A Shifted Composition of the Lung Microbiota Conditions the Antifungal Response of Immunodeficient Mice
Abstract
:1. Introduction
2. Results
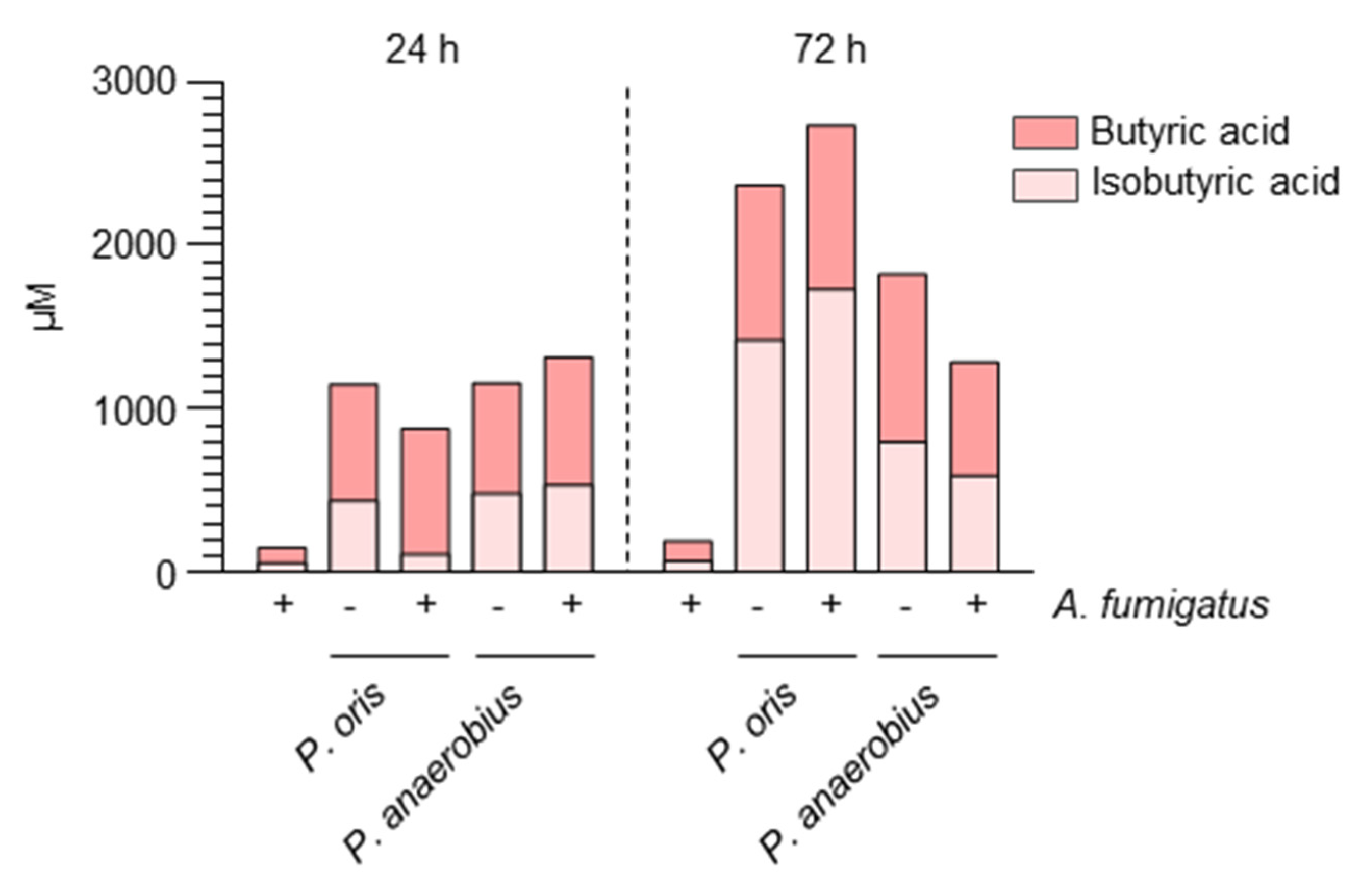
2.1. Aspergillus Infection Is Associated with the Production of SCFAs
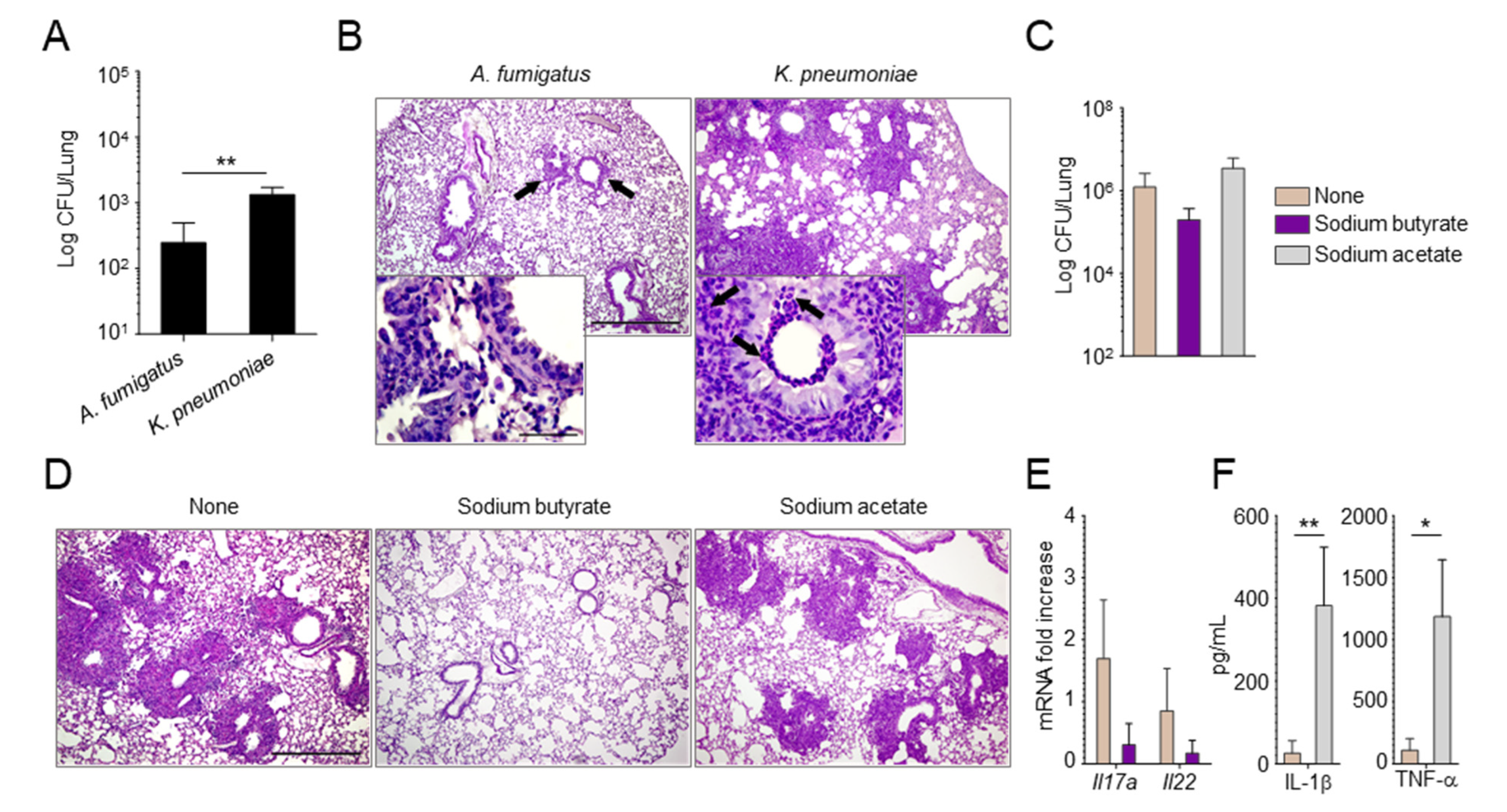
2.2. Manipulating the Composition of the Microbiome or Their Products Alters the Susceptibility to Aspergillus Infection
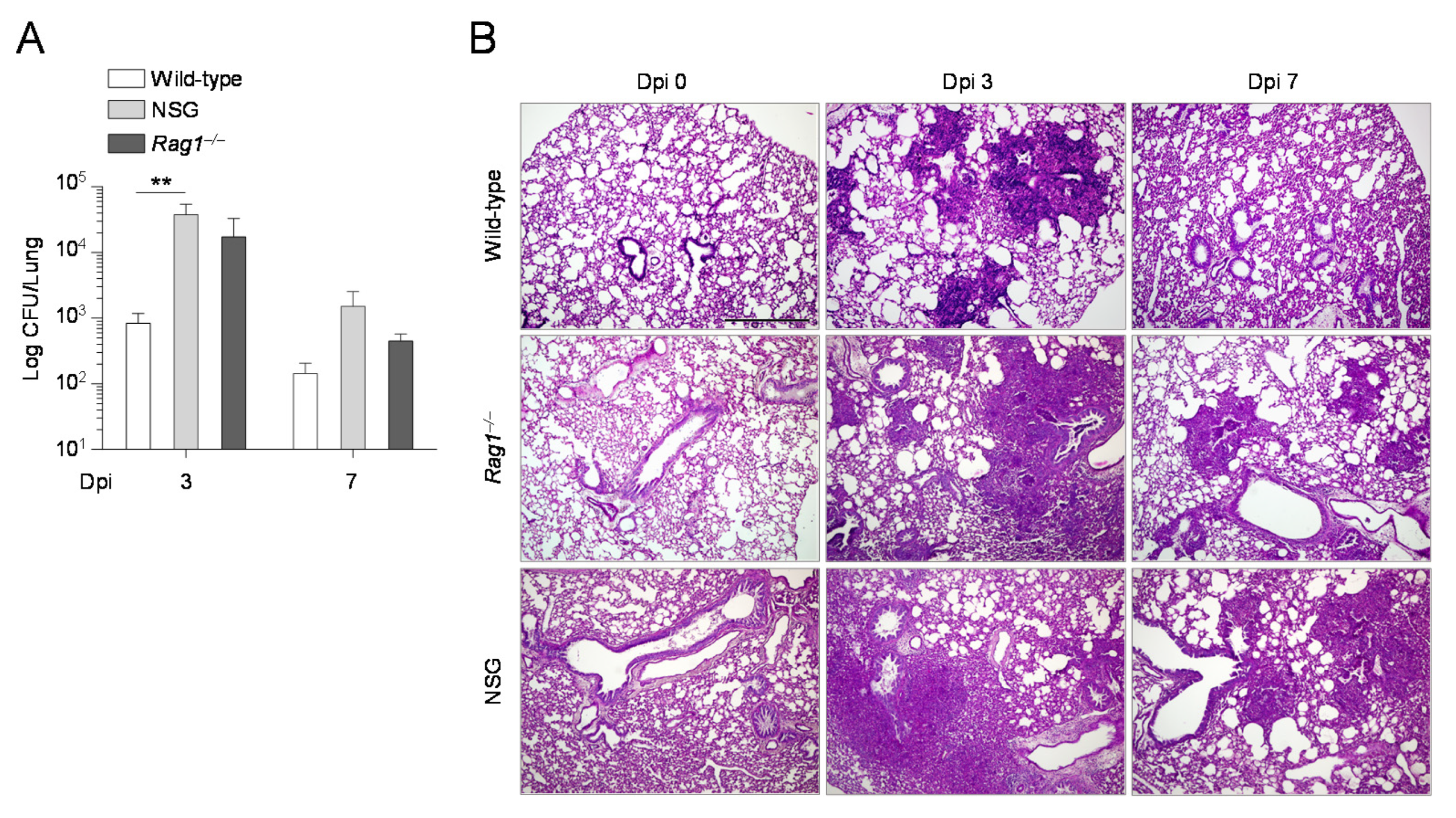
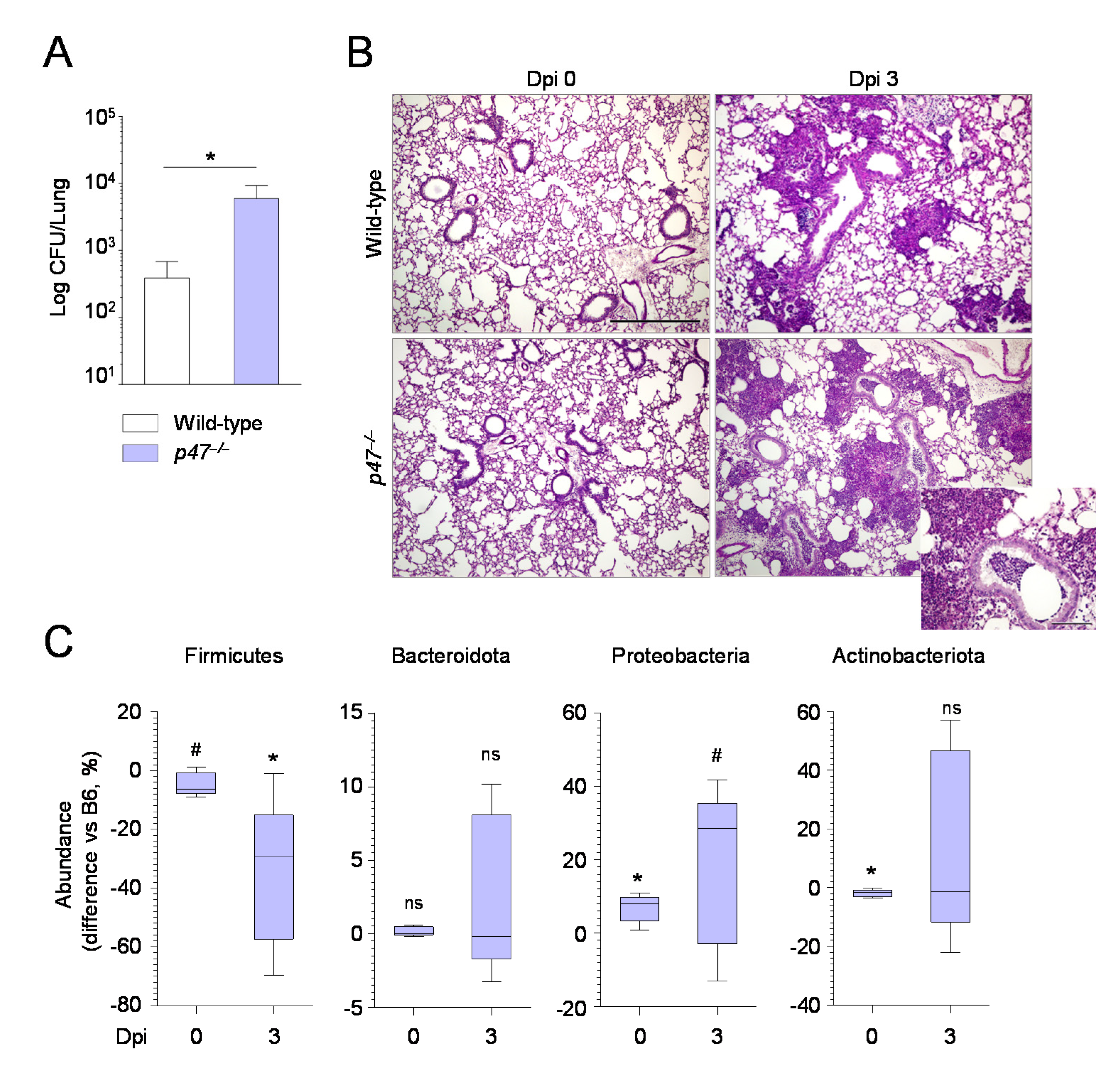
2.3. The Composition of the Lung Microbiome and the Levels of SCFAs Are Shaped by the Immune System
3. Discussion
4. Materials and Methods
4.1. In Vitro Co-Cultures
4.2. Mice, Infections and Treatments
4.3. Real-Time PCR
4.4. ELISA
4.5. Quantification of SCFA
4.6. Metagenomics
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pflughoeft, K.J.; Versalovic, J. Human microbiome in health and disease. Annu. Rev. Pathol. 2012, 7, 99–122. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Kayama, H.; Okumura, R.; Takeda, K. Interaction Between the Microbiota, Epithelia, and Immune Cells in the Intestine. Annu. Rev. Immunol 2020, 38, 23–48. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 2021, 18, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef] [Green Version]
- Yip, W.; Hughes, M.R.; Li, Y.; Cait, A.; Hirst, M.; Mohn, W.W.; McNagny, K.M. Butyrate Shapes Immune Cell Fate and Function in Allergic Asthma. Front. Immunol. 2021, 12, 628453. [Google Scholar] [CrossRef]
- Ostaff, M.J.; Stange, E.F.; Wehkamp, J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 2013, 5, 1465–1483. [Google Scholar] [CrossRef]
- Pabst, O.; Slack, E. IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 2020, 13, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Barnes, P.D.; Marr, K.A. Aspergillosis: Spectrum of disease, diagnosis, and treatment. Infect. Dis. Clin. N. Am. 2006, 20, 545–561. [Google Scholar] [CrossRef]
- van de Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latge, J.P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef]
- Kolwijck, E.; van de Veerdonk, F.L. The potential impact of the pulmonary microbiome on immunopathogenesis of Aspergillus-related lung disease. Eur. J. Immunol. 2014, 44, 3156–3165. [Google Scholar] [CrossRef]
- Goncalves, S.M.; Lagrou, K.; Duarte-Oliveira, C.; Maertens, J.A.; Cunha, C.; Carvalho, A. The microbiome-metabolome crosstalk in the pathogenesis of respiratory fungal diseases. Virulence 2017, 8, 673–684. [Google Scholar] [CrossRef] [Green Version]
- Costantini, C.; Nunzi, E.; Spolzino, A.; Palmieri, M.; Renga, G.; Zelante, T.; Englmaier, L.; Coufalikova, K.; Spacil, Z.; Borghi, M.; et al. Pharyngeal microbial signatures are predictive of the risk of fungal pneumonia in hematologic patients. Infect. Immun. 2021, 19, 105. [Google Scholar] [CrossRef]
- Castellano-Gonzalez, G.; McGuire, H.M.; Luciani, F.; Clancy, L.E.; Li, Z.; Avdic, S.; Hughes, B.; Singh, M.; Fazekas de St Groth, B.; Renga, G.; et al. Rapidly expanded partially HLA DRB1-matched fungus-specific T cells mediate in vitro and in vivo antifungal activity. Blood Adv. 2020, 4, 3443–3456. [Google Scholar] [CrossRef] [PubMed]
- Marsland, B.J.; Gollwitzer, E.S. Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 2014, 14, 827–835. [Google Scholar] [CrossRef] [PubMed]
- de Luca, A.; Smeekens, S.P.; Casagrande, A.; Iannitti, R.; Conway, K.L.; Gresnigt, M.S.; Begun, J.; Plantinga, T.S.; Joosten, L.A.; van der Meer, J.W.; et al. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 3526–3531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Dwyer, D.N.; Zhou, X.; Wilke, C.A.; Xia, M.; Falkowski, N.R.; Norman, K.C.; Arnold, K.B.; Huffnagle, G.B.; Murray, S.; Erb-Downward, J.R.; et al. Lung Dysbiosis, Inflammation, and Injury in Hematopoietic Cell Transplantation. Am. J. Respir. Crit. Care Med. 2018, 198, 1312–1321. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Suez, J.; Elinav, E. The interplay between the innate immune system and the microbiota. Curr. Opin. Immunol. 2014, 26, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sparks, J.B.; Karyala, S.V.; Settlage, R.; Luo, X.M. Host adaptive immunity alters gut microbiota. ISME J. 2015, 9, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Man, S.M. Inflammasomes in the gastrointestinal tract: Infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 721–737. [Google Scholar] [CrossRef]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iannitti, R.G.; Napolioni, V.; Oikonomou, V.; De Luca, A.; Galosi, C.; Pariano, M.; Massi-Benedetti, C.; Borghi, M.; Puccetti, M.; Lucidi, V.; et al. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nat. Commun. 2016, 7, 10791. [Google Scholar] [CrossRef]
- Nogueira, M.F.; Pereira, L.; Jenull, S.; Kuchler, K.; Lion, T. Klebsiella pneumoniae prevents spore germination and hyphal development of Aspergillus species. Sci. Rep. 2019, 9, 218. [Google Scholar] [CrossRef] [Green Version]
- Yildiz, S.; Pereira Bonifacio Lopes, J.P.; Berge, M.; Gonzalez-Ruiz, V.; Baud, D.; Kloehn, J.; Boal-Carvalho, I.; Schaeren, O.P.; Schotsaert, M.; Hathaway, L.J.; et al. Respiratory tissue-associated commensal bacteria offer therapeutic potential against pneumococcal colonization. Elife 2020, 9, e53581. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Puccetti, M.; Xiroudaki, S.; Ricci, M.; Giovagnoli, S. Postbiotic-Enabled Targeting of the Host-Microbiota-Pathogen Interface: Hints of Antibiotic Decline? Pharmaceutics 2020, 12, 624. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021, 2, 7. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, S.; Mande, S.S. Diet, Microbiota and Gut-Lung Connection. Front. Microbiol. 2018, 9, 2147. [Google Scholar] [CrossRef] [PubMed]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, A.; Iannitti, R.G.; Bozza, S.; Beau, R.; Casagrande, A.; D’Angelo, C.; Moretti, S.; Cunha, C.; Giovannini, G.; Massi-Benedetti, C.; et al. CD4(+) T cell vaccination overcomes defective cross-presentation of fungal antigens in a mouse model of chronic granulomatous disease. J. Clin. Investig. 2012, 122, 1816–1831. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Reali, G.; Femminella, M.; Nunzi, E.; Valocchi, D. Genomics as a service: A joint computing and networking perspective. Comput. Netw. 2018, 145, 27–51. [Google Scholar] [CrossRef] [Green Version]
- Femminella, M.; Pergolesi, M.; Reali, G. Performance Evaluation of Edge Cloud Computing System for Big Data Applications. In Proceedings of the 2016 5th IEEE International Conference on Cloud Networking (Cloudnet), Pisa, Italy, 3–5 October 2016; pp. 170–175. [Google Scholar]
- Felicetti, L.; Femminella, M.; Reali, G.; Liò, P. A Molecular Communication System in Blood Vessels for Tumor Detection. In Proceedings of the ACM The First Annual International Conference on Nanoscale Computing and Communication, Atlanta, GA, USA, 6–9 May 2014; p. 21. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P.; Minchin, P.R.; Belbin, L. Compositional dissimilarity as a robust measure of ecological distance. Vegetatio 1987, 69, 57–68. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbo, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunzi, E.; Renga, G.; Palmieri, M.; Pieraccini, G.; Pariano, M.; Stincardini, C.; D’Onofrio, F.; Santarelli, I.; Bellet, M.M.; Bartoli, A.; et al. A Shifted Composition of the Lung Microbiota Conditions the Antifungal Response of Immunodeficient Mice. Int. J. Mol. Sci. 2021, 22, 8474. https://doi.org/10.3390/ijms22168474
Nunzi E, Renga G, Palmieri M, Pieraccini G, Pariano M, Stincardini C, D’Onofrio F, Santarelli I, Bellet MM, Bartoli A, et al. A Shifted Composition of the Lung Microbiota Conditions the Antifungal Response of Immunodeficient Mice. International Journal of Molecular Sciences. 2021; 22(16):8474. https://doi.org/10.3390/ijms22168474
Chicago/Turabian StyleNunzi, Emilia, Giorgia Renga, Melissa Palmieri, Giuseppe Pieraccini, Marilena Pariano, Claudia Stincardini, Fiorella D’Onofrio, Ilaria Santarelli, Marina Maria Bellet, Andrea Bartoli, and et al. 2021. "A Shifted Composition of the Lung Microbiota Conditions the Antifungal Response of Immunodeficient Mice" International Journal of Molecular Sciences 22, no. 16: 8474. https://doi.org/10.3390/ijms22168474
APA StyleNunzi, E., Renga, G., Palmieri, M., Pieraccini, G., Pariano, M., Stincardini, C., D’Onofrio, F., Santarelli, I., Bellet, M. M., Bartoli, A., Costantini, C., & Romani, L. (2021). A Shifted Composition of the Lung Microbiota Conditions the Antifungal Response of Immunodeficient Mice. International Journal of Molecular Sciences, 22(16), 8474. https://doi.org/10.3390/ijms22168474





