglia, L.; Cingolani, C.; Conti, A.; Quattrini, I.; Manara, M.C.; Tognon, M.; Picci, P.; Benassi, M.S. miRNA expression profile in human osteosarcoma: Role of miR-1 and miR-133b in proliferation and cell cycle control. Int. J. Oncol. 2013, 42, 667–675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Ito, Y.; Sugito, N.; Kumazaki, M.; Shinohara, H.; Yamada, N.; Nakagawa, Y.; Sugiyama, T.; Futamura, M.; Otsuki, Y.; et al. Organ-specific PTB1-associated microRNAs determine expression of pyruvate kinase isoforms. Sci. Rep. 2015, 5, 8647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Li, G. MicroRNA-133b inhibits proliferation and invasion of ovarian cancer cells through Akt and Erk1/2 inactivation by targeting epidermal growth factor receptor. Int. J. Clin. Exp. Pathol. 2015, 8, 10605–10614. [Google Scholar]
- Lu, Q.; Wu, D.; Xu, B.; Qian, W.; Li, P.; Tao, J.; Yin, C.; Zhang, W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol. Rep. 2012, 27, 1967–1975. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, D.; Tao, J.; Qu, P.; Zhou, Z.; Hou, J. MicroRNA-133 inhibits cell proliferation, migration and invasion by targeting epidermal growth factor receptor and its downstream effector proteins in bladder cancer. Scand. J. Urol. 2013, 47, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shao, X.; Gao, W.; Zhang, Z.; Liu, P.; Wang, R.; Huang, P.; Yin, Y.; Shu, Y. MicroRNA-133b inhibits the growth of non-small-cell lung cancer by targeting the epidermal growth factor receptor. FEBS J. 2012, 279, 3800–3812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Pan, H.; Zhou, Y.; Zhou, J.; Fan, Y.; Qü, P. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol. Med. Rep. 2014, 9, 2491–2498. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Lei, X.; Qin, Y.; Zhang, X.; Jin, H.; Wang, C.; Wang, X.; Li, G.; Tan, C.; Su, J. MicroRNA-133b inhibits cell migration and invasion by targeting matrix metalloproteinase 14 in glioblastoma. Oncol. Lett. 2015, 10, 2781–2786. [Google Scholar] [CrossRef]
- Fazi, F.; Rosa, A.; Fatica, A.; Gelmetti, V.; De Marchis, M.L.; Nervi, C.; Bozzoni, I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 2005, 123, 819–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Gronostajski, R.M. Roles of the NFI/CTF gene family in transcription and development. Gene 2000, 249, 31–45. [Google Scholar] [CrossRef]
- Bauernfeind, F.; Rieger, A.; Schildberg, F.A.; Knolle, P.A.; Schmid-Burgk, J.; Hornung, V. NLRP3 Inflammasome Activity Is Negatively Controlled by miR-223. J. Immunol. 2012, 189, 4175–4181. [Google Scholar] [CrossRef] [Green Version]
- Calvente, C.J.; Tameda, M.; Johnson, C.D.; Del Pilar, H.; Lin, Y.C.; Adronikou, N.; Jeu, X.D.M.D.; Llorente, C.; Boyer, J.; Feldstein, A.E. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J. Clin. Investig. 2019, 129, 4091–4109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorhoi, A.; Iannaccone, M.; Farinacci, M.; Faé, K.C.; Schreiber, J.; Moura-Alves, P.; Nouailles, G.; Mollenkopf, H.-J.; Oberbeck-Müller, D.; Jörg, S.; et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Investig. 2013, 123, 4836–4848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Feng, D.; Li, M.; Gao, Y.; Ramirez, T.; Cao, H.; Kim, S.-J.; Yang, Y.; Cai, Y.; Ju, C.; et al. Hepatic mitochondrial DNA/Toll-like receptor 9/MicroRNA-223 forms a negative feedback loop to limit neutrophil overactivation and acetaminophen hepatotoxicity in mice. Hepatology 2017, 66, 220–234. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Wang, H.; Liu, Y.; Song, Y.; Lai, L.; Han, Q.; Cao, X.; Wang, Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1beta production in macrophages by targeting STAT3. PLoS ONE 2012, 7, e42971. [Google Scholar]
- Pulikkan, J.A.; Dengler, V.; Peramangalam, P.S.; Zada, A.A.P.; Müller-Tidow, C.; Bohlander, S.K.; Tenen, D.G.; Behre, G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood 2010, 115, 1768–1778. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Yang, Z.; Deng, Z.; Zhou, Y.; Gong, Q.; Zhao, R.; Chen, T. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life 2018, 70, 536–546. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xu, Y.; Deng, Y.; Li, H. MicroRNA-223 Regulates Cardiac Fibrosis After Myocardial Infarction by Targeting RASA1. Cell. Physiol. Biochem. 2018, 46, 1439–1454. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Y.; Chen, Q.; Qiu, N.; Zhao, Y.; You, X. MiR-223 inhibited cell metastasis of human cervical cancer by modulating epithelial-mesenchymal transition. Int. J. Clin. Exp. Pathol. 2015, 8, 11224–11229. [Google Scholar]
- Nian, W.; Ao, X.; Wu, Y.; Huang, Y.; Shao, J.; Wang, Y.; Chen, Z.; Chen, F.; Wang, D. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase. Oncol. Lett. 2013, 6, 359–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masciarelli, S.; Fontemaggi, G.; Di Agostino, S.; Donzelli, S.; Carcarino, E.; Strano, S.; Blandino, G. Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 2014, 33, 1601–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rindi, G. The ENETS Guidelines: The New TNM Classification System. Tumori J. 2010, 96, 806–809. [Google Scholar] [CrossRef]
- Zatelli, M.C.; Grossrubatscher, E.M.; Guadagno, E.; Sciammarella, C.; Faggiano, A.; Colao, A. Circulating tumor cells and miRNAs as prognostic markers in neuroendocrine neoplasms. Endocr. Relat. Cancer 2017, 24, R223–R237. [Google Scholar] [CrossRef] [PubMed]
- Vicentini, C.; Fassan, M.; D’Angelo, E.; Corbo, V.; Silvestris, N.; Nuovo, G.J.; Scarpa, A. Clinical Application of MicroRNA Testing in Neuroendocrine Tumors of the Gastrointestinal Tract. Molecules 2014, 19, 2458–2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberg, K.; Modlin, I.M.; De Herder, W.; Pavel, M.; Klimstra, D.; Frilling, A.; Metz, D.C.; Heaney, A.; Kwekkeboom, D.J.; Strosberg, J.R.; et al. Consensus on biomarkers for neuroendocrine tumour disease. Lancet Oncol. 2015, 16, e435–e446. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.C.; Pavel, M.; Phan, A.T.; Kulke, M.H.; Hoosen, S.; Peter, J.S.; Cherfi, A.; Öberg, K.E. Chromogranin A and Neuron-Specific Enolase as Prognostic Markers in Patients with Advanced pNET Treated with Everolimus. J. Clin. Endocrinol. Metab. 2011, 96, 3741–3749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kidd, M.; Bodei, L.; Modlin, I.M. Chromogranin A: Any relevance in neuroendocrine tumors? Curr. Opin. Endocrinol. Diabetes Obes. 2016, 23, 28–37. [Google Scholar] [CrossRef]
- Bowden, M.; Zhou, C.; Zhang, S.; Brais, L.; Rossi, A.; Naudin, L.; Thiagalingam, A.; Sicinska, E.; Kulke, M.H. Profiling of metastatic small intestine neuroendocrine tumors reveals characteristic miRNAs detectable in plasma. Oncotarget 2017, 8, 54331–54344. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-C.; Essaghir, A.; Martijn, C.; Lloyd, R.V.; Demoulin, J.-B.; Öberg, K.; Giandomenico, V. Global microRNA profiling of well-differentiated small intestinal neuroendocrine tumors. Mod. Pathol. 2013, 26, 685–696. [Google Scholar] [CrossRef]
- Li, S.-C.; Khan, M.; Caplin, M.; Meyer, T.; Öberg, K.; Giandomenico, V. Somatostatin Analogs Treated Small Intestinal Neuroendocrine Tumor Patients Circulating MicroRNAs. PLoS ONE 2015, 10, e0125553. [Google Scholar] [CrossRef] [PubMed]
- Malczewska, A.; Frampton, A.E.; Mato Prado, M.; Ameri, S.; Dabrowska, A.F.; Zagorac, S.; Clift, A.K.M.; Kos-Kudła, B.; Faiz, O.; Stebbing, J.; et al. Circulating MicroRNAs in Small-bowel Neuroendocrine Tumors: A Potential Tool for Diagnosis and Assessment of Effectiveness of Surgical Resection. Ann. Surg. 2021, 174, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.; Moore, A.R.; Parsons, B.; O’Hara, A.; Boyce, M.; Dockray, G.J.; Varro, A.; Pritchard, D.M. Gastrin-induced miR-222 promotes gastric tumor development by suppressing p27kip1. Oncotarget 2016, 7, 45462–45478. [Google Scholar] [CrossRef] [Green Version]
- Malczewska, A.; Kidd, M.; Matar, S.; Kos-Kudła, B.; Modlin, I.M. A Comprehensive Assessment of the Role of miRNAs as Biomarkers in Gastroenteropancreatic Neuroendocrine Tumors. Neuroendocrinology 2018, 107, 73–90. [Google Scholar] [CrossRef]
- Li, A.; Yu, J.; Kim, H.; Wolfgang, C.L.; Canto, M.I.; Hruban, R.H.; Goggins, M. MicroRNA Array Analysis Finds Elevated Serum miR-1290 Accurately Distinguishes Patients with Low-Stage Pancreatic Cancer from Healthy and Disease Controls. Clin. Cancer Res. 2013, 19, 3600–3610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kövesdi, A.; Kurucz, P.A.; Nyírő, G.; Darvasi, O.; Patócs, A.; Butz, H. Circulating miRNA Increases the Diagnostic Accuracy of Chromogranin A in Metastatic Pancreatic Neuroendocrine Tumors. Cancers 2020, 12, 2488. [Google Scholar] [CrossRef]
- Wang, M.; Xia, X.; Chu, W.; Xia, L.; Meng, T.; Liu, L.; Liu, Y. Roles of miR-186 and PTTG1 in colorectal neuroendocrine tumors. Int. J. Clin. Exp. Med. 2015, 8, 22149–22157. [Google Scholar] [PubMed]
- Kasinski, A.; Slack, F.J. miRNA-34 Prevents Cancer Initiation and Progression in a Therapeutically Resistant K-ras and p53-Induced Mouse Model of Lung Adenocarcinoma. Cancer Res. 2012, 72, 5576–5587. [Google Scholar] [CrossRef] [Green Version]
- Pramanik, D.; Campbell, N.; Karikari, C.; Chivukula, R.; Kent, O.; Mendell, J.T.; Maitra, A. Restitution of Tumor Suppressor MicroRNAs Using a Systemic Nanovector Inhibits Pancreatic Cancer Growth in Mice. Mol. Cancer Ther. 2011, 10, 1470–1480. [Google Scholar] [CrossRef] [Green Version]
- Park, J.K.; Kogure, T.; Nuovo, G.J.; Jiang, J.; He, L.; Kim, J.H.; Phelps, M.A.; Papenfuss, T.L.; Croce, C.M.; Patel, T.; et al. miR-221 Silencing Blocks Hepatocellular Carcinoma and Promotes Survival. Cancer Res. 2011, 71, 7608–7616. [Google Scholar] [CrossRef] [Green Version]
- Goedeke, L.; Salerno, A.; Ramírez, C.; Guo, L.; Allen, R.M.; Yin, X.; Langley, S.R.; Esau, C.; Wanschel, A.; Fisher, E.A.; et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol. Med. 2014, 6, 1133–1141. [Google Scholar] [CrossRef]
- Hullinger, T.G.; Montgomery, R.L.; Seto, A.G.; Dickinson, B.A.; Semus, H.M.; Lynch, J.M.; Dalby, C.M.; Robinson, K.; Stack, C.; Latimer, P.A.; et al. Inhibition of miR-15 Protects Against Cardiac Ischemic Injury. Circ. Res. 2012, 110, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, Y.; Wang, W.; Chang, B.H.J.; Danesh, F.R. MicroRNA-29c Is a Signature MicroRNA under High Glucose Conditions That Targets Sprouty Homolog 1, and Its in Vivo Knockdown Prevents Progression of Diabetic Nephropathy. J. Biol. Chem. 2011, 286, 11837–11848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Ree, M.H.; van der Meer, A.J.; van Nuenen, A.C.; de Bruijne, J.; Ottosen, S.; Janssen, H.L.; Kootstra, N.A.; Reesink, H.W. Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment. Pharmacol. Ther. 2016, 43, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Anastasiadou, E.; Seto, A.G.; Beatty, X.; Hermreck, M.; Gilles, M.-E.; Stroopinsky, D.; Pinter-Brown, L.C.; Pestano, L.; Marchese, C.; Avigan, D.; et al. Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin. Cancer Res. 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.-K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.-L.; Kim, T.-Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.-S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Lanford, R.E.; Hildebrandt-Eriksen, E.S.; Petri, A.; Persson, R.; Lindow, M.; Munk, M.E.; Kauppinen, S.; Ørum, H. Therapeutic Silencing of MicroRNA-122 in Primates with Chronic Hepatitis C Virus Infection. Science 2010, 327, 198–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, H.L.A.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; Van Der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| miR on Chr. | Potential Functions |
|---|---|
| 34a/b/c on Chr. 11 |
|
| miR-143/miR-145 on Chr. 5 | |
| 193a/193b on Chr. 16/17 |
|
| 375 on Chr. 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geisler, L.; Mohr, R.; Lambrecht, J.; Knorr, J.; Jann, H.; Loosen, S.H.; Özdirik, B.; Luedde, T.; Hammerich, L.; Tacke, F.; et al. The Role of miRNA in the Pathophysiology of Neuroendocrine Tumors. Int. J. Mol. Sci. 2021, 22, 8569. https://doi.org/10.3390/ijms22168569
Geisler L, Mohr R, Lambrecht J, Knorr J, Jann H, Loosen SH, Özdirik B, Luedde T, Hammerich L, Tacke F, et al. The Role of miRNA in the Pathophysiology of Neuroendocrine Tumors. International Journal of Molecular Sciences. 2021; 22(16):8569. https://doi.org/10.3390/ijms22168569
Chicago/Turabian StyleGeisler, Lukas, Raphael Mohr, Joeri Lambrecht, Jana Knorr, Henning Jann, Sven H. Loosen, Burcin Özdirik, Tom Luedde, Linda Hammerich, Frank Tacke, and et al. 2021. "The Role of miRNA in the Pathophysiology of Neuroendocrine Tumors" International Journal of Molecular Sciences 22, no. 16: 8569. https://doi.org/10.3390/ijms22168569
APA StyleGeisler, L., Mohr, R., Lambrecht, J., Knorr, J., Jann, H., Loosen, S. H., Özdirik, B., Luedde, T., Hammerich, L., Tacke, F., Wree, A., Hellberg, T., & Roderburg, C. (2021). The Role of miRNA in the Pathophysiology of Neuroendocrine Tumors. International Journal of Molecular Sciences, 22(16), 8569. https://doi.org/10.3390/ijms22168569









