Functional Characterization of Multiple Ehrlichia chaffeensis Sodium (Cation)/Proton Antiporter Genes Involved in the Bacterial pH Homeostasis
Abstract
1. Introduction
2. Results
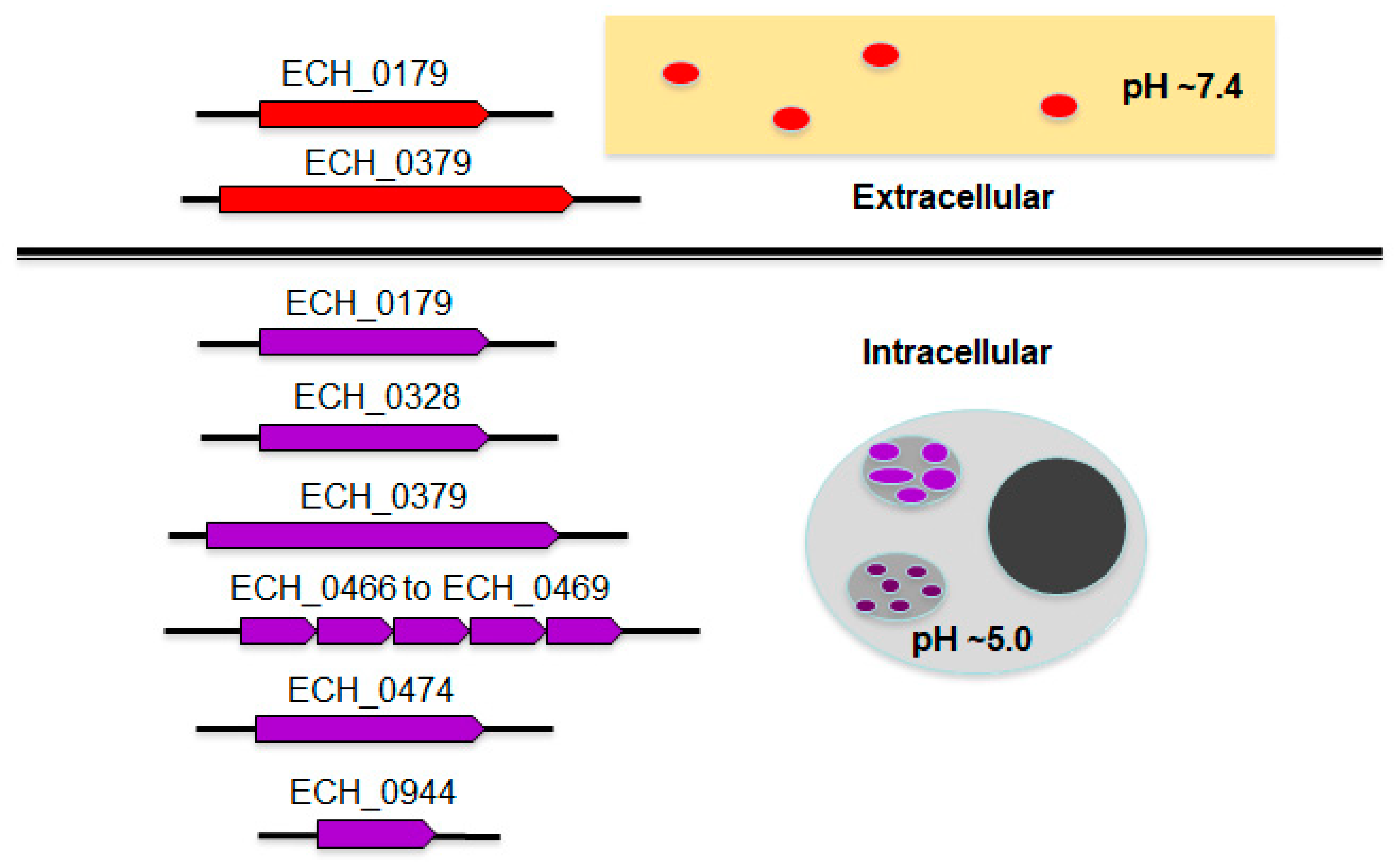
2.1. The E. chaffeensis Genome Has 10 Protein Coding Sequences Distributed across 6 Loci
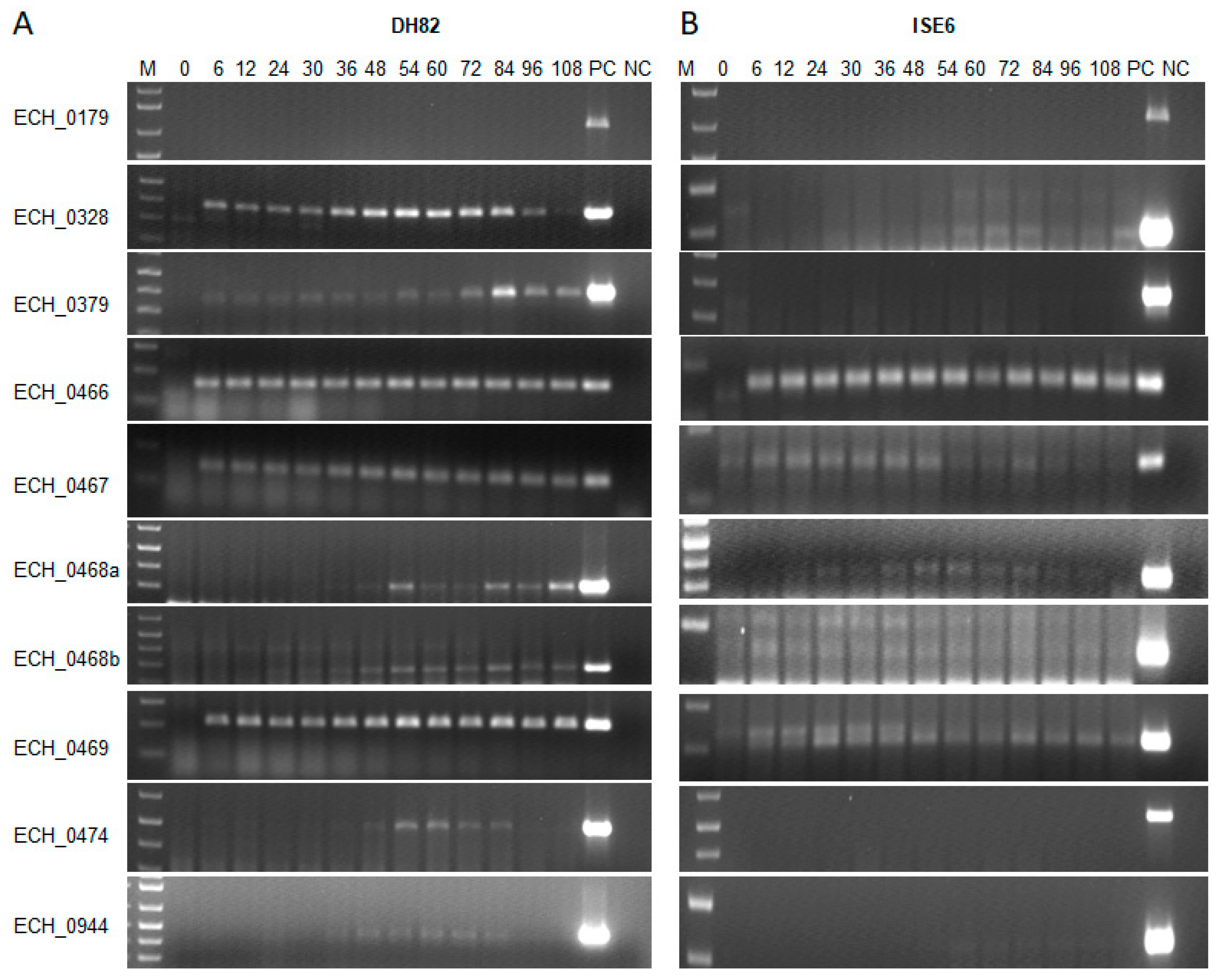
2.2. E. chaffeensis Antiporter Transcription Assessed In Vitro
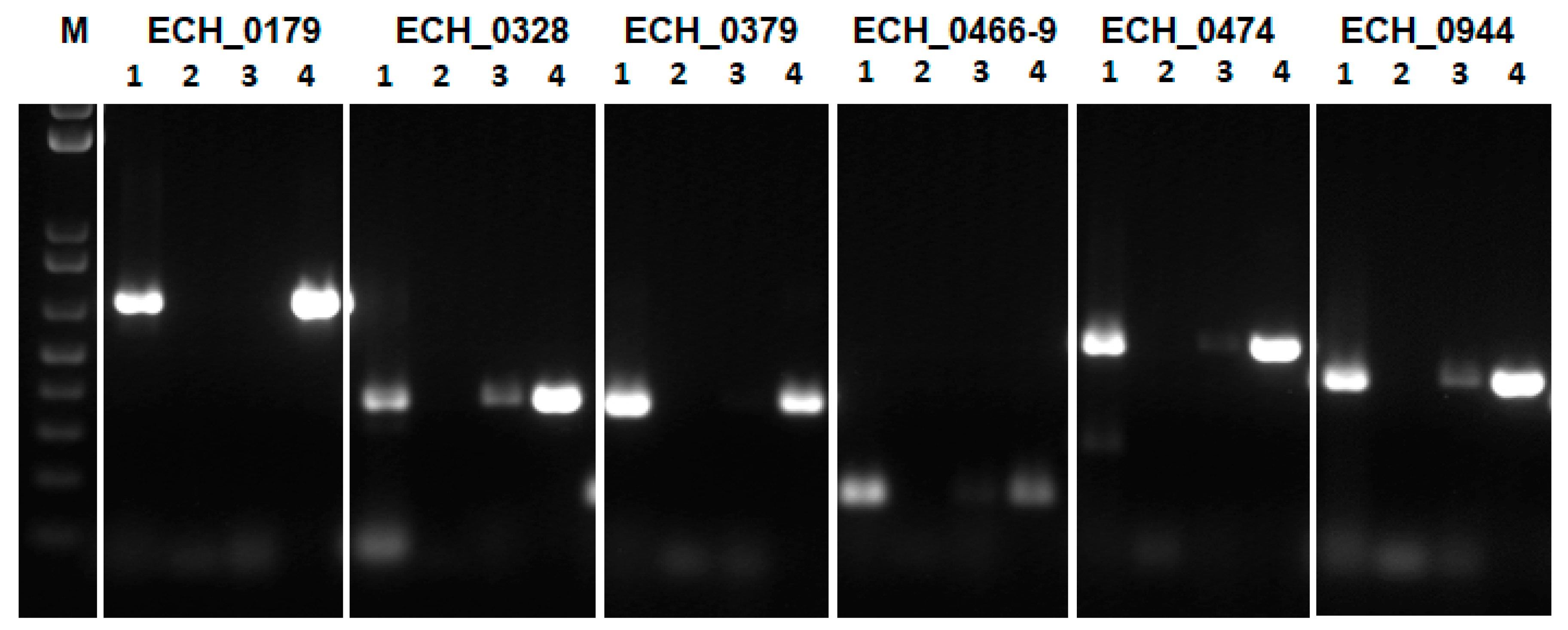
2.3. Transcription of E. chaffeensis Antiporter Genes Assessed in the Antiporter Function Deficient E. coli Mutant (EP432)
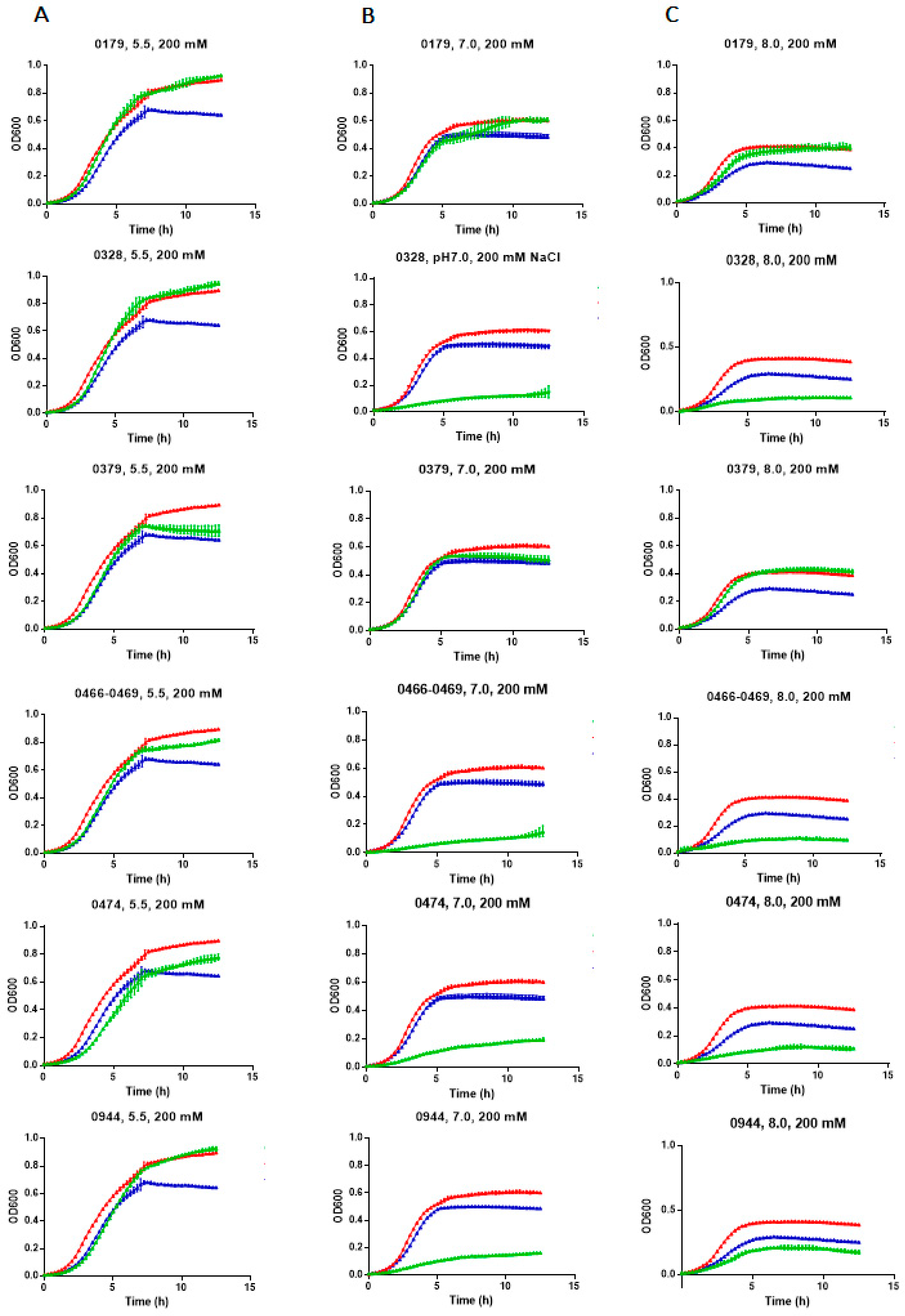
2.4. E. chaffeensis Antiporter Proteins Functionally Complement Antiporter Deficiency in E. coli
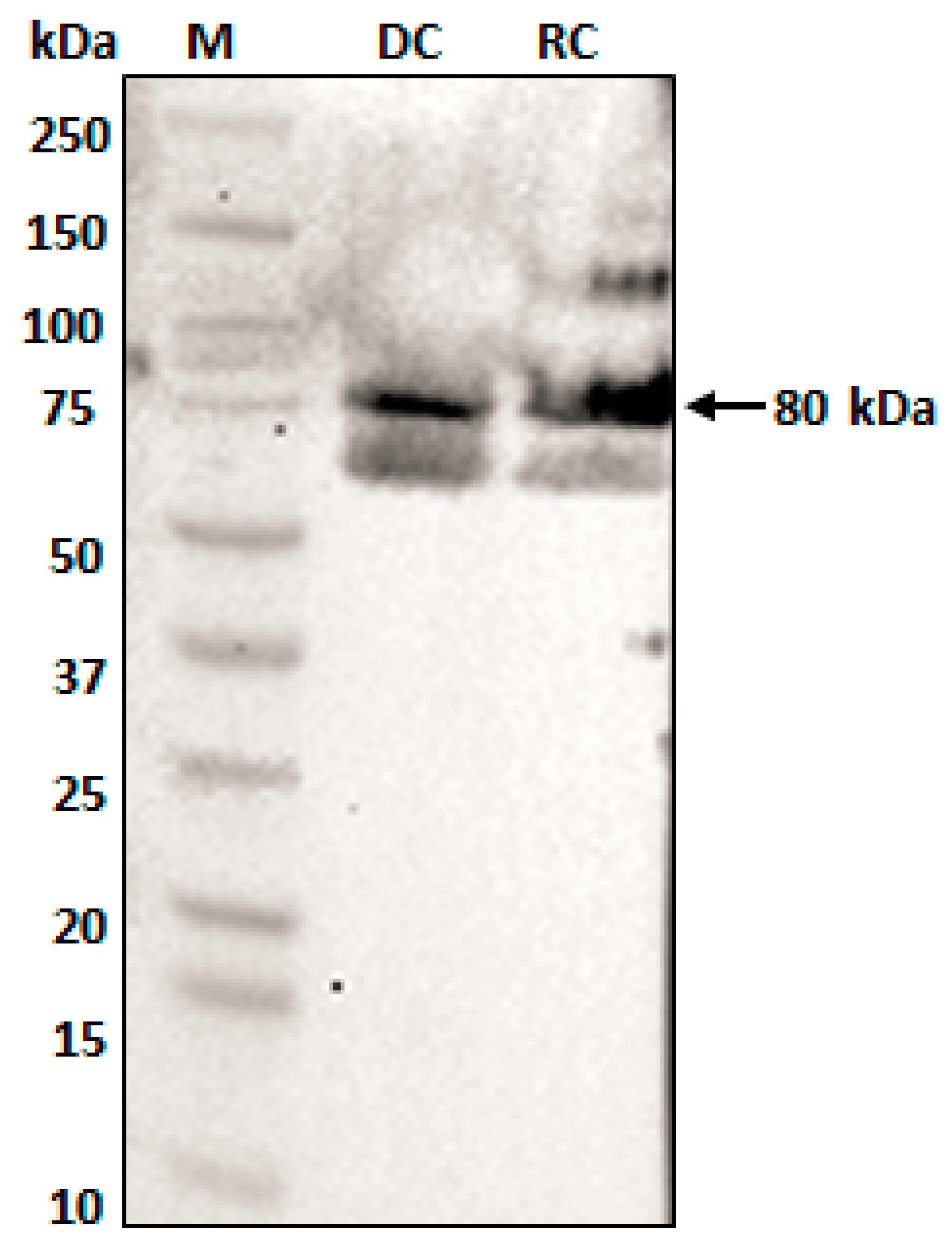
2.5. Native ECH_0379 Formed a Dimer and Expressed in Both Replicating (RC) and Infectious (DC) Forms and Similarly It Forms Dimers When Expressed as a Recombinant Protein
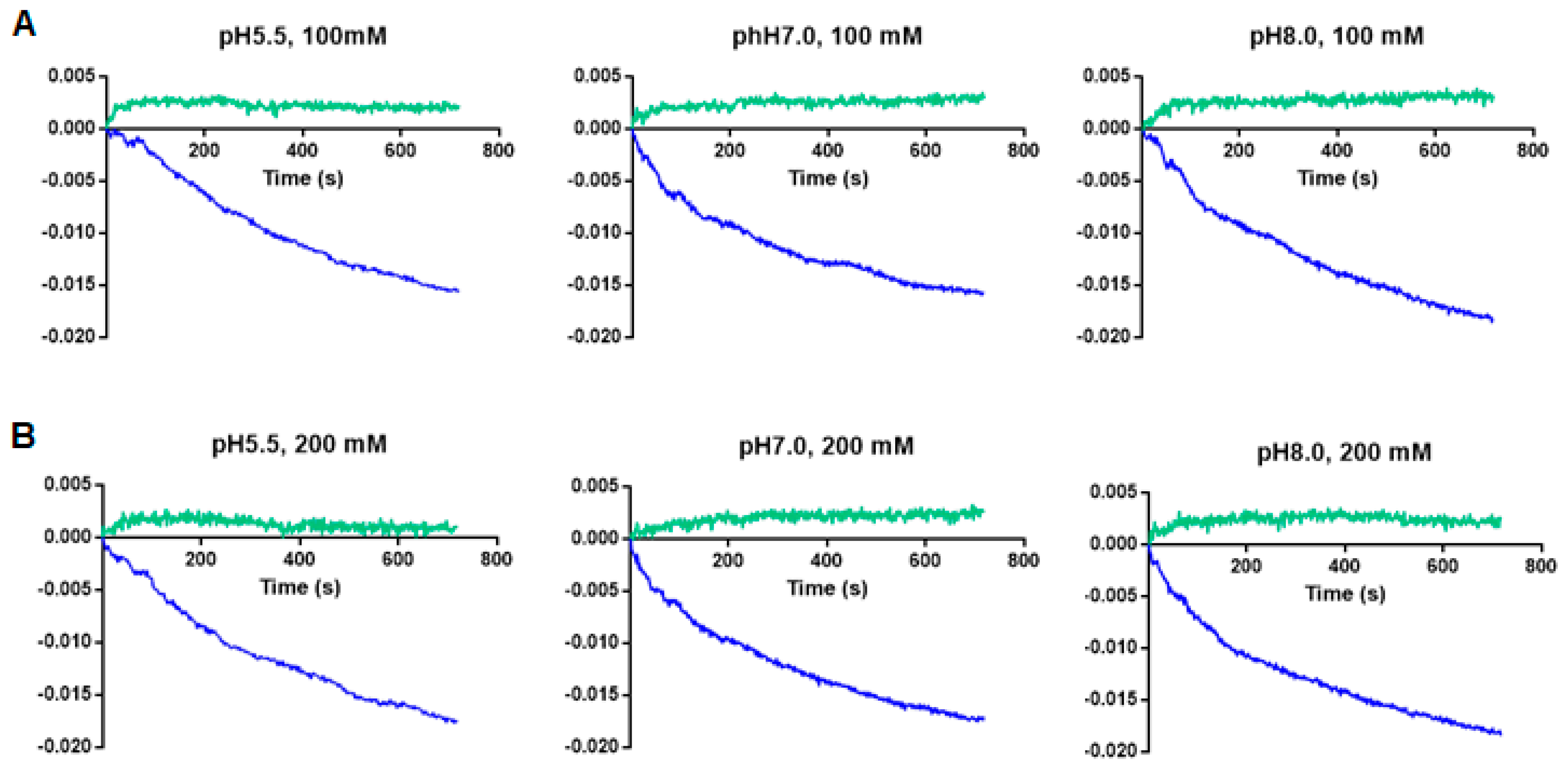
2.6. Proteoliposome Prepared with the Recombinant ECH_0379 Protein Assessed in Diffusion Analysis
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. Cultivation of E. chaffeensis
4.3. RNA Isolation
4.4. Gene Expression Levels Determined by Semi-Quantitative PCR
4.5. Constructing the Recombinant Plasmids
4.6. Growth Complementation under NaCl Stress
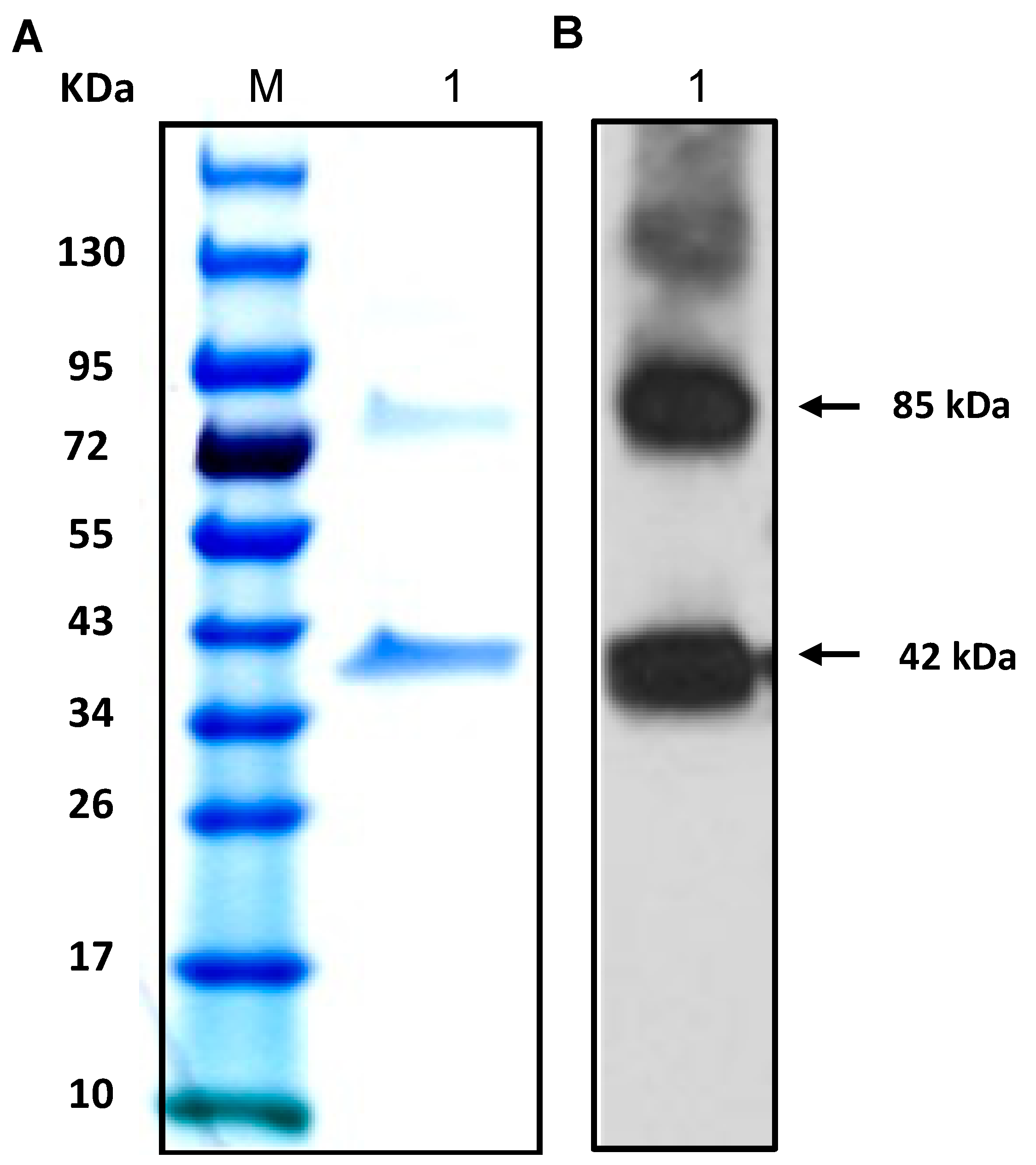
4.7. Overexpression and Purification of the Protein Encoded by ECH_0379
4.8. Western Blot Analysis to Detect ECH_0379 Protein Prepared in E. coli as Well as the Native Protein Expressed in RC and DC Purified Fractions of E. Chaffeensis
4.9. Proteoliposome Diffusion Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barnewall, R.E.; Rikihisa, Y.; Lee, E.H. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 1997, 65, 1455–1461. [Google Scholar] [CrossRef]
- Thomas, R.J.; Dumler, J.S.; Carlyon, J.A. Current management of human granulocytic anaplasmosis, human monocytic ehrlichiosis and Ehrlichia ewingii ehrlichiosis. Expert Rev. Anti Infect. Ther. 2009, 7, 709–722. [Google Scholar] [CrossRef]
- Rikihisa, Y. Molecular Pathogenesis of Ehrlichia chaffeensis Infection. Annu. Rev. Microbiol. 2015, 69, 283–304. [Google Scholar] [CrossRef]
- Ewing, S.A.; Dawson, J.E.; Kocan, A.A.; Barker, R.W.; Warner, C.K.; Panciera, R.J.; Fox, J.C.; Kocan, K.M.; Blouin, E.F. Experimental transmission of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) among white-tailed deer by Amblyomma americanum (Acari: Ixodidae). J. Med. Entomol. 1995, 32, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, J.M.; Davidson, W.R.; Stallknecht, D.E.; Dawson, J.E.; Howerth, E.W. Isolation of Ehrlichia chaffeensis from wild white-tailed deer (Odocoileus virginianus) confirms their role as natural reservoir hosts. J. Clin. Microbiol. 1997, 35, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Popov, V.L.; Gao, S.; Walker, D.H.; Yu, X.J. The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 2007, 9, 610–618. [Google Scholar] [CrossRef]
- Dedonder, S.E.; Cheng, C.; Willard, L.H.; Boyle, D.L.; Ganta, R.R. Transmission electron microscopy reveals distinct macrophage- and tick cell-specific morphological stages of Ehrlichia chaffeensis. PLoS ONE 2012, 7, e36749. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, J.E.; Sharma, S. Physiology, Alkalosis, Metabolic; StatPearls: Treasure Island, FL, USA, 2018. [Google Scholar]
- Padan, E.; Venturi, M.; Gerchman, Y.; Dover, N. Na(+)/H(+) antiporters. Biochim. Biophys. Acta 2001, 1505, 144–157. [Google Scholar] [CrossRef]
- Uria-Avellanal, C.; Robertson, N.J. Na(+)/H(+) exchangers and intracellular pH in perinatal brain injury. Transl. Stroke Res. 2014, 5, 79–98. [Google Scholar] [CrossRef]
- Cheng, C.; Nair, A.D.S.; Indukuri, V.V.; Gong, S.; Felsheim, R.F.; Jaworski, D.; Munderloh, U.G.; Ganta, R.R. Targeted and Random Mutagenesis of Ehrlichia chaffeensis for the Identification of Genes Required for in Vivo Infection. PLoS Pathog. 2013, 9, e1003171. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, L.; Liu, H.; Cheng, C.; Ganta, R.R. A genetic system for targeted mutations to disrupt and restore genes in the obligate bacterium, Ehrlichia chaffeensis. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Zhang, T.; Kedzierska-Mieszkowska, S.; Liu, H.; Cheng, C.; Ganta, R.R.; Zolkiewski, M. Aggregate-reactivation activity of the molecular chaperone ClpB from Ehrlichia chaffeensis. PLoS ONE 2013, 8, e62454. [Google Scholar] [CrossRef]
- Pinner, E.; Kotler, Y.; Padan, E.; Schuldiner, S. Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J. Biol. Chem. 1993, 268, 1729–1734. [Google Scholar] [CrossRef]
- Mesbah, N.M.; Cook, G.M.; Wiegel, J. The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na(K)/H antiporters. Mol. Microbiol. 2009, 74, 270–281. [Google Scholar] [CrossRef]
- Lentes, C.J.; Mir, S.H.; Boehm, M.; Ganea, C.; Fendler, K.; Hunte, C. Molecular characterization of the Na+/H+-antiporter NhaA from Salmonella Typhimurium. PLoS ONE 2014, 9, e101575. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dzioba-Winogrodzki, J.; Winogrodzki, O.; Krulwich, T.A.; Boin, M.A.; Häse, C.C.; Dibrov, P. The Vibrio cholerae Mrp system: Cation/proton antiport properties and enhancement of bile salt resistance in a heterologous host. J. Mol. Microbiol. Biotechnol. 2009, 16, 176–186. [Google Scholar] [CrossRef]
- Lee, C.; Yashiro, S.; Dotson, D.L.; Uzdavinys, P.; Iwata, S.; Sansom, M.S.P.; von Ballmoos, C.; Beckstein, O.; Drew, D.; Cameron, A.D. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J. Gen. Physiol. 2014, 144, 529–544. [Google Scholar] [CrossRef]
- Paulino, C.; Wöhlert, D.; Kapotova, E.; Yildiz, Ö.; Kühlbrandt, W. Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. Elife 2014, 26, e03583. [Google Scholar] [CrossRef]
- Masrati, G.; Dwivedi, M.; Rimon, A.; Gluck-Margolin, Y.; Kessel, A.; Ashkenazy, H.; Mayrose, I.; Padan, E.; Ben-Tal, N. Broad phylogenetic analysis of cation/proton antiporters reveals transport determinants. Nat. Commun. 2018, 9, 4205. [Google Scholar] [CrossRef]
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335. [Google Scholar] [CrossRef] [PubMed]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef]
- Fuster, D.G.; Alexander, R.T. Traditional and emerging roles for the SLC9 Na+/H+ exchangers. Pflüg. Arch. Eur. J. Physiol. 2014, 466, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Bakker, E.P.; Mangerich, W.E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J. Bacteriol. 1981, 147, 820–826. [Google Scholar] [CrossRef]
- Csonka, L.N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 1989, 53, 121–147. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Tokuda, H.; Unemoto, T. K+/H+ antiporter functions as a regulator of cytoplasmic pH in a marine bacterium, Vibrio alginolyticus. Biochim. Et Biophys. Acta BBA Biomembr. 1984, 776, 330–336. [Google Scholar] [CrossRef]
- Nakamura, T.; Kawasaki, S.; Unemoto, T. Roles of K+ and Na+ in pH homeostasis and growth of the marine bacterium Vibrio alginolyticus. J. Gen. Microbiol. 1992, 138, 1271–1276. [Google Scholar] [CrossRef]
- Zilberstein, D.; Agmon, V.; Schuldiner, S.; Padan, E. The sodium/proton antiporter is part of the pH homeostasis mechanism in Escherichia coli. J. Biol. Chem. 1982, 257, 3687–3691. [Google Scholar] [CrossRef]
- Corratgé-Faillie, C.; Jabnoune, M.; Zimmermann, S.; Véry, A.A.; Fizames, C.; Sentenac, H. Potassium and sodium transport in non-animal cells: The Trk/Ktr/HKT transporter family. Cell. Mol. Life Sci. 2010, 67, 2511–2532. [Google Scholar] [CrossRef]
- Dawson, J.E.; Biggie, K.L.; Warner, C.K.; Cookson, K.; Jenkins, S.; Levine, J.F.; Olson, J.G. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human monocytic ehrlichiosis, in dogs, from southeast Virginia. Am. J. Vet. Res. 1996, 57, 1175–1179. [Google Scholar] [PubMed]
- Paddock, C.D.; Childs, J.E. Ehrlichia chaffeensis: A prototypical emerging pathogen. Clin. Microbiol. Rev. 2003, 16, 37–64. [Google Scholar] [CrossRef]
- Cheng, C.; Nair, A.D.S.; Jaworski, D.C.; Ganta, R.R. Mutations in Ehrlichia chaffeensis Causing Polar Effects in Gene Expression and Differential Host Specificities. PLoS ONE 2015, 10, e0132657. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Häse, C.C.; Barquera, B. Role of sodium bioenergetics in Vibrio cholerae. Biochim. Biophys. Acta BBA Bioenerg. 2001, 1505, 169–178. [Google Scholar] [CrossRef]
- Miller, C.J.; Drasar, B.S.; Feachem, R.G. Response of toxigenic Vibrio cholerae 01 to physico-chemical stresses in aquatic environments. J. Hyg. 1984, 93, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Vimont, S.; Berche, P. NhaA, an Na(+)/H(+) antiporter involved in environmental survival of Vibrio cholerae. J. Bacterial. 2000, 182, 2937–2944. [Google Scholar] [CrossRef]
- Orlowski, J.; Grinstein, S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflug. Arch. 2004, 447, 549–565. [Google Scholar] [CrossRef]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta BBA Biomembr. 2005, 1717, 67–88. [Google Scholar] [CrossRef]
- Biggs, H.M.; Behravesh, C.B.; Bradley, K.K.; Dahlgren, F.S.; Drexler, N.A.; Dumler, J.S.; Folk, S.M.; Kato, C.Y.; Lash, R.R.; Levin, M.M.; et al. Diagnosis and Management of Tickborne Rickettsial Diseases: Rocky Mountain Spotted Fever and Other Spotted Fever Group Rickettsioses, Ehrlichioses, and Anaplasmosis—United States. MMWR Recomm. Rep. 2016, 65, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Koza, D.J.; Nsiah, Y.A. Synthesis and biological evaluation of 9-substituted tetracycline derivatives. Bioorganic Med. Chem. Lett. 2002, 12, 2163–2165. [Google Scholar] [CrossRef]
- Padan, E. The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem Sci. 2008, 33, 435–443. [Google Scholar] [CrossRef]
- Drew, D.; Boudker, O. Shared Molecular Mechanisms of Membrane Transporters. Annu. Rev. Biochem. 2016, 85, 543–572. [Google Scholar] [CrossRef]
- Munderloh, U.G.; Jauron, S.D.; Fingerle, V.; Leitritz, L.; Hayes, S.F.; Hautman, J.M.; Nelson, C.M.; Huberty, B.W.; Kurtti, T.J.; Ahlstrand, G.G.; et al. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J. Clin. Microbiol. 1999, 37, 2518–2524. [Google Scholar] [CrossRef]
- Sirigireddy, K.R.; Ganta, R.R. Multiplex detection of Ehrlichia and Anaplasma species pathogens in peripheral blood by real-time reverse transcriptase-polymerase chain reaction. J. Mol. Diagn. 2005, 7, 308–316. [Google Scholar] [CrossRef]
- Eedunuri, V.K.; Zhang, Y.; Cheng, C.; Chen, L.; Liu, H.; Omsland, A.; Boyle, D.; Ganta, R.R. Protein and DNA synthesis demonstrated in cell-free Ehrlichia chaffeensis organisms in axenic medium. Sci. Rep. 2018, 8, 1–12. [Google Scholar]
- Nikaido, H.; Rosenberg, E.Y. Porin channels in Escherichia coli: Studies with liposomes reconstituted from purified proteins. J. Bacteriol. 1983, 153, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H.; Nikaido, K.; Harayama, S. Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem. 1991, 266, 770–779. [Google Scholar] [CrossRef]
- Huang, H.; Wang, X.; Kikuchi, T.; Kumagai, Y.; Rikihisa, Y. Porin activity of Anaplasma phagocytophilum outer membrane fraction and purified P44. J. Bacteriol. 2007, 189, 1998–2006. [Google Scholar] [CrossRef][Green Version]
- Kumagai, Y.; Huang, H.; Rikihisa, Y. Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. J. Bacteriol. 2008, 190, 3597–3605. [Google Scholar] [CrossRef]
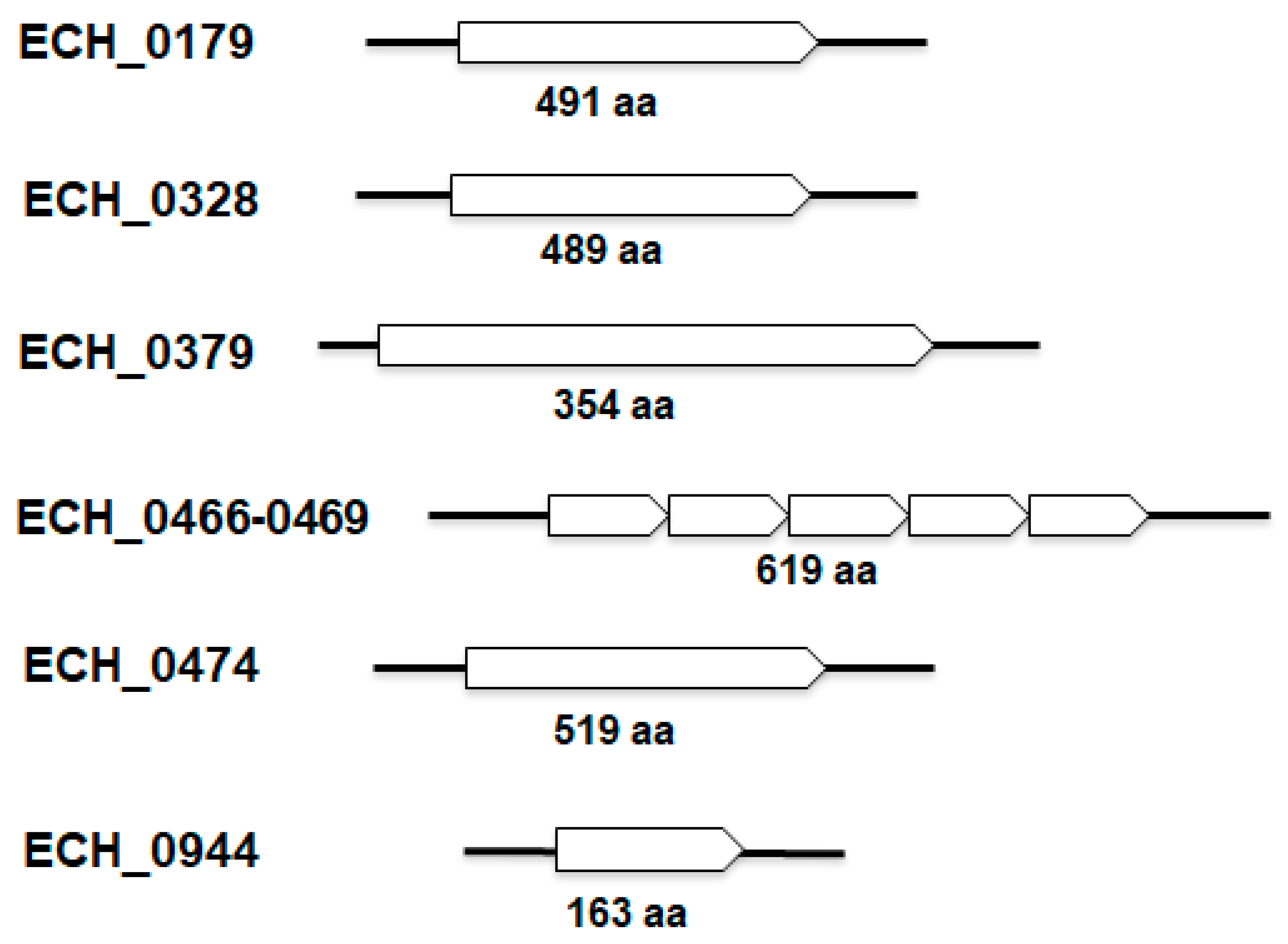
| Old_Locus_Tag * | New_locus_Tag $ | Length of AA # | Function Annotation |
|---|---|---|---|
| ECH_0179 | ECH_RS00755 | 491 | cation/proton antiporter |
| ECH_0328 | ECH_RS01335 | 489 | cation/proton antiporter subunit D |
| ECH_0379 | ECH_RS01545 | 353 | hypothetical protein (sodium/proton antiporter; Cheng et al. 2013) |
| ECH_0466 | ECH_RS01920 | 89 | cation/proton antiporter subunit F |
| ECH_0467 | ECH_RS01925 | 99 | cation/proton antiporter subunit G |
| ECH_0468a (5′) | ECH_RS01930 | 181 | DUF4040 domain-containing protein/sodium/proton antiporter subunit B (E. ruminantium) |
| ECH_0468b (3′) | ECH_RS01935 | 139 | cation/proton antiporter subunit B |
| ECH_0469 | ECH_RS01940 | 111 | cation/proton antiporter subunit C |
| ECH_0474 | ECH_RS01960 | 519 | cation/proton antiporter subunit D |
| ECH_0944 | ECH_RS03855 | 163 | sodium/proton antiporter subunit E |
| Gene | Primer Name | Sequence | Size |
|---|---|---|---|
| ECH_0179 | RRG2107 | Forward: 5′-GGCTATACAAGTTGGGTTGTTGT-3′ | 672 bp |
| RRG2108 | Reverse: 5′-CACACATACACCACAGATAGACCT-3′ | ||
| ECH_0328 | RRG2068 | Forward: 5′-GCATGCGATATCATTTGGAA-3′ | 370 bp |
| RRG2069 | Reverse: 5′-GAATTGGAAAAGCCGCATTA-3′ | ||
| ECH_0379 | RRG1276 | Forward: 5′-CTAAGGTTGTAGGGAATGCAACC-3′ | 374 bp |
| RRG1277 | Reverse: 5′-ACAAGGTAAGTACCTTGCTTGCTC-3′ | ||
| ECH_0466 | RRG2054 | Forward: 5′-TGCTGCAAATTTGTTTGGAA-3′ | 165 bp |
| RRG2055 | Reverse: 5′-TCTCCAAAAGAACCATGAAGA-3′ | ||
| ECH_0467 | RRG2056 | Forward: 5′-TGGACTTGCTATGCGATCTG-3′ | 151 bp |
| RRG2057 | Reverse: 5′-TCAGTCAGCATCATCTTACCTTT -3′ | ||
| ECH_0468a | RRG2062 | Forward: 5′-TTTGATGGCATTGTTGCATT-3′ | 332 bp |
| RRG2063 | Reverse: 5′-CTCGAAAACTTGCTAAAATTGC-3′ | ||
| ECH_0468b | RRG2060 | Forward: 5′-CAGGCTGGTGTTATTGTTGC-3′ | 259 bp |
| RRG2061 | Reverse: 5′-TACACACAGTCATGCCCACA-3′ | ||
| ECH_0469 | RRG2058 | Forward: 5′-GGTTATAGGTTTGTATGTTACTACTGC-3′ | 213 bp |
| RRG2059 | Reverse: 5′-ATTGCAACCCCAACAACAAT-3′ | ||
| ECH_0474 | RRG2109 | Forward: 5′-GGCATCTGGTGGGTTTTTAGG -3′ | 525 bp |
| RRG2110 | Reverse: 5′-GCAGAACATACTGCCTCTACTG-3′ | ||
| ECH_0944 | RRG2064 | Forward: 5′-GGTTTGCCCTATCAGGGTATC-3′ | 426bp |
| RRG2065 | Reverse: 5′-CACCAGACATTGACTCTTCATCT-3′ |
| Target | Primer | Sequence # | Amplicon Size | Vector |
|---|---|---|---|---|
| ECH-0179 | RRG2097 | Forward: 5′-tgacg-GGATCC-TAGTGCCATTGGAGTATATGTAAG-3′ | 1976 bp | pBluescript II SK(+) |
| RRG2098 | Reverse: 5′-tgacg-GTCGAC-TTTACTTTAATTTTAATAAAGCTGCTG-3′ | |||
| ECH_0328 | RRG2099 | Forward: 5′-tgacg-GTCGAC-TCTTAATCTATAAGCGGCATATGC-3′ | 1893 bp | pBluescript II SK(+) |
| RRG2100 | Reverse: 5′-tgacg-GGATCC-AATATAGGAACAACTAAACAAACTAC-3′ | |||
| ECH_0379 | PRG2131 | Forward: 5′-tgacg-GGATCC-GTTTTTTAGCATCCTTTGTGTTAAAAG-3′ | 1543 bp | pBluescript II SK(+) |
| PRG2132 | Reverse: 5′-tgacg-GTCGAC-ATATCGACAAGCAATTGATACAGAG-3′ | |||
| ECH_0466, to 0469 | RRG2101 | Forward: 5′-tgacg-GGATCC- ATGTAGAATTCACAGAGCTTTAGC-3′ | 2291 bp | pBluescript II SK(+) |
| RRG2102 | Reverse: 5′-tgacg-GTCGAC-ATTATGATTTGACACTAGACTTACAC-3′ | |||
| ECH_0474 | RRG2103 | Forward: 5′-tgacg-GGATCC-TGTTGTTGATATATATGTTCAGTATG-3′ | 1883 bp | pBluescript II SK(+) |
| RRG2104 | Reverse: 5′-tgacg-GTCGAC -TTACTCTGCATCTTTTGGATTACA-3′ | |||
| ECH-0944 | RRG2105 | Forward: 5′-tgacg-GGATCC-GTAATACAACCCCAGTTATACAGA-3′ | 828 bp | pBluescript II SK(+) |
| RRG2106 | Reverse: 5′-tgacg-GTCGAC-TAATCACTTGTCAGATTTAATGACA-3′ | |||
| nhaA | PRG2158 | Forward: 5′-tgacg-GTCGAC-GCTCATTTCTCTCCCTGATAACA-3′ | 1817 bp | pBluescript II SK(+) |
| PRG2159 | Reverse: 5′-tgacg-CTGCAG-TGCTCTCTTCTCCTTGACCTT AC-3′ | |||
| ECH_0379 | RRG1397 | Forward: 5′-gag-GCTAGC-ATGATAATAAAGTTTGGGGTTGAAAATCTGATC-3′ | 1056 bp | pET-28a(+) |
| RRG1398 | Reverse: 5′-cga-CTCGAG-CTATAAATCTACACTTTCTTCAACAATATTAATAC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, L.; Liu, H.; Alizadeh, K.; Juarez-Rodriguez, M.D.; Ganta, R.R. Functional Characterization of Multiple Ehrlichia chaffeensis Sodium (Cation)/Proton Antiporter Genes Involved in the Bacterial pH Homeostasis. Int. J. Mol. Sci. 2021, 22, 8420. https://doi.org/10.3390/ijms22168420
Wei L, Liu H, Alizadeh K, Juarez-Rodriguez MD, Ganta RR. Functional Characterization of Multiple Ehrlichia chaffeensis Sodium (Cation)/Proton Antiporter Genes Involved in the Bacterial pH Homeostasis. International Journal of Molecular Sciences. 2021; 22(16):8420. https://doi.org/10.3390/ijms22168420
Chicago/Turabian StyleWei, Lanjing, Huitao Liu, Kimia Alizadeh, Maria D. Juarez-Rodriguez, and Roman R. Ganta. 2021. "Functional Characterization of Multiple Ehrlichia chaffeensis Sodium (Cation)/Proton Antiporter Genes Involved in the Bacterial pH Homeostasis" International Journal of Molecular Sciences 22, no. 16: 8420. https://doi.org/10.3390/ijms22168420
APA StyleWei, L., Liu, H., Alizadeh, K., Juarez-Rodriguez, M. D., & Ganta, R. R. (2021). Functional Characterization of Multiple Ehrlichia chaffeensis Sodium (Cation)/Proton Antiporter Genes Involved in the Bacterial pH Homeostasis. International Journal of Molecular Sciences, 22(16), 8420. https://doi.org/10.3390/ijms22168420






