alfaNET: A Database of Alfalfa-Bacterial Stem Blight Protein–Protein Interactions Revealing the Molecular Features of the Disease-causing Bacteria
Abstract
1. Introduction
2. Results
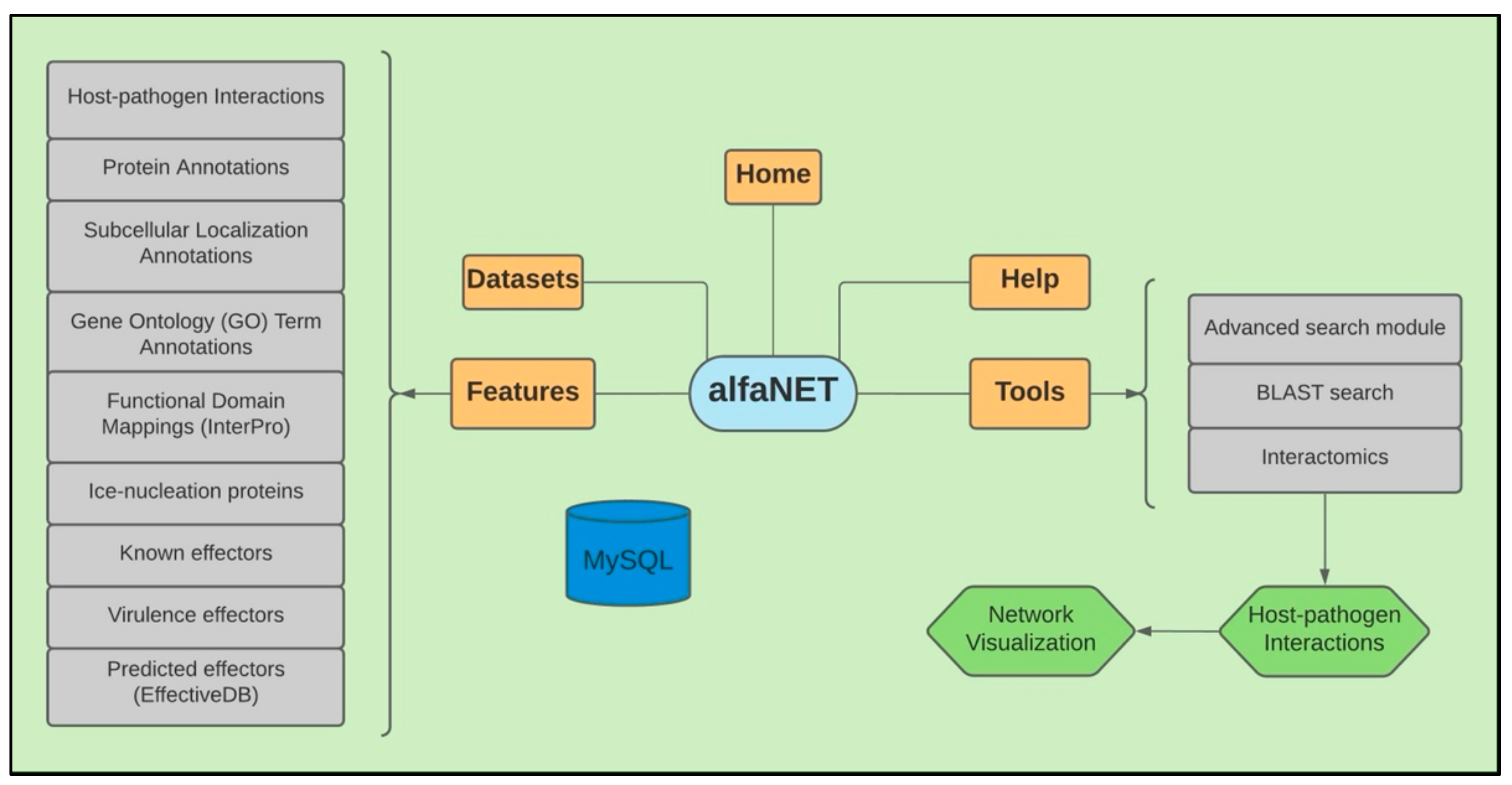
Database Architecture and Implementation
3. Discussion
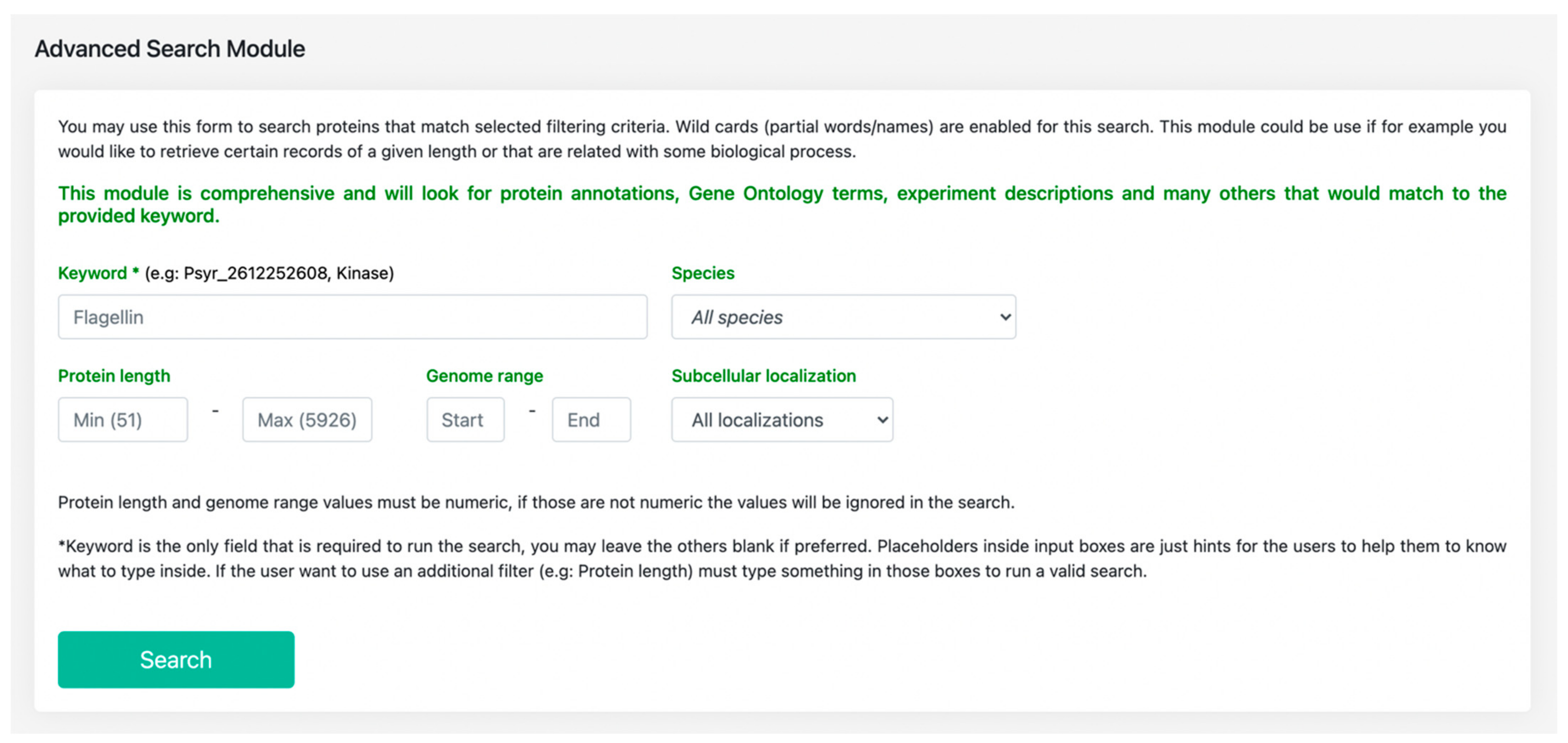
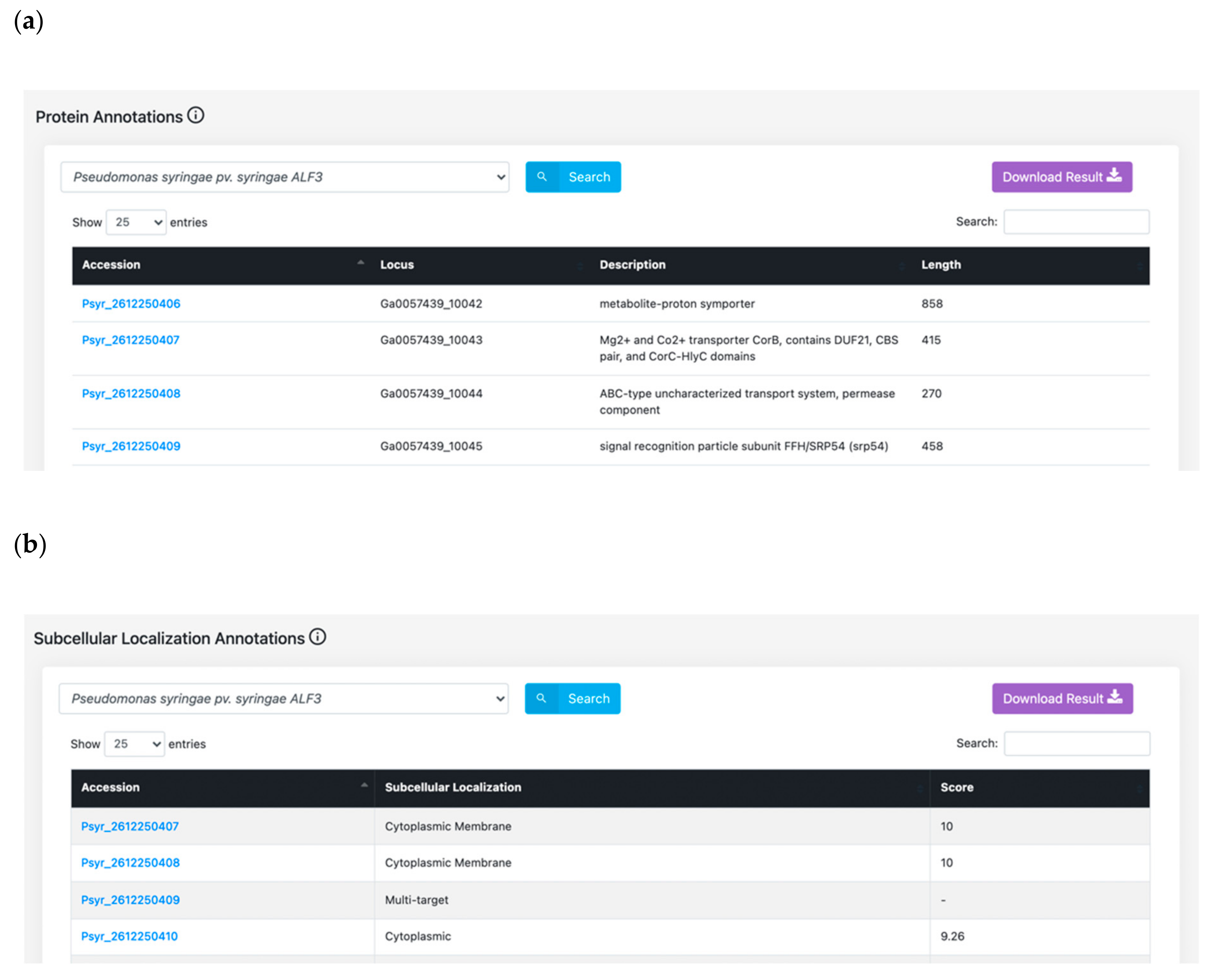
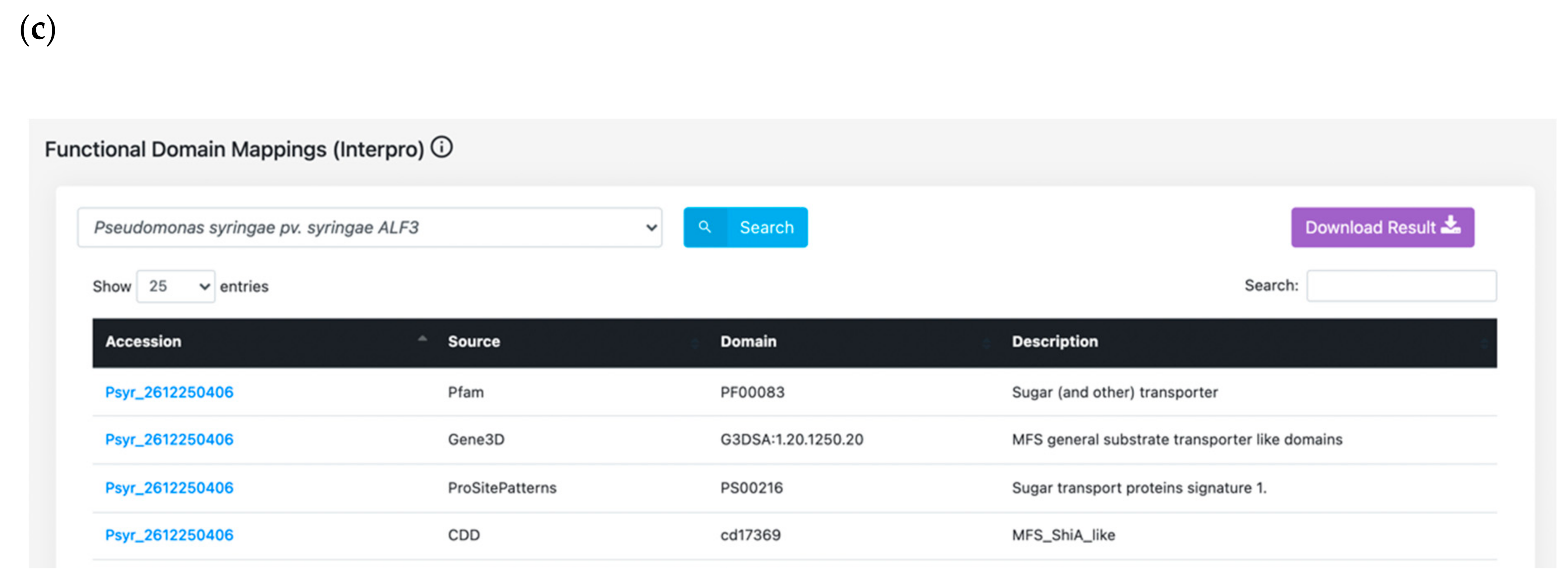
3.1. Features Search: A Central Resource to Retrieve P. syringae Annotations
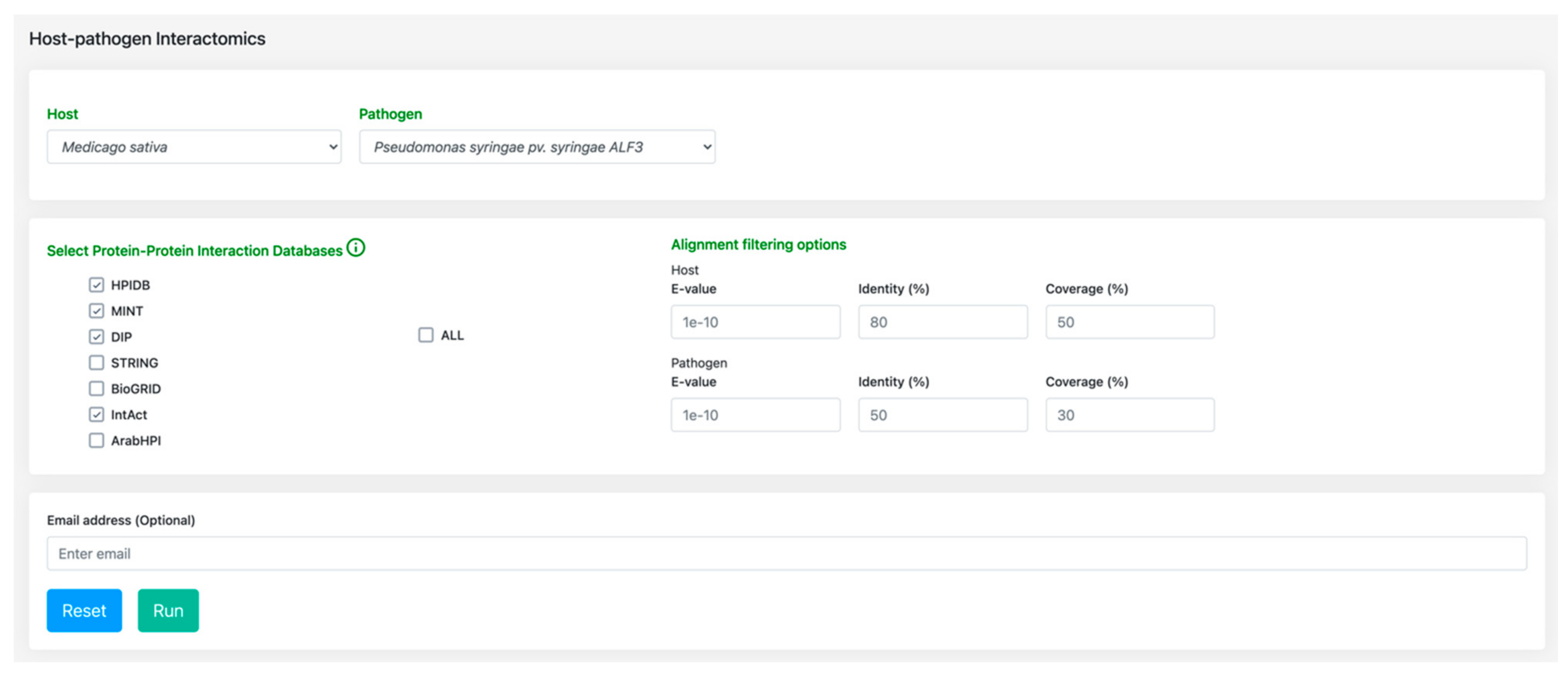
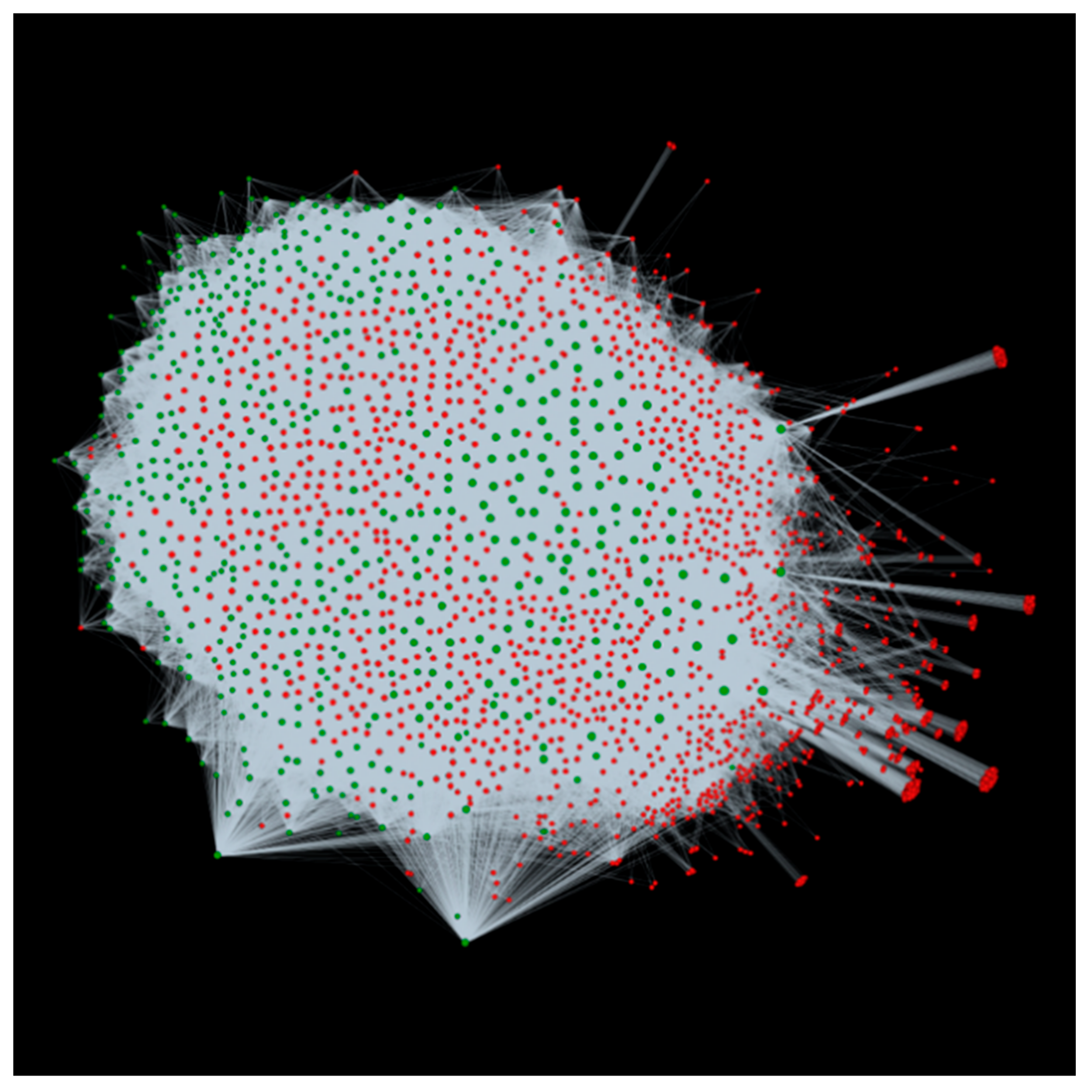
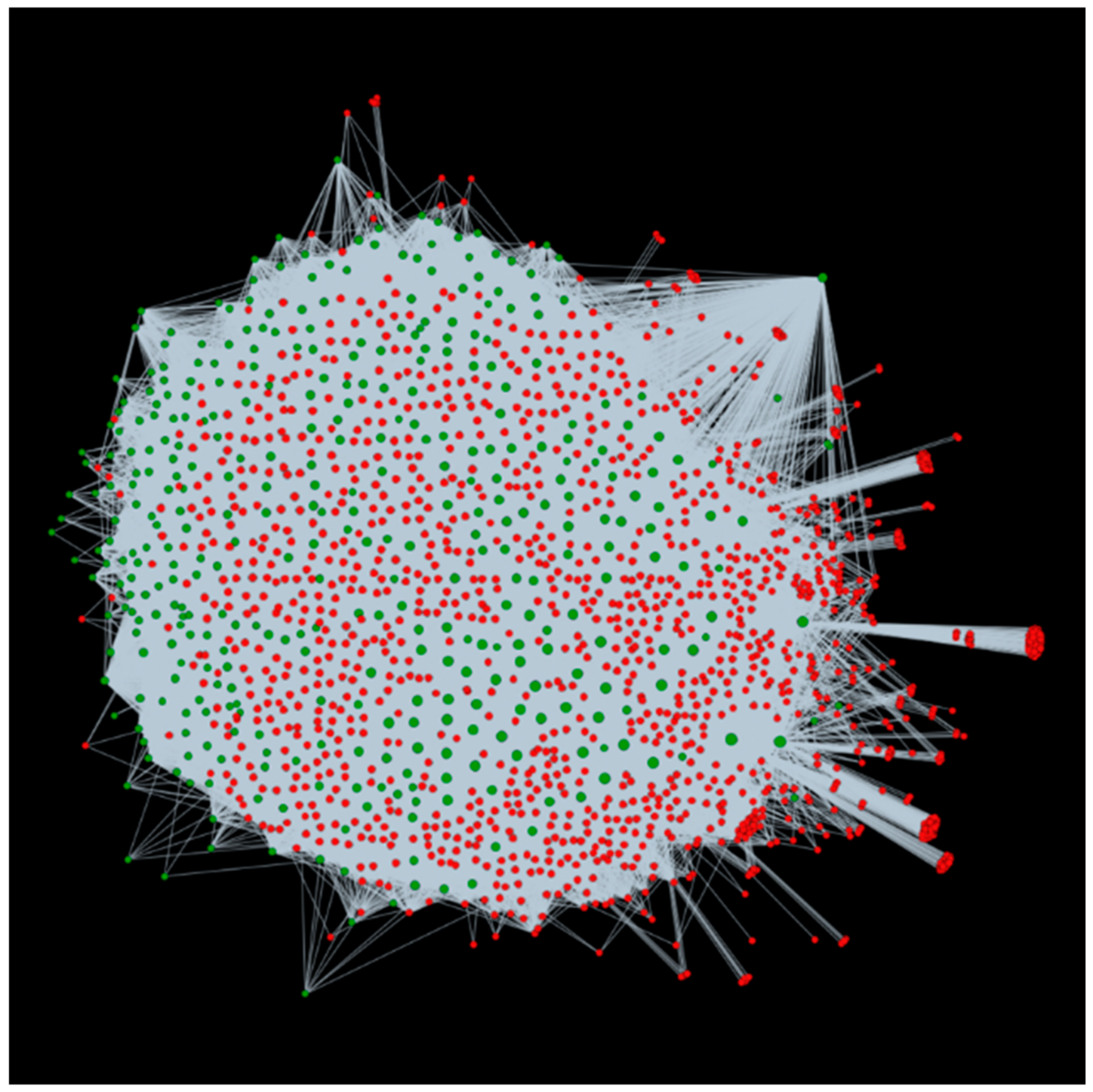
3.2. Host-Pathogen Interactome: Towards a Better Understanding of Bacterial Stem Blight Infection Mechanisms
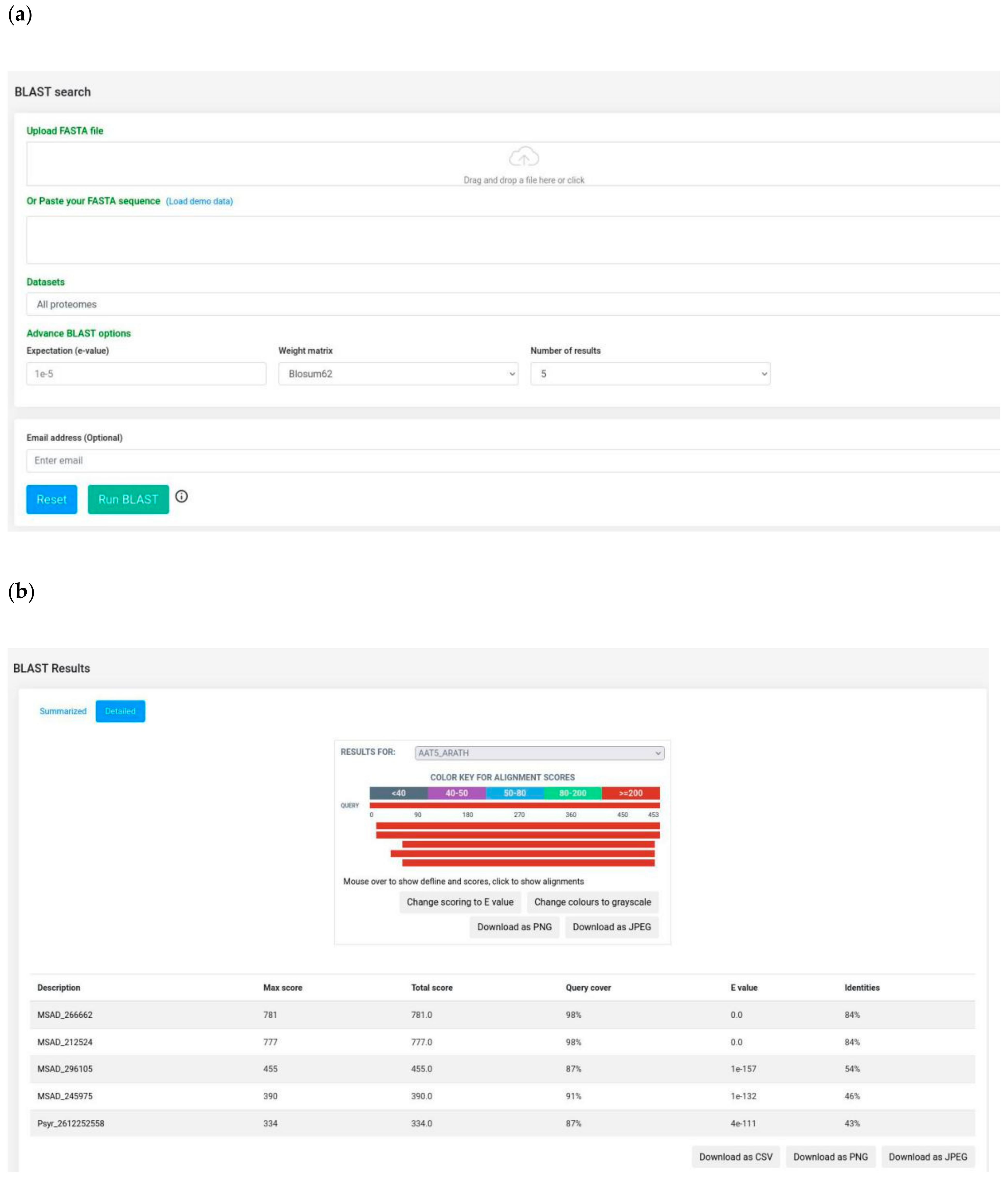
3.3. BLAST Server and Bulk Data Download
3.4. Applicability of alfaNET: A Case Study on Resistant and Susceptible Plant Responses in Medicago sativa to Bacterial Stem Blight
3.5. Limitations and Future Development
4. Materials and Methods
4.1. Data Source and Processing
4.2. Host-Pathogen Interactome Comparison Tool
4.3. Protein Annotation
4.4. Dataset Collection for P. syringae Effectors
4.4.1. Known T3SS Effectors
4.4.2. Virulence Effectors
4.4.3. Predicted Effectors
4.5. Dataset Collection for Ice-Nucleation Proteins
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernandez, A.; Sheaffer, C.; Tautges, N.; Putnam, D.; Hunter, M. Alfalfa, Wildlife & the Environment, 2nd ed.; National Alfalfa and Forage Alliance: St. Paul, MN, USA, 2019; Volume 34. [Google Scholar]
- Rafińska, K.; Pomastowski, P.; Wrona, O.; Górecki, R.; Buszewski, B. Medicago sativa as a source of secondary metabolites for agriculture and pharmaceutical industry. Phytochem. Lett. 2017, 20, 520–539. [Google Scholar] [CrossRef]
- Nicholson, R.L.; Hammerschmidt, R. Phenolic Compounds and Their Role in Disease Resistance. Annu. Rev. Phytopathol. 1992, 30, 369–389. [Google Scholar] [CrossRef]
- Harighi, B. Occurrence of alfalfa bacterial stem blight disease in Kurdistan province, Iran. J. Phytopathol. 2007, 155, 593–595. [Google Scholar] [CrossRef]
- Proud Sponsor of Midwest Forage Association. Forage Focus; 2017; Technical Report. [Google Scholar]
- Saint-Vincent, P.M.B.; Ridout, M.; Engle, N.L.; Lawrence, T.J.; Yeary, M.L.; Tschaplinski, T.J.; Newcombe, G.; Pelletier, D.A. Isolation, characterization, and pathogenicity of two Pseudomonas syringae pathovars from populus trichocarpa seeds. Microorganisms 2020, 8, 1137. [Google Scholar] [CrossRef]
- Li, Q.; Yan, Q.; Chen, J.; He, Y.; Wang, J.; Zhang, H.; Yu, Z.; Li, L. Molecular characterization of an ice nucleation protein variant (InaQ) from Pseudomonas syringae and the analysis of its transmembrane transport activity in Escherichia coli. Int. J. Biol. Sci. 2012, 8, 1097–1108. [Google Scholar] [CrossRef]
- Harrison, J.; Dornbusch, M.R.; Samac, D.; Studholme, D.J. Draft genome sequence of Pseudomonas syringae pv. syringae ALF3 isolated from alfalfa. Genome Announc. 2016, 4, 2015–2016. [Google Scholar] [CrossRef]
- Quirino, B.F.; Bent, A.F. Deciphering host resistance and pathogen virulence: The Arabidopsis/Pseudomonas interaction as a model. Mol. Plant Pathol. 2003, 4, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. Bacterial pathogens in plants: Life up against the wall. Plant Cell 1996, 8, 1683–1698. [Google Scholar] [CrossRef]
- Samac, D.; Studholme, D.J.; Ao, S. Characterization of the bacterial stem blight pathogen of alfalfa, Pseudomonas syringae pv. syringae ALF3. Phytopathology 2014, 104, 102. [Google Scholar]
- Nemchinov, L.G.; Shao, J.; Lee, M.N.; Postnikova, O.A.; Samac, D.A. Resistant and susceptible responses in alfalfa (Medicago sativa) to bacterial stem blight caused by Pseudomonas syringae pv. syringae. PLoS ONE 2017, 12, e0189781. [Google Scholar] [CrossRef] [PubMed]
- Loaiza, C.D.; Duhan, N.; Lister, M.; Kaundal, R. In silico prediction of host–pathogen protein interactions in melioidosis pathogen Burkholderia pseudomallei and human reveals novel virulence factors and their targets. Brief. Bioinform. 2020, 22, bbz162. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, 581–591. [Google Scholar] [CrossRef]
- Memišević, V.; Kumar, K.; Zavaljevski, N.; DeShazer, D.; Wallqvist, A.; Reifman, J. DBSecSys 2.0: A database of Burkholderia mallei and Burkholderia pseudomallei secretion systems. BMC Bioinform. 2016, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Zhang, D.; Ban, R.; Ma, X.; Chen, D.; Li, G.; Liu, J.; Wisniewski, M.; Droby, S.; Liu, Y. PCPPI: A comprehensive database for the prediction of Penicillium-crop protein-protein interactions. Database 2017, 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Lam, D.K.W.; Fleming, L.; Lo, R.; Whiteside, M.D.; Yu, N.Y.; Hancock, R.E.W.; Brinkman, F.S.L. Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011, 39, 596–600. [Google Scholar] [CrossRef]
- Li, J.; Dai, X.; Liu, T.; Zhao, P.X. LegumeIP: An integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Res. 2012, 40, 1221–1229. [Google Scholar] [CrossRef]
- Blanco-Míguez, A.; Fdez-Riverola, F.; Sánchez, B.; Lourenço, A. BlasterJS: A novel interactive JavaScript visualisation component for BLAST alignment results. PLoS ONE 2018, 13, e0205286. [Google Scholar] [CrossRef]
- Jacomy, A. Sigmajs. 2012. Available online: http://sigmajs.org/ (accessed on 1 August 2021).
- Wei, L.; Xing, P.; Zeng, J.; Chen, J.X.; Su, R.; Guo, F. Improved prediction of protein–protein interactions using novel negative samples, features, and an ensemble classifier. Artif. Intell. Med. 2017, 83, 67–74. [Google Scholar] [CrossRef]
- He, Y.; Wu, L.; Liu, X.; Zhang, X.; Jiang, P.; Ma, H. Yeast two-hybrid screening for proteins that interact with PFT in wheat. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Free, R.B.; Hazelwood, L.A.; Sibley, D.R. Identifying novel protein-protein interactions using co-immunoprecipitation and mass spectroscopy. Curr. Protoc. Neurosci. 2009, 46, 5–28. [Google Scholar] [CrossRef]
- Ding, Z.; Kihara, D. Computational identification of protein-protein interactions in model plant proteomes. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Zhang, H.; Li, W.; Dai, Y. Prediction of Protein-Protein Interactions with Local Weight-Sharing Mechanism in Deep Learning. Biomed Res. Int. 2020, 2020, 5072520. [Google Scholar] [CrossRef]
- Saha, S.; Sengupta, K.; Chatterjee, P.; Basu, S.; Nasipuri, M. Analysis of protein targets in pathogen-host interaction in infectious diseases: A case study on Plasmodium falciparum and Homo sapiens interaction network. Brief. Funct. Genomic. 2018, 17, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Boschiero, C.; Dong, Y.; Zhao, P.X. LegumeIP V3: From models to crops—An integrative gene discovery platform for translational genomics in legumes. Nucleic Acids Res. 2020, 49, D1472–D1479. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.S.; Weirick, T.; Kaundal, R. Predicting genome-scale Arabidopsis-Pseudomonas syringae interactome using domain and interolog-based approaches. BMC Bioinform. 2014, 15, S13. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Kerrien, S.; Abbani, S.; Aranda, B.; Bhate, J.; Bridge, A.; Briganti, L.; Brinkman, F.S.L.; Cesareni, G.; Chatr-aryamontri, A.; et al. Exchange Consortium (IMEx). Nat. Methods 2012, 9, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Salwinski, L.; Miller, C.S.; Smith, A.J.; Pettit, F.K.; Bowie, J.U.; Eisenberg, D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004, 32, 449–451. [Google Scholar] [CrossRef]
- Kerrien, S.; Aranda, B.; Breuza, L.; Bridge, A.; Broackes-Carter, F.; Chen, C.; Duesbury, M.; Dumousseau, M.; Feuermann, M.; Hinz, U.; et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012, 40, 841–846. [Google Scholar] [CrossRef]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the molecular interaction database: 2012 Update. Nucleic Acids Res. 2012, 40, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K.; O’Donnell, L.; Oster, S.; Theesfeld, C.; Sellam, A.; et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017, 45, D369–D379. [Google Scholar] [CrossRef] [PubMed]
- Ammari, M.G.; Gresham, C.R.; McCarthy, F.M.; Nanduri, B. HPIDB 2.0: A curated database for host-pathogen interactions. Database 2016, 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Castillo-Lara, S.; Abril, J.F. PlanNET: Homology-based predicted interactome for multiple planarian transcriptomes. Bioinformatics 2018, 34, 1016–1023. [Google Scholar] [CrossRef]
- Yu, H.; Luscombe, N.M.; Lu, H.X.; Zhu, X.; Xia, Y.; Han, J.D.J.; Bertin, N.; Chung, S.; Vidal, M.; Gerstein, M. Annotation transfer between genomes: Protein-protein interrologs and protein-DNA regulogs. Genome Res. 2004, 14, 1107–1118. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Cenk Sahinalp, S.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Jehl, M.A.; Arnold, R.; Rattei, T. Effective-A database of predicted secreted bacterial proteins. Nucleic Acids Res. 2011, 39, 591–595. [Google Scholar] [CrossRef][Green Version]
- Shames, S.R.; Finlay, B.B. Bacterial effector interplay: A new way to view effector function. Trends Microbiol. 2012, 20, 214–219. [Google Scholar] [CrossRef]
- Feil, H.; Feil, W.S.; Chain, P.; Larimer, F.; DiBartolo, G.; Copeland, A.; Lykidis, A.; Trong, S.; Nolan, M.; Goltsman, E.; et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 2005, 102, 11064–11069. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kataria, R.; Kaundal, R. alfaNET: A Database of Alfalfa-Bacterial Stem Blight Protein–Protein Interactions Revealing the Molecular Features of the Disease-causing Bacteria. Int. J. Mol. Sci. 2021, 22, 8342. https://doi.org/10.3390/ijms22158342
Kataria R, Kaundal R. alfaNET: A Database of Alfalfa-Bacterial Stem Blight Protein–Protein Interactions Revealing the Molecular Features of the Disease-causing Bacteria. International Journal of Molecular Sciences. 2021; 22(15):8342. https://doi.org/10.3390/ijms22158342
Chicago/Turabian StyleKataria, Raghav, and Rakesh Kaundal. 2021. "alfaNET: A Database of Alfalfa-Bacterial Stem Blight Protein–Protein Interactions Revealing the Molecular Features of the Disease-causing Bacteria" International Journal of Molecular Sciences 22, no. 15: 8342. https://doi.org/10.3390/ijms22158342
APA StyleKataria, R., & Kaundal, R. (2021). alfaNET: A Database of Alfalfa-Bacterial Stem Blight Protein–Protein Interactions Revealing the Molecular Features of the Disease-causing Bacteria. International Journal of Molecular Sciences, 22(15), 8342. https://doi.org/10.3390/ijms22158342






