NGS Analysis Confirms Common TP53 and RB1 Mutations, and Suggests MYC Amplification in Ocular Adnexal Sebaceous Carcinomas
Abstract
1. Introduction
2. Results
2.1. Clinicopathologic Characteristics
2.2. Mutations in Sebaceous Carcinoma
2.3. Copy Number Variations in Sebaceous Carcinoma
2.4. MYC Fluorescence In Situ Hybridization (FISH)
2.5. MYC Immunohistochemistry
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. DNA Extraction and Next-Generation Sequencing
4.3. MYC Fluorescence In Situ Hybridization
4.4. MYC Immunohistochemistry
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Orr, C.; Yazdanie, F.; Shinder, R. Current review of sebaceous cell carcinoma. Curr. Opin. Ophthalmol. 2018, 29, 445–450. [Google Scholar] [CrossRef]
- Dasgupta, T.; Wilson, L.; Yu, J. A retrospective review of 1349 cases of sebaceous carcinoma. Cancer 2009, 115, 158–165. [Google Scholar] [CrossRef]
- Shields, J.; Demirci, H.; Marr, B.; Eagle, R., Jr.; Shields, C. Sebaceous carcinoma of the eyelids: Personal experience with 60 cases. Ophthalmology 2004, 111, 2151–2157. [Google Scholar] [CrossRef]
- Shields, J.; Saktanasate, J.; Lally, S.; Carrasco, J.; Shields, C. Sebaceous carcinoma of the ocular region: The 2014 professor Winifred Mao lecture. Asia Pac. J. Ophthalmol. 2015, 4, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Killian, J.; Baum, C.; Otley, C.; Roenigk, R.; Arpey, C.; Weaver, A.; Brewer, J. Characteristics of sebaceous carcinoma and early outcomes of treatment using Mohs micrographic surgery versus wide local excision: An update of the Mayo clinic experience over the past 2 decades. Derm. Surg. 2014, 40, 241–246. [Google Scholar] [CrossRef]
- Stagner, A.; Jakobiec, F. Updates on the molecular pathology of selected ocular and ocular adnexal tumors: Potential targets for future therapy. Semin. Ophthalmol. 2016, 31, 188–196. [Google Scholar] [CrossRef]
- Shalin, S.C.; Lyle, S.; Calonje, E.; Lazar, A.J.F. Sebaceous neoplasia and the Muir-Torre syndrome: Important connections with clinical implications. Histopathology 2010, 56, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Sa, H.-S.; Rubin, M.L.; Xu, S.; Ning, J.; Tetzlaff, M.; Sagiv, O.; Kandl, T.J.; Esmaeli, B. Prognostic factors for local recurrence, metastasis and survival for sebaceous carcinoma of the eyelid: Observations in 100 patients. Br. J. Ophthalmol. 2019, 103, 980. [Google Scholar] [CrossRef]
- Tripathi, R.; Chen, Z.; Li, L.; Bordeaux, J.S. Incidence and survival of sebaceous carcinoma in the United States. J. Am. Acad. Derm. 2016, 75, 1210–1215. [Google Scholar] [CrossRef]
- Bell, W.; Singh, K.; Rajan, A.; Eberhart, C. Expression of P16 and P53 in intraepithelial periocular sebaceous carcinoma. Ocul. Oncol. Pathol. 2016, 2, 71–75. [Google Scholar] [CrossRef]
- Tetzlaff, M.; Curry, J.; Ning, J.; Sagiv, O.; Kandi, T.; Peng, B.; Bell, D.; Routbort, M.; Hudgens, C.; Ivan, D.; et al. Distinct biological types of ocular adnexal sebaceous carcinoma: HPV-driven and virus-negative tumors arise through nonoverlapping molecular-genetic alterations. Clin. Cancer Res. 2019, 15, 1280–1290. [Google Scholar] [CrossRef] [PubMed]
- Kiyosaki, K.; Nakada, C.; Hijiya, N.; Tsukamoto, Y.; Matsuura, K.; Nakatsuka, K.; Daa, T.; Yokoyama, S.; Imaizumi, M.; Moriyama, M. Analysis of P53 mutations and the expression of P53 and P21WAF1/CIP1protein in 15 cases of sebaceous carcinoma of the eyelid. Investig. Opthalmol. Vis. Sci. 2010, 51, 7. [Google Scholar] [CrossRef]
- Singh, G.; Weinstock, B.; Wu, A. Clinical and immunohistochemical features of sebaceous carcinoma: Focusing on the P53 tumor suppressor. J. Clin. Exp. Ophthalmol. 2017, 8, 1. [Google Scholar] [CrossRef]
- Cabral, E.S.; Auerbach, A.; Killian, J.K.; Barrett, T.L.; Cassarino, D.S. Distinction of benign sebaceous proliferations from sebaceous carcinomas by immunohistochemistry. Am. J. Dermatol. 2006, 28, 465–471. [Google Scholar] [CrossRef]
- Ali-Ridha, A.N.; Brownstein, S.; Jiang, K.; Milman, T.; Burns, B.; Blanco, P.; Farmer, J. Immunohistochemical analysis of sebaceous cell carcinoma in comparison to both basal cell carcinoma and squamous cell carcinoma. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5432. [Google Scholar]
- Bao, Y.; Selfridge, J.E.; Wang, J.; Zhao, Y.; Cui, J.; Guda, K.; Wang, Z.; Zhu, Y. Mutations in TP53, ZNF750, and RB1 typify ocular sebaceous carcinoma. J. Genet. Genom. 2019, 46, 315–318. [Google Scholar] [CrossRef]
- Shalin, S.C.; Sakharpe, A.; Lyle, S.; Lev, D.; Calonje, E.; Lazar, A.J. P53 staining correlates with tumor type and location in sebaceous neoplasms. Am. J. Dermatopathol. 2012, 34, 129–138. [Google Scholar] [CrossRef]
- Gonzalez-Fernandez, F.; Kaltreider, S.A.; Patnaik, B.D.; Retief, J.D.; Bao, Y.; Newman, S.; Stoler, M.H.; Levine, P.A. Sebaceous carcinoma: Tumor progression through mutational inactivation of P53. Ophthalmology 1998, 105, 497–506. [Google Scholar] [CrossRef]
- Hussain, R.; Matthews, J.; Dubovy, S.; Thompson, J.; Wang, G. UV-independent P53 mutations in sebaceous carcinoma of the eyelid. Ophthalmic Plast. Reconstr. Surg. 2014, 30, 392–395. [Google Scholar] [CrossRef]
- Buitrago-Pérez, A.; Garaulet, G.; Vázquez-Carballo, A.; Paramio, J.M.; García-Escudero, R. Molecular signature of HPV-induced carcinogenesis: PRb, P53 and gene expression profiling. Curr. Genom. 2009, 10, 26–34. [Google Scholar] [CrossRef]
- Moore, R.; Zhang, X.; Allison, D.; Rooper, L.; Campbell, A.; Eberhart, C. High-risk human papillomavirus and ZEB1 in ocular adnexal sebaceous carcinoma. J. Cutan. Pathol. 2021, 48, 1027–1033. [Google Scholar] [CrossRef]
- Bladen, J.; Wang, J.; Sangaralingam, A.; Moosajee, M.; Fitchett, C.; Chelala, C.; Beaconsfield, M.; O’Toole, E.; Philpott, M.; Ezra, D. MicroRNA and transcriptome analysis in periocular sebaceous gland carcinoma. Sci. Rep. 2018, 8, 7531. [Google Scholar] [CrossRef]
- Mulay, K.; Aggarwal, E.; White, V. Periocular sebaceous gland carcinoma: A comprehensive review. Saudi J. Ophthalmol. 2013, 27, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Tetzlaff, M.; Singh, R.; Seviour, E.; Curry, J.; Hudgens, C.; Bell, D.; Wimmer, D.; Ning, J.; Czerniak, B.; Zhang, L.; et al. Next-generation sequencing identifies high frequency of mutations in potentially clinically actionable genes in sebaceous carcinoma. J. Pathol. 2016, 240, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Tomonari, M.; Shimada, M.; Nakada, Y.; Yamamoto, I.; Itoh, M.; Koike, Y.; Kobayashi, A.; Miki, J.; Yamada, H.; Kimura, T.; et al. Muir–Torre syndrome: Sebaceous carcinoma concurrent with colon cancer in a kidney transplant recipient. A case report. BMC Nephrol. 2019, 20, 394. [Google Scholar] [CrossRef]
- Marazza, G.; Masouyé, I.; Taylor, S.; Prins, C.; Gaudin, T.; Saurat, J.-H.; French, L.E. An illustrative case of Muir-Torre syndrome. Arch. Dermatol. 2006, 142, 1039–1042. [Google Scholar] [CrossRef]
- Hare, H.H.; Mahendraker, N.; Sarwate, S.; Tangella, K. Muir-Torre syndrome: A rare but important disorder. Cutis 2008, 82, 252–256. [Google Scholar]
- Chen, Q.; Wang, M.; Xu, Z.; Wang, M.; Jin, S.; Tian, S.; Xiao, S. Muir-Torre syndrome with a frame-shift mutation in the MSH2 gene: A rare case report and literature review. Int. J. Gynecol. Pathol. 2020, 39, 136–140. [Google Scholar] [CrossRef]
- Suspiro, A.; Fidalgo, P.; Cravo, M.; Albuquerque, C.; Ramalho, E.; Leitão, C.N.; Costa Mira, F. The Muir-Torre syndrome: A rare variant of hereditary nonpolyposis colorectal cancer associated with HMSH2 mutation. Am. J. Gastroenterol. 1998, 93, 1572–1574. [Google Scholar] [CrossRef]
- Perera, S.; Ramyar, L.; Mitri, A.; Pollett, A.; Gallinger, S.; Speevak, M.D.; Aronson, M.; Bapat, B. A novel complex mutation in MSH2 contributes to both Muir-Torre and Lynch syndrome. J. Hum. Genet. 2010, 55, 37–41. [Google Scholar] [CrossRef]
- Morales-Burgos, A.; Sánchez, J.L.; Figueroa, L.D.; De Jesús-Monge, W.E.; Cruz-Correa, M.R.; González-Keelan, C.; Nazario, C.M. MSH-2 and MLH-1 protein expression in Muir Torre syndrome-related and sporadic sebaceous neoplasms. P. R. Health Sci. J. 2008, 27, 322–327. [Google Scholar]
- Entius, M.M.; Keller, J.J.; Drillenburg, P.; Kuypers, K.C.; Giardiello, F.M.; Offerhaus, G.J. Microsatellite instability and expression of HMLH-1 and HMSH-2 in sebaceous gland carcinomas as markers for Muir-Torre syndrome. Clin. Cancer Res. 2000, 6, 1784–1789. [Google Scholar]
- North, J.; Golovato, J.; Vaske, C.; Sanborn, J.; Nguyen, A.; Wu, W.; Goode, B.; Stevers, M.; McMullen, K.; Perez White, B.; et al. Cell of origin and mutation pattern define three clinically distinct classes of sebaceous carcinoma. Nat. Commun. 2018, 9, 1894. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Johnson, A.; Holla, V.; Bailey, A.M.; Brusco, L.; Chen, K.; Routbort, M.; Patel, K.P.; Zeng, J.; Kopetz, S.; et al. A decision support framework for genomically informed investigational cancer therapy. J. Natl. Cancer Inst. 2015, 107, djv098. [Google Scholar] [CrossRef]
- Xu, S.; Moss, T.; Rubin, M.; Ning, J.; Eterovic, A.; Yu, H.; Jia, R.; Fan, X.; Tetzlaff, M.; Esmaeli, B. Whole-exome sequencing for ocular adnexal sebaceous carcinoma suggests PCDH15 as a novel mutation associated with metastasis. Mod. Pathol. 2020, 33, 1256–1263. [Google Scholar] [CrossRef]
- Gratton, R.; Tricarico, P.; Moltrasio, C.; Lima Estevão de Oliveira, A.; Brandão, L.; Marzano, A.; Zupin, L.; Crovella, S. Pleiotropic role of notch signaling in human skin diseases. Int. J. Mol. Sci. 2020, 21, 4214. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C.; Lowry, W.; Pasolli, H.; Fuchs, E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006, 20, 3022–3035. [Google Scholar] [CrossRef] [PubMed]
- Aubin-Houzelstein, G. Notch signaling and the developing hair follicle. Adv. Exp. Med. Biol. 2012, 727, 142–160. [Google Scholar] [CrossRef]
- Zhang, Y.; Lam, O.; Nguyen, M.-T.T.; Ng, G.; Pear, W.S.; Ai, W.; Wang, I.-J.; Kao, W.W.-Y.; Liu, C.-Y. Mastermind-like transcriptional co-activator-mediated notch signaling is indispensable for maintaining conjunctival epithelial identity. Development 2013, 140, 594. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-Y.; Kao, C.-H.; Lin, K.-C.; Kaartinen, V.; Yang, L.-T. Notch signaling regulates late-stage epidermal differentiation and maintains postnatal hair cycle homeostasis. PLoS ONE 2011, 6, e15842. [Google Scholar] [CrossRef]
- Pan, Y.; Lin, M.-H.; Tian, X.; Cheng, H.-T.; Gridley, T.; Shen, J.; Kopan, R. γ-Secretase functions through notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell 2004, 7, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wen, H.; Brayton, C.; Das, P.; Smithson, L.A.; Fauq, A.; Fan, X.; Crain, B.J.; Price, D.L.; Golde, T.E.; et al. Epidermal growth factor receptor and notch pathways participate in the tumor suppressor function of γ-secretase. J. Biol. Chem. 2007, 282, 32264–32273. [Google Scholar] [CrossRef]
- Kumar, A.; Kumari, N.; Nallabelli, N.; Prasad, R. Pathogenic and therapeutic role of H3K4 family of methylases and demethylases in cancers. Indian J. Clin. Biochem. 2019, 34, 123–132. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Lee, S.; Yang, Q.-H.; Lee, D.; Lee, S.-K.; Roeder, R.; Lee, J. A Tumor suppressive coactivator complex of P53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl. Acad. Sci. USA 2009, 106, 8513–8518. [Google Scholar] [CrossRef] [PubMed]
- Kudithipudi, S.; Jeltsch, A. Role of somatic cancer mutations in human protein lysine methyltransferases. Biochim. Biophys. Acta BBA Rev. Cancer 2014, 1846, 366–379. [Google Scholar] [CrossRef]
- Weirich, S.; Kudithipudi, S.; Kycia, I.; Jeltsch, A. Somatic cancer mutations in the MLL3-SET domain alter the catalytic properties of the enzyme. Clin. Epigenet. 2015, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-W.; Cheong, C.; Sohn, Y.-C.; Goo, Y.-H.; Oh, W.; Park, J.; Joe, S.; Kang, H.-S.; Kim, D.-K.; Kee, C.; et al. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: Implication for posterior lenticonus with cataract. Mol. Cell. Biol. 2002, 22, 8409–8414. [Google Scholar] [CrossRef][Green Version]
- Lin, D.-C.; Hao, J.-J.; Nagata, Y.; Xu, L.; Shang, L.; Meng, X.; Sato, Y.; Okuno, Y.; Varela, A.M.; Ding, L.-W.; et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 467–473. [Google Scholar] [CrossRef]
- Watt, F.M.; Frye, M.; Benitah, S.A. MYC in mammalian epidermis: How can an oncogene stimulate differentiation? Nat. Rev. Cancer 2008, 8, 234–242. [Google Scholar] [CrossRef]
- Bull, J.J.; Mïller-Röver, S.; Patel, S.V.; Chronnell, C.M.T.; McKay, I.A.; Philpott, M.P. Contrasting localization of c-Myc with other Myc superfamily transcription factors in the human hair follicle and during the hair growth cycle. J. Investig. Dermatol. 2001, 116, 617–622. [Google Scholar] [CrossRef]
- Coffey, R.J., Jr.; Bascom, C.C.; Sipes, N.J.; Graves-Deal, R.; Weissman, B.E.; Moses, H.L. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor beta. Mol. Cell. Biol. 1988, 8, 3088–3093. [Google Scholar] [CrossRef]
- Hashiro, M.; Matsumoto, K.; Okumura, H.; Hashimoto, K.; Yoshikawa, K. Growth inhibition of human keratinocytes by antisense C-Myc oligomer is not coupled to induction of differentiation. Biochem. Biophys. Res. Commun. 1991, 174, 287–292. [Google Scholar] [CrossRef]
- Gandarillas, A.; Watt, F.M. C-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 1997, 11, 2869–2882. [Google Scholar] [CrossRef]
- Pelengaris, S.; Littlewood, T.; Khan, M.; Elia, G.; Evan, G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell 1999, 3, 565–577. [Google Scholar] [CrossRef]
- Waikel, R.; Wang, X.; Roop, D. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene 1999, 18, 4870–4878. [Google Scholar] [CrossRef][Green Version]
- Cottle, D.; Kretzschmar, K.; Schweiger, P.; Quist, S.; Gollnick, H.; Natsuga, K.; Aoyagi, S.; Watt, F. C-MYC-induced sebaceous gland differentiation is controlled by an androgen receptor/P53 axis. Cell Rep. 2013, 3, 427–441. [Google Scholar] [CrossRef]
- Horsley, V.; O’Carroll, D.; Tooze, R.; Ohinata, Y.; Saitou, M.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; Fuchs, E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 2006, 126, 597–609. [Google Scholar] [CrossRef]
- Fuchs, E. Skin stem cells: Rising to the surface. J. Cell Biol. 2008, 180, 273–284. [Google Scholar] [CrossRef]
- Feldman, A.; Mukha, D.; Maor, I.; Sedov, E.; Koren, E.; Yosefzon, Y.; Shlomi, T.; Fuchs, Y. Blimp1+ cells generate functional mouse sebaceous gland organoids in vitro. Nat. Commun. 2019, 10, 2348. [Google Scholar] [CrossRef]
- Boukamp, P. Non-melanoma skin cancer: What drives tumor development and progression? Carcinogenesis 2005, 26, 1657–1667. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, J.; Choung, H.-K.; Lee, M.; Khwarg, S. Expression of Shh and Wnt signaling pathway proteins in eyelid sebaceous gland carcinoma: Clinicopathologic study. Investig. Ophthalmol. Vis. Sci. 2013, 54, 370–377. [Google Scholar] [CrossRef]
- Martín-Subero, J.; Odero, M.; Hernandez, R.; Cigudosa, J.; Agirre, X.; Saez, B.; Sanz-García, E.; Ardanaz, M.; Novo, F.; Gascoyne, R.; et al. Amplification of IGH/MYC fusion in clinically aggressive IGH/BCL2-positive germinal center B-cell lymphomas. Genes Chromosom. Cancer 2005, 43, 414–423. [Google Scholar] [CrossRef]
- Rao, N.A.; Hidayat, L.C.A.A.; McLean, L.C.I.W.; Zimmerman, L.E. Sebaceous carcinomas of the ocular adnexa: A clinicopathologic study of 104 cases, with five-year follow-up data. Hum. Pathol. 1982, 13, 113–122. [Google Scholar] [CrossRef]
- Arnold, I.; Watt, F.M. C-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 2001, 11, 558–568. [Google Scholar] [CrossRef]
- Braun, K.M. Manipulation of stem cell proliferation and lineage commitment: Visualisation of label-retaining cells in wholemounts of mouse epidermis. Development 2003, 130, 5241–5255. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.; Pelengaris, S.; Hendrix, S.; Chronnell, C.; Khan, M.; Philpott, M. Ectopic expression of C-Myc in the skin affects the hair growth cycle and causes an enlargement of the sebaceous gland. Br. J. Dermatol. 2005, 152, 1125–1133. [Google Scholar] [CrossRef]
- Frye, M. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development 2003, 130, 2793–2808. [Google Scholar] [CrossRef]
- Ehrmann, C.; Schneider, M.R. Genetically modified laboratory mice with sebaceous glands abnormalities. Cell. Mol. Life Sci. 2016, 73, 4623–4642. [Google Scholar] [CrossRef]
- Ni, Q.; Zhao, J.; Gao, Y.; Qin, D.; Chen, X.; Ainiwaer, X. Prediction of potential drugs and targets based on meibomian gland dysfunction module classification to guide individualized treatment. J. Cell. Biochem. 2019, 120, 14813–14821. [Google Scholar] [CrossRef]
- Lee, L. Novel Gene Biomarkers Associated with Meibomian Gland Dysfunction. Ph.D. Thesis, University of New South Wales, Sydney, Australia, 2016. [Google Scholar]
- Tawfik, H.A.; Abdulhafez, M.H.; Fouad, Y.A.; Dutton, J.J. Embryologic and fetal development of the human eyelid. Ophthalmic Plast. Reconstr. Surg. 2016, 32, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, G.K.; Mutton, L.N.; Khalili, M.; McMullin, R.P.; Hicks, J.L.; Bianchi-Frias, D.; Horn, L.A.; Kulac, I.; Moubarek, M.S.; Nelson, P.S.; et al. Combined MYC activation and Pten loss are sufficient to create genomic instability and lethal metastatic prostate cancer. Cancer Res. 2016, 76, 283. [Google Scholar] [CrossRef] [PubMed]
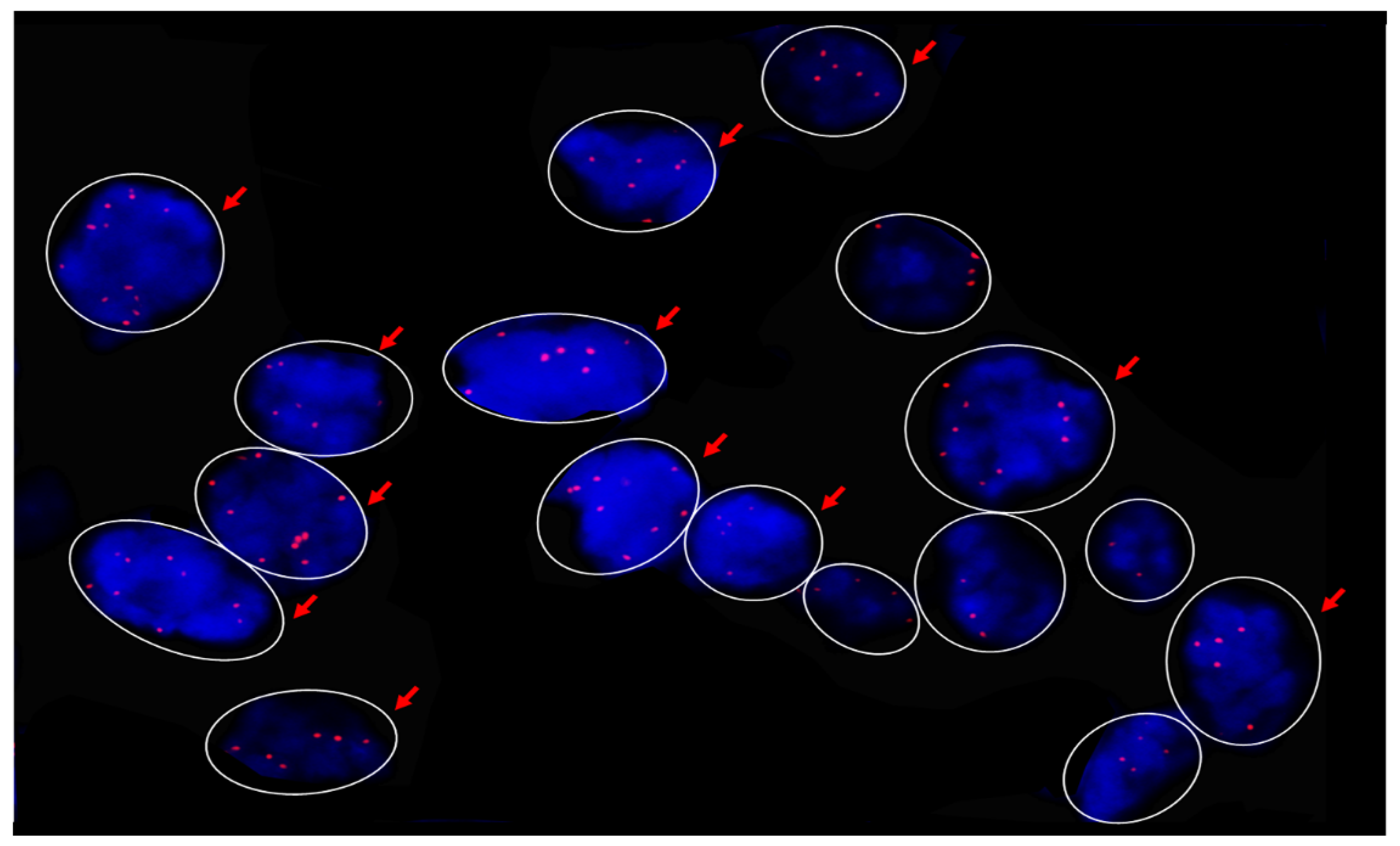
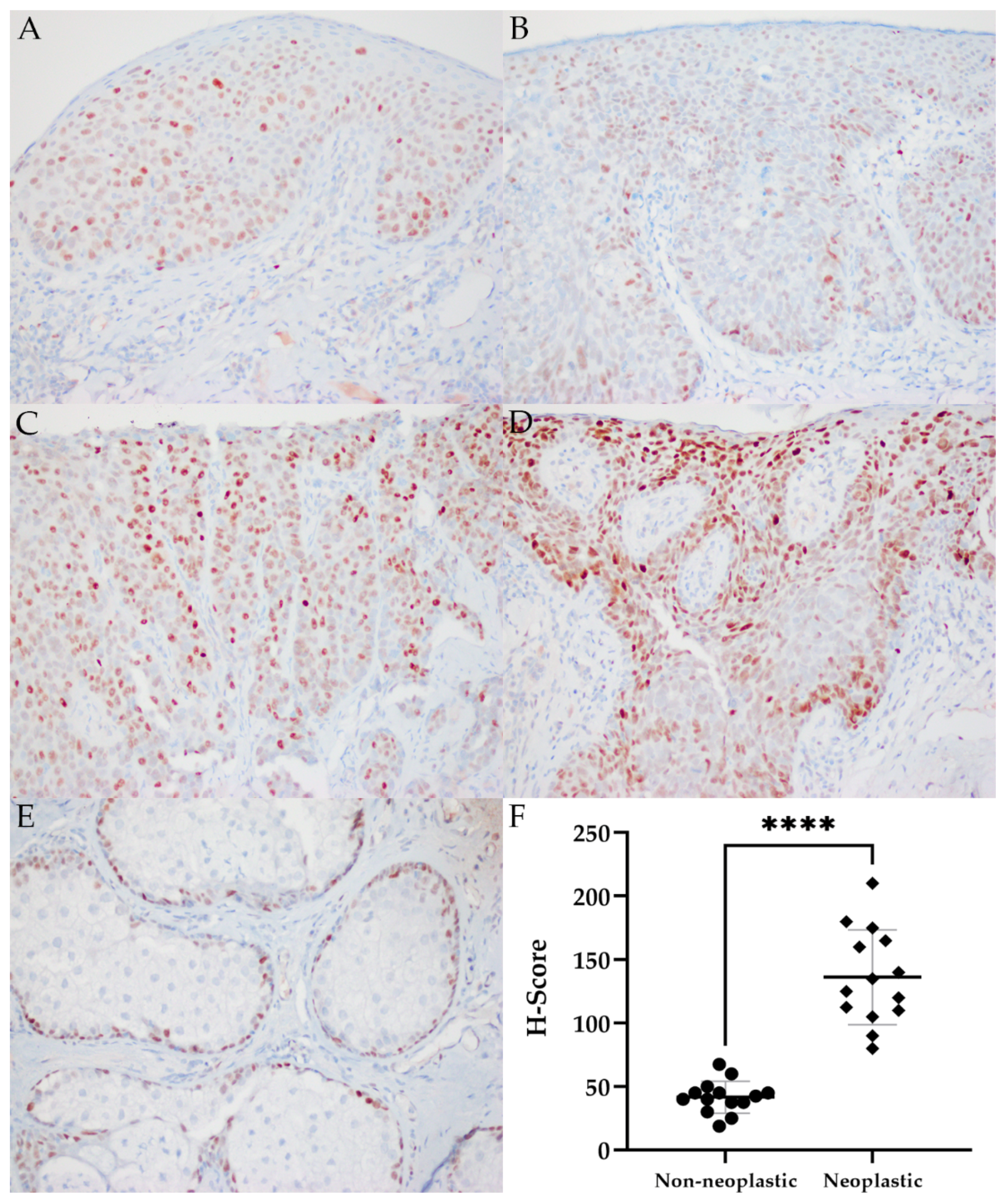

| Case Number | Sex | Age (years) | Tumor Type | Tumor Location | Tumor Laterality | Epithelial Involvement | T Stage | History of Malignancy |
|---|---|---|---|---|---|---|---|---|
| 1 | F | 54 | OA | Upper lid | Left | None | T1 | None |
| 2 | M | 77 | OA | Upper lid | Right | Intraepithelial | T1 | None |
| 3 | M | 83 | OA | Upper lid | Left | Combined | T1 | Non-cutaneous |
| 4 | F | 80 | OA | Upper lid | Right | Combined | T1 | None |
| 5 | M | 87 | OA | Upper lid | Left | Combined | T1 | None |
| 6 | F | 60 | OA | Upper lid | Right | Combined | T1 | None |
| 7 | F | 82 | OA | Upper lid | Left | Combined | T2 | Cutaneous |
| 8 | M | 83 | OA | Upper lid | Right | Subepithelial | T1 | Non-cutaneous |
| 9 | F | 90 | OA | Upper lid | Right | Subepithelial | T2 | None |
| 10 | F | 55 | OA | Lower lid | Right | Combined | T1 | Cutaneous |
| 11 | M | 43 | OA | Upper lid | Right | None | T1 | Non-cutaneous |
| 12 | F | 58 | EO | Lateral breast | Left | None | T1 | Multiple 1 |
| 13 | M | 87 | EO | Post-auricular neck | Left | None | T1 | Cutaneous |
| Case Number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutations in Clinically Actionable Genes | |||||||||||||
| ABL1 | p.E197K | ||||||||||||
| ATM | p.Q2433Pfs*11 | p.I2669Yfs*6 | |||||||||||
| BRCA1 | p.K1487I | p.R1726G | NE | ||||||||||
| BRCA2 | p.L2510_Y2511insKTCN | NE | |||||||||||
| EGFR | p.P772S | p.Q976Pfs*9 | |||||||||||
| FGFR2 | p.S702L | ||||||||||||
| FGFR3 | p.F384L | p.R728W | |||||||||||
| HRAS | p.P167Rfs*50, p.P167R | p.P167Rfs*50 | |||||||||||
| IDH2 | p.V335I | ||||||||||||
| MLH1 | p.E694X | ||||||||||||
| MSH2 | NE | p.Q409Rfs*4, p.Q409R, p.Q409H, p.I679T | |||||||||||
| MSH6 | p.V526L | NE | |||||||||||
| MTOR | p.L93Qfs*28 | p.P677S | NE | ||||||||||
| NF1 | p.K1915T | NE | p.I679Dfs*20, p.C1924Wfs*3 | ||||||||||
| NTRK1 | p.H467Q | NE | |||||||||||
| NTRK3 | p.R169C | p.Y604H | NE | ||||||||||
| PALB2 | p.S417Y | p.R1117Sfs*8 | NE | ||||||||||
| PDGFRA | p.E241X | ||||||||||||
| PIK3CA | p.V101fs*0 | ||||||||||||
| PMS2 | p.V397I | p.S418F | p.L236Sfs*3 | p.L236delinsYLLKKIM | NE | ||||||||
| POLD1 | NE | p.A242T | |||||||||||
| PTCH2 | NE | p.G1023S, p.E48del | |||||||||||
| PTEN | p.P281A | ||||||||||||
| RB1 | p.? (Unknown) | p.? (Unknown) | p.Q354Efs*5 | p.W75X, p.R358X | p.? (Unknown) | p.E30X | p.R500I | ||||||
| RET | p.A349V | ||||||||||||
| ROS1 | p.F1300L | p.D2344N | p.L2337F | NE | |||||||||
| SMO | p.L23_G24insL | ||||||||||||
| TERT | p.A279T | NE | |||||||||||
| TP53 | p.R273C | p.G266R | p.? (Unknown) | p.C277F | p.R273C | p.G245S | p.L257P | p.? (Unknown) | p.E339X | p.R196* | |||
| TSC1 | p.G274S | NE | |||||||||||
| TSC2 | p.G440S | NE | |||||||||||
| Genes with Frequent Mutations (≥ 25%) | |||||||||||||
| KMT2C | p.A1685S | p.S888T | NE | p.K2797Rfs*25 | p.K2797Rfs*25 | ||||||||
| MNX1 | p.A134_G135insAA | p.A174del | p.A134_G135insAA | p.A134_G135insAA | p.A134_G135insAA | p.A134_G135insAA | NE | ||||||
| NOTCH1 | p.G1320Afs*124 | p.Y550fs*0 | NE | p.R203C | p.L1531Cfs*48, p.P1443Afs*35 | ||||||||
| PCLO | p.G1750E | p.P2128Q | NE | p.E2925D | |||||||||
| PTPRT | p.R1209Q | p.R260W | NE | p.P1094Rfs*5 | |||||||||
| MYC Gain of Copy | |||||||||||||
| Mean log2 Ratio | <0.5 | NE | <0.5 | 0.833 | 0.8959 | 0.5483 | 0.8178 | 1.3315 | 1.4416 | <0.5 | NE | 0.7733 | 0.6051 |
| Variable | All Cases (n = 11) n (%) | No MYC Copy Gain (n = 3) n (%) | MYC Copy Gain (n = 8) n (%) | p Value |
|---|---|---|---|---|
| Sex | ||||
| Male Female | 4 (36) 7 (64) | 1 (33) 2 (67) | 3 (37) 5 (63) | 1.00 |
| Age (years), mean ± SD | 73 ± 13.8 | 64 ± 16.4 | 76 ± 12.3 | 0.53 |
| Tumor site Ocular adnexal Extraocular | 9 (82) 2 (18) | 3 (100) 0 (0) | 6 (75) 2 (25) | 1.00 |
| Tumor location (OA: n = 9) Upper lid Lower lid | 7 (78) 2 (22) | 2 (67) 1 (33) | 5 (83) 1 (17) | 1.00 |
| Tumor laterality (OA: n = 9) Right Left | 5 (56) 4 (44) | 1 (33) 2 (67) | 4 (67) 2 (33) | 0.52 |
| Variable | All Cases (n = 11) n (%) | No MYC Copy Gain (n = 3) n (%) | MYC Copy Gain (n = 8) n (%) | p Value |
|---|---|---|---|---|
| MYC expression (IHC) | ||||
| Low High | 5 (45) 6 (55) | 1 (33) 2 (67) | 4 (50) 4 (50) | 1.00 |
| Tumor size (mm), mean ± SD | 7.7 ± 2.6 | 6.5 ± 0.7 | 8.0 ± 2.9 | 0.50 |
| TP53 Mutation WT | 8 (73) 3 (27) | 2 (67) 1 (33) | 6 (75) 2 (25) | 1.00 |
| RB1 Mutation WT | 6 (55) 5 (45) | 2 (67) 1 (33) | 4 (50) 4 (50) | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peterson, C.; Moore, R.; Hicks, J.L.; Morsberger, L.A.; De Marzo, A.M.; Zou, Y.; Eberhart, C.G.; Campbell, A.A. NGS Analysis Confirms Common TP53 and RB1 Mutations, and Suggests MYC Amplification in Ocular Adnexal Sebaceous Carcinomas. Int. J. Mol. Sci. 2021, 22, 8454. https://doi.org/10.3390/ijms22168454
Peterson C, Moore R, Hicks JL, Morsberger LA, De Marzo AM, Zou Y, Eberhart CG, Campbell AA. NGS Analysis Confirms Common TP53 and RB1 Mutations, and Suggests MYC Amplification in Ocular Adnexal Sebaceous Carcinomas. International Journal of Molecular Sciences. 2021; 22(16):8454. https://doi.org/10.3390/ijms22168454
Chicago/Turabian StylePeterson, Cornelia, Robert Moore, Jessica L. Hicks, Laura A. Morsberger, Angelo M. De Marzo, Ying Zou, Charles G. Eberhart, and Ashley A. Campbell. 2021. "NGS Analysis Confirms Common TP53 and RB1 Mutations, and Suggests MYC Amplification in Ocular Adnexal Sebaceous Carcinomas" International Journal of Molecular Sciences 22, no. 16: 8454. https://doi.org/10.3390/ijms22168454
APA StylePeterson, C., Moore, R., Hicks, J. L., Morsberger, L. A., De Marzo, A. M., Zou, Y., Eberhart, C. G., & Campbell, A. A. (2021). NGS Analysis Confirms Common TP53 and RB1 Mutations, and Suggests MYC Amplification in Ocular Adnexal Sebaceous Carcinomas. International Journal of Molecular Sciences, 22(16), 8454. https://doi.org/10.3390/ijms22168454






