Regulation Network of Colorectal-Cancer-Specific Enhancers in the Progression of Colorectal Cancer
Abstract
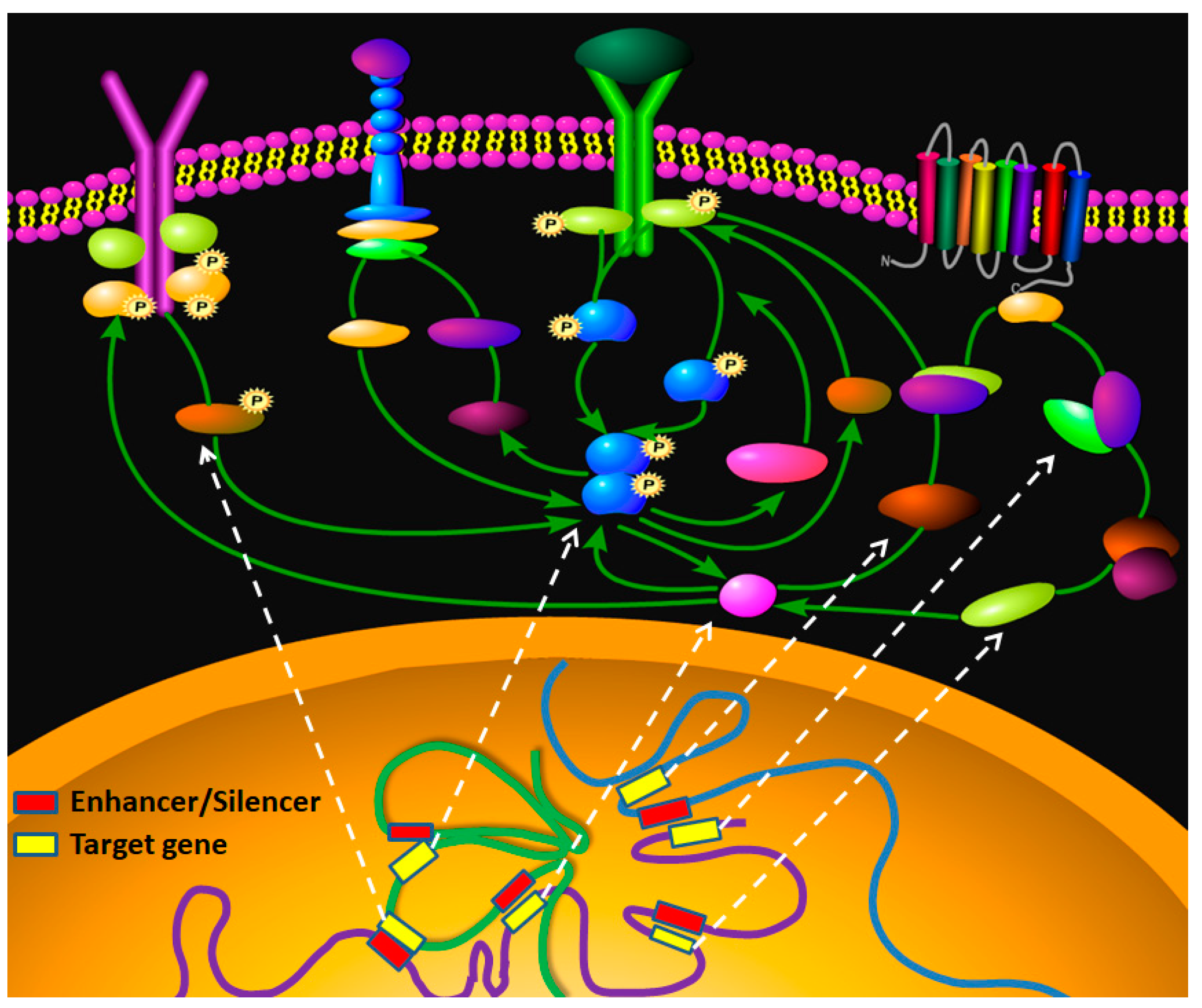
:1. Introduction
2. Results
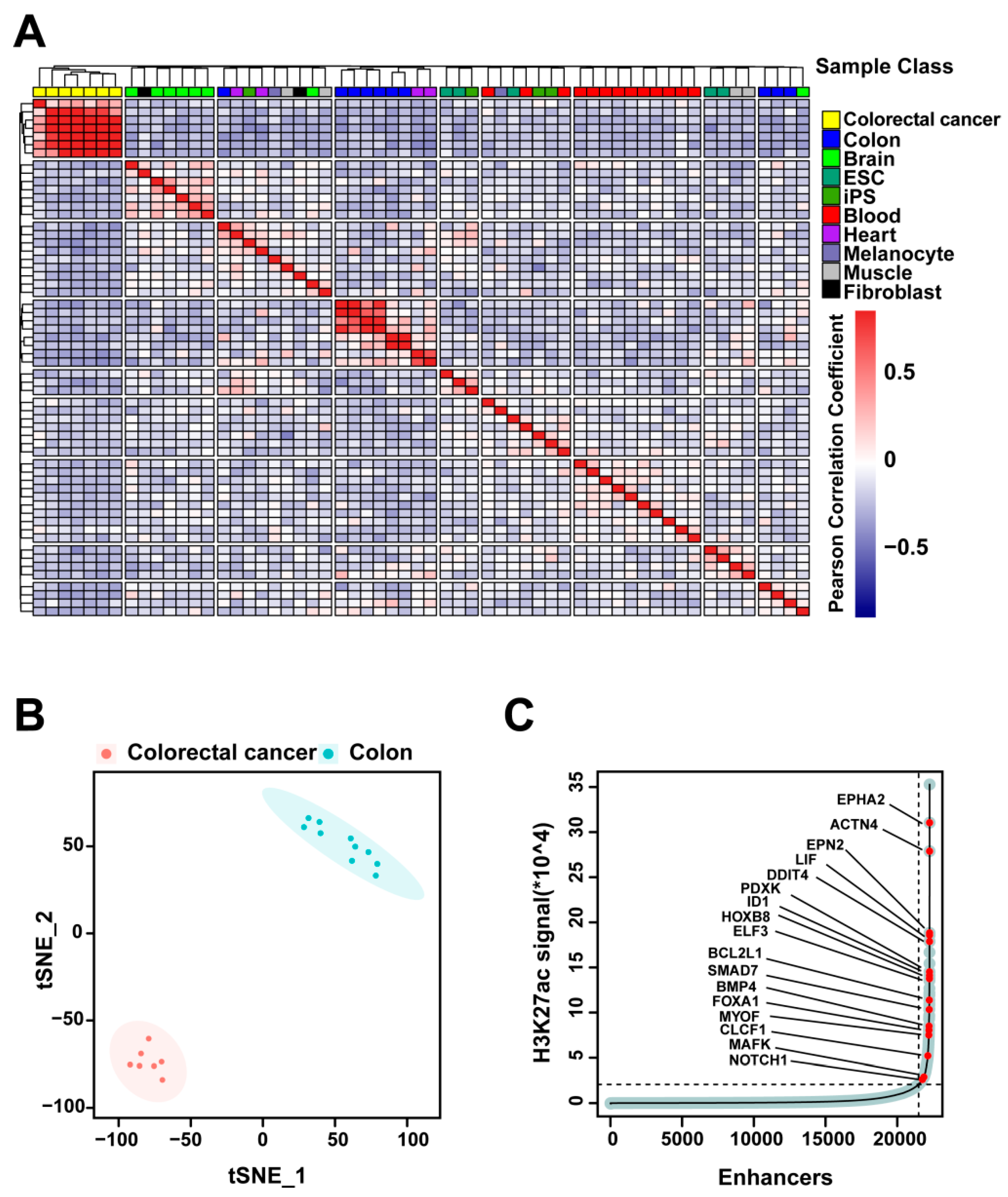
2.1. H3K27ac Profiles Define Colorectal-Cancer-Specific Enhancers
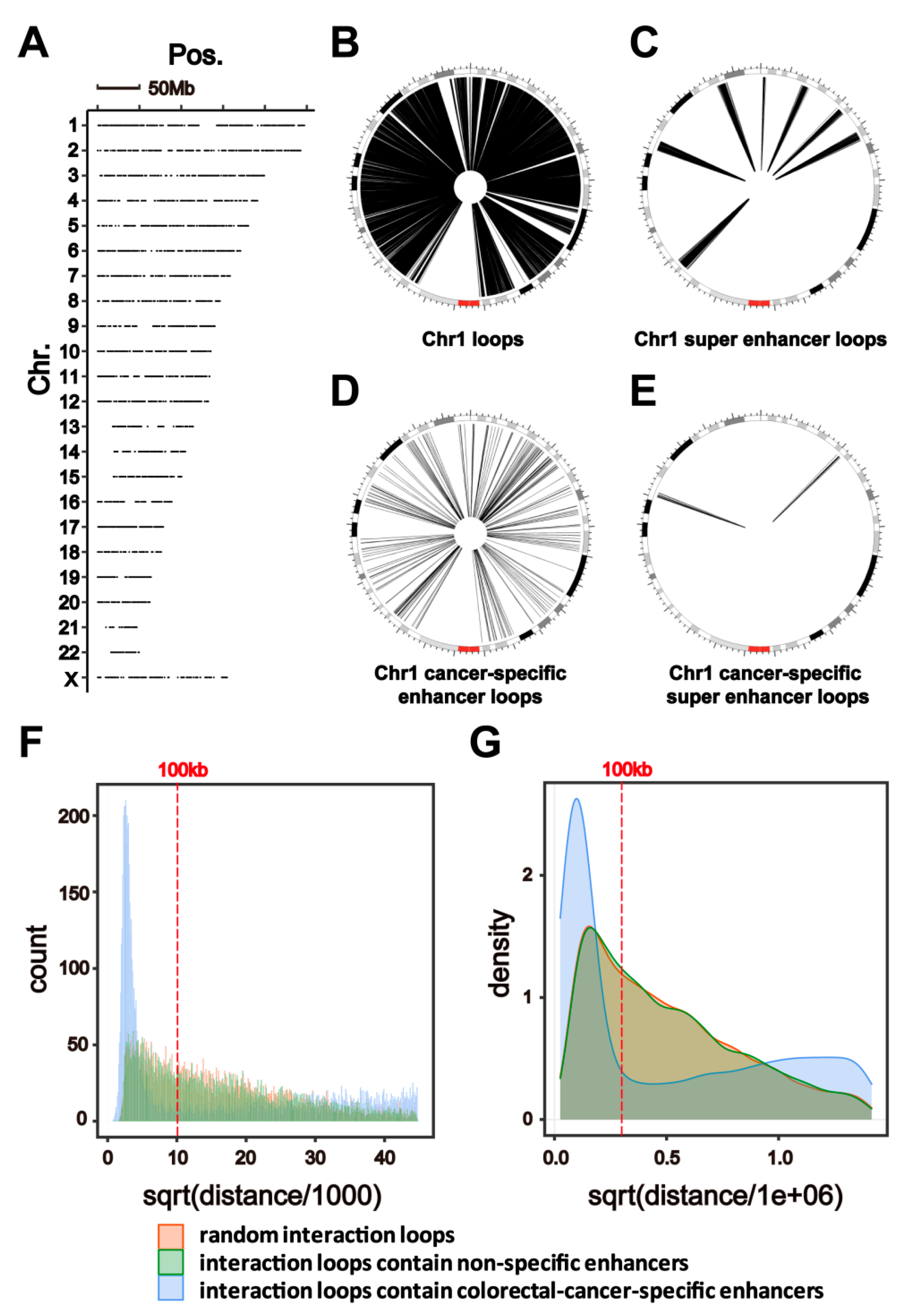
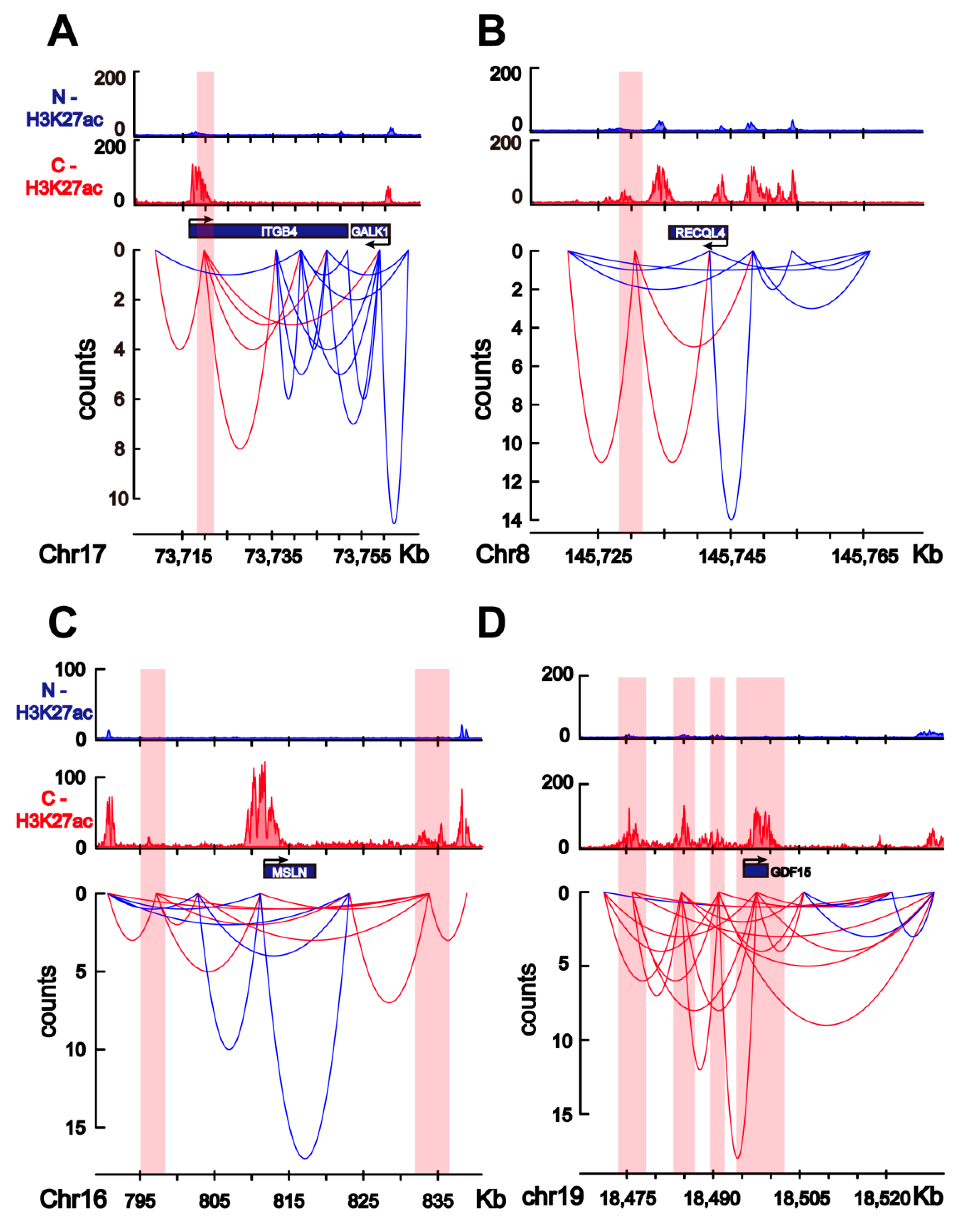
2.2. HiChIP Identifies Chromatin Interactions Containing Colorectal-Cancer-Specific Enhancers
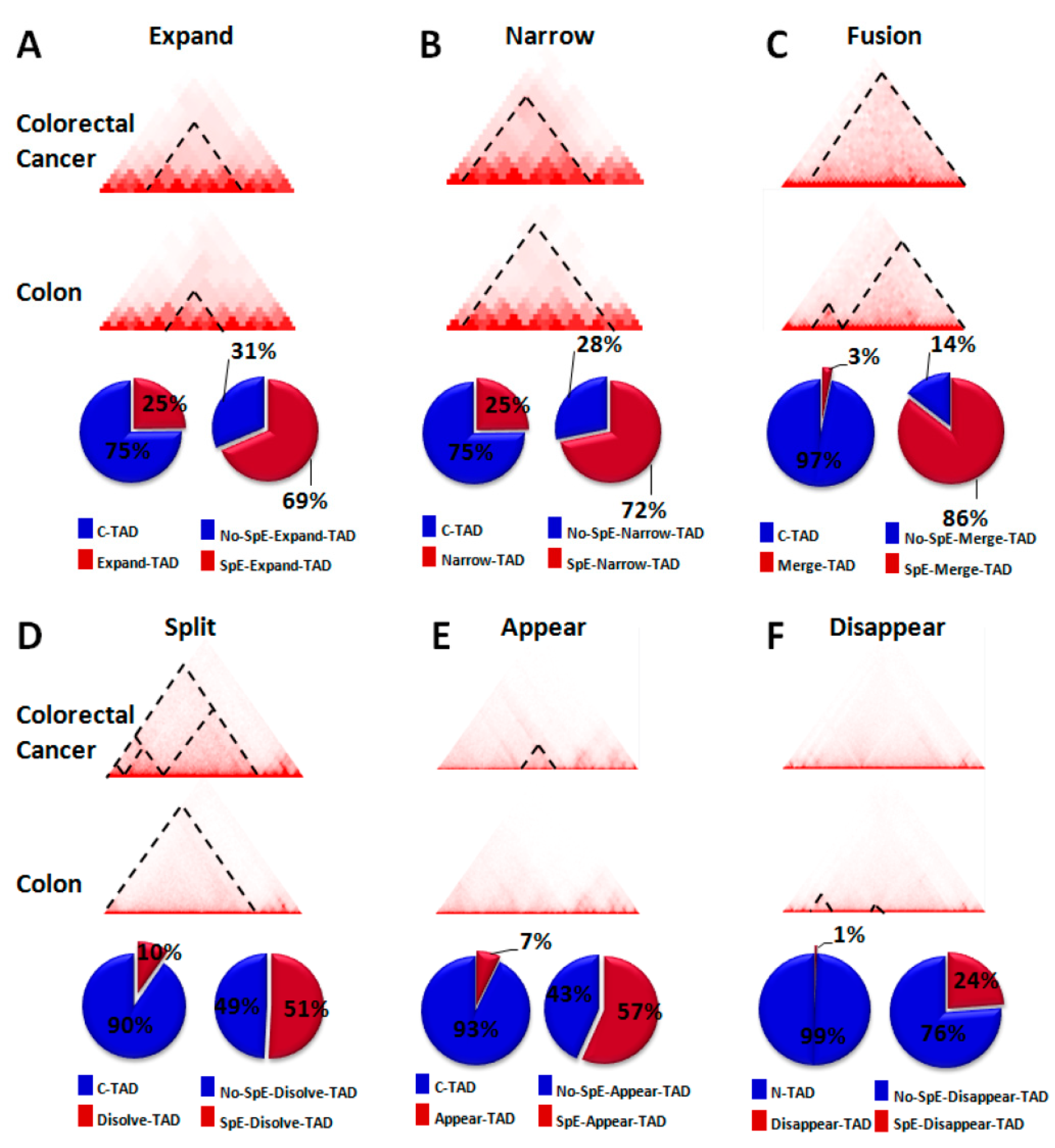
2.3. Change in TAD Boundaries in Colorectal Cancer Cells Compared to Normal Colon Cells
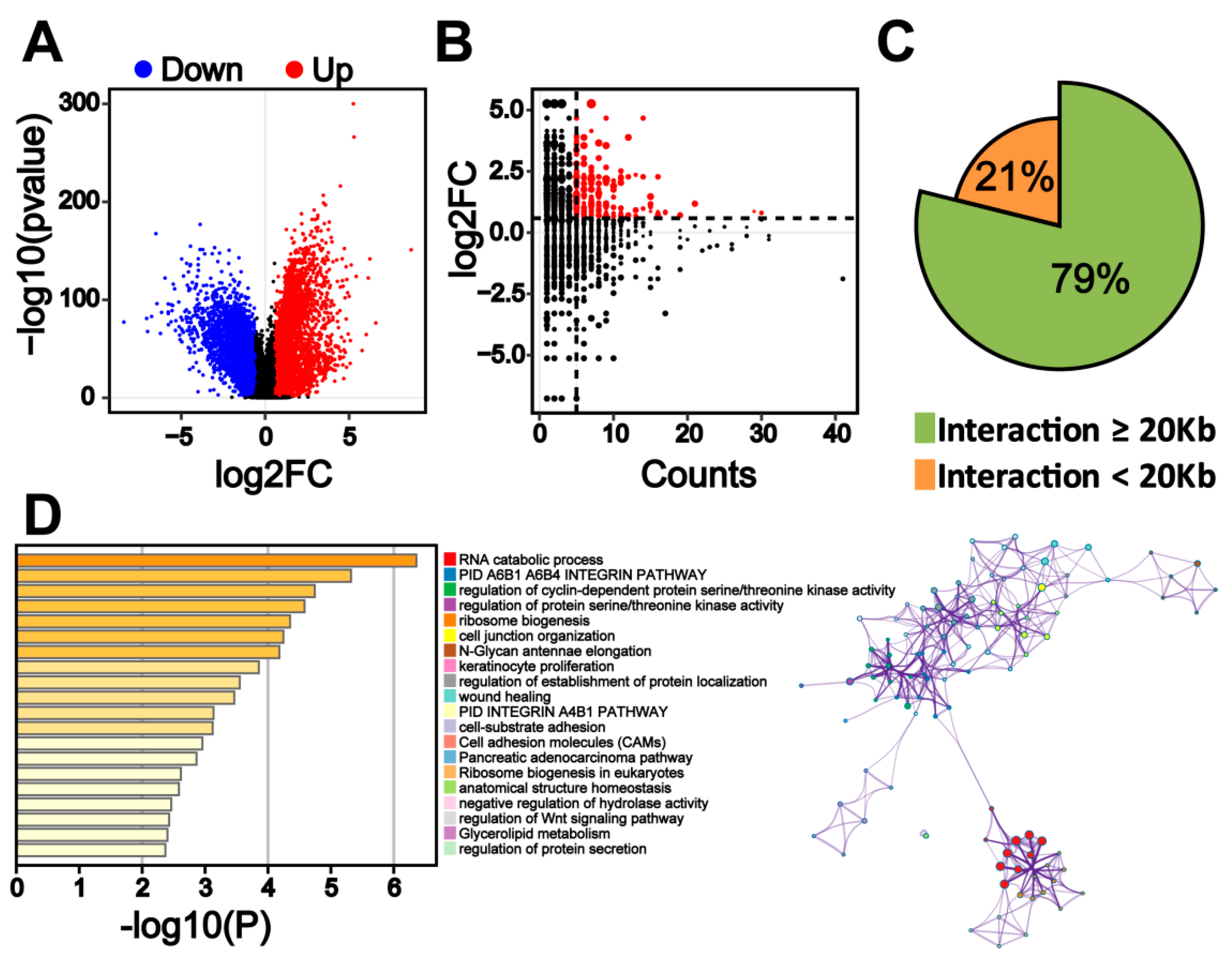
2.4. Transcriptome Change in Colorectal Cancer Cells Is Associated with Colorectal-Specific Enhancers
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. HiChIP
4.3. ChIP-Seq Data Processing and Super Enhancer Analysis
4.4. Unsupervised Hierarchical Clustering Analysis
4.5. t-SNE Analysis
4.6. HiChIP Data Processing
4.7. Hi-C Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [Green Version]
- Fearon, E.R. Molecular genetics of colorectal cancer. Ann. Rev. Pathol. 2011, 6, 479–507. [Google Scholar] [CrossRef]
- Wei, Z.; Gao, F.; Kim, S.; Yang, H.; Lyu, J.; An, W.; Wang, K.; Lu, W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 2013, 13, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Su, G.; Guo, D.; Chen, J.; Liu, M.; Zheng, J.; Wang, W.; Zhao, X.; Yin, Q.; Zhang, L.; Zhao, Z.; et al. A distal enhancer maintaining Hoxa1 expression orchestrates retinoic acid-induced early ESCs differentiation. Nucleic Acids Res. 2019, 47, 6737–6752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonev, B.; Mendelson Cohen, N.; Szabo, Q.; Fritsch, L.; Papadopoulos, G.L.; Lubling, Y.; Xu, X.; Lv, X.; Hugnot, J.P.; Tanay, A.; et al. Multiscale 3D genome rewiring during mouse neural development. Cell 2017, 171, 557–572.e24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; He, A.; Chen, B.; Bi, J.; Chen, J.; Guo, D.; Qian, Y.; Wang, W.; Shi, T.; Zhao, Z.; et al. A HOTAIR regulatory element modulates glioma cell sensitivity to temozolomide through long-range regulation of multiple target genes. Genome Res. 2020, 30, 155–163. [Google Scholar] [CrossRef]
- Qian, Y.; Zhang, L.; Cai, M.; Li, H.; Xu, H.; Yang, H.; Zhao, Z.; Rhie, S.K.; Farnham, P.J.; Shi, J.; et al. The prostate cancer risk variant rs55958994 regulates multiple gene expression through extreme long-range chromatin interaction to control tumor progression. Sci. Adv. 2019, 5, eaaw6710. [Google Scholar] [CrossRef] [Green Version]
- Kloetgen, A.; Thandapani, P.; Ntziachristos, P.; Ghebrechristos, Y.; Nomikou, S.; Lazaris, C.; Chen, X.; Hu, H.; Bakogianni, S.; Wang, J.; et al. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat. Genet. 2020, 52, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Rhie, S.K.; Perez, A.A.; Lay, F.D.; Schreiner, S.; Shi, J.; Polin, J.; Farnham, P.J. A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome. Nat. Commun. 2019, 10, 4154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, S.E.; Reyes, A.; Qi, Y.; Adriaens, C.; Hegazi, E.; Pelka, K.; Chen, J.H.; Zou, L.S.; Drier, Y.; Hecht, V.; et al. Large-scale topological changes restrain malignant progression in colorectal cancer. Cell 2020, 182, 1474–1489.e23. [Google Scholar] [CrossRef] [PubMed]
- Berman, B.P.; Weisenberger, D.J.; Aman, J.F.; Hinoue, T.; Ramjan, Z.; Liu, Y.; Noushmehr, H.; Lange, C.P.; van Dijk, C.M.; Tollenaar, R.A.; et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2011, 44, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Mack, S.C.; Pajtler, K.W.; Chavez, L.; Okonechnikov, K.; Bertrand, K.C.; Wang, X.; Erkek, S.; Federation, A.; Song, A.; Lee, C. Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 2018, 553, 101–105. [Google Scholar] [CrossRef]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [Green Version]
- Roadmap Epigenomics, C.; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Akhtar-Zaidi, B.; Cowper-Sal-Lari, R.; Corradin, O.; Saiakhova, A.; Bartels, C.F.; Balasubramanian, D.; Myeroff, L.; Lutterbaugh, J.; Jarrar, A.; Kalady, M.F.; et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 2012, 336, 736–739. [Google Scholar] [CrossRef] [Green Version]
- Thandapani, P. Super-enhancers in cancer. Pharmacol. Ther. 2019, 199, 129–138. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, A.M.; Bachelder, R.E.; Rabinovitz, I.; O’Connor, K.L.; Tani, T.; Shaw, L.M. The metastatic odyssey: The integrin connection. Surg. Oncol. Clin. N. Am. 2001, 10, 313–328. [Google Scholar] [CrossRef]
- Li, M.; Jiang, X.; Wang, G.; Zhai, C.; Liu, Y.; Li, H.; Zhang, Y.; Yu, W.; Zhao, Z. ITGB4 is a novel prognostic factor in colon cancer. J. Cancer 2019, 10, 5223–5233. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.N.; Li, H.; Yao, H.; Liu, X.; Li, L.; Leung, K.S.; Kung, H.F.; Lin, M.C. Adapalene inhibits the activity of cyclin-dependent kinase 2 in colorectal carcinoma. Mol. Med. Rep. 2015, 12, 6501–6508. [Google Scholar] [CrossRef] [Green Version]
- Cam, W.R.; Masaki, T.; Shiratori, T.Y.; Kato, N.; Okamoto, M.; Yamaji, Y.; Igarashi, K.; Sano, T.; Omata, M. Activation of cyclin E-dependent kinase activity in colorectal cancer. Digest. Dis. Sci. 2001, 46, 2187–2198. [Google Scholar] [CrossRef]
- Martinez-Jimenez, F.; Muinos, F.; Sentis, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef]
- Zhunussova, G.; Afonin, G.; Abdikerim, S.; Jumanov, A.; Perfilyeva, A.; Kaidarova, D.; Djansugurova, L. Mutation spectrum of cancer-associated genes in patients with early onset of colorectal cancer. Front. Oncol. 2019, 9, 673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, S.; Tsunoda, T.; Riku, M.; Ito, H.; Inoko, A.; Murakami, H.; Ebi, M.; Ogasawara, N.; Pastan, I.; Kasugai, K.; et al. Diffuse mesothelin expression leads to worse prognosis through enhanced cellular proliferation in colorectal cancer. Oncol. Lett. 2020, 19, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Wallin, U.; Glimelius, B.; Jirstrom, K.; Darmanis, S.; Nong, R.Y.; Ponten, F.; Johansson, C.; Pahlman, L.; Birgisson, H. Growth differentiation factor 15: A prognostic marker for recurrence in colorectal cancer. Br. J. Cancer 2011, 104, 1619–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Ma, N.; Zheng, X.; Li, X.; Ma, X.; Hu, J.; Cao, B. GDF15 repression contributes to 5-fluorouracil resistance in human colon cancer by regulating epithelial-mesenchymal transition and apoptosis. BioMed Res. Int. 2020, 2020, 2826010. [Google Scholar] [CrossRef]
- Zheng, X.; Ma, N.; Wang, X.; Hu, J.; Ma, X.; Wang, J.; Cao, B. Exosomes derived from 5-fluorouracil-resistant colon cancer cells are enriched in GDF15 and can promote angiogenesis. J. Cancer 2020, 11, 7116–7126. [Google Scholar] [CrossRef]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing chromosome conformation. Science 2002, 295, 1306–1311. [Google Scholar] [CrossRef] [Green Version]
- Belaghzal, H.; Dekker, J.; Gibcus, J.H. Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 2017, 123, 56–65. [Google Scholar] [CrossRef]
- Mumbach, M.R.; Rubin, A.J.; Flynn, R.A.; Dai, C.; Khavari, P.A.; Greenleaf, W.J.; Chang, H.Y. HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 2016, 13, 919–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Layseca, P.; Streuli, C.H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. J. Int. Soc. Matrix Biol. 2014, 34, 144–153. [Google Scholar] [CrossRef]
- Wilkinson, E.J.; Woodworth, A.M.; Parker, M.; Phillips, J.L.; Malley, R.C.; Dickinson, J.L.; Holloway, A.F. Epigenetic regulation of the ITGB4 gene in prostate cancer. Exp. Cell Res. 2020, 392, 112055. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.L.; O’Connor, K.L. Clinical significance of the integrin alpha6beta4 in human malignancies. Lab. Investig. J. Tech. Methods Pathol. 2015, 95, 976–986. [Google Scholar] [CrossRef] [Green Version]
- Ni, H.; Dydensborg, A.B.; Herring, F.E.; Basora, N.; Gagne, D.; Vachon, P.H.; Beaulieu, J.F. Upregulation of a functional form of the beta4 integrin subunit in colorectal cancers correlates with c-Myc expression. Oncogene 2005, 24, 6820–6829. [Google Scholar] [CrossRef] [Green Version]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ helicases in DNA repair, recombination, and replication. Ann. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, D.; Zhao, Y.; Balajee, A.S. Human RecQL4 helicase plays multifaceted roles in the genomic stability of normal and cancer cells. Cancer Lett. 2018, 413, 1–10. [Google Scholar] [CrossRef]
- Guo, L.; Li, Y.; Zhao, C.; Peng, J.; Song, K.; Chen, L.; Zhang, P.; Ma, H.; Yuan, C.; Yan, S.; et al. RECQL4, negatively regulated by miR-10a-5p, facilitates cell proliferation and invasion via MAFB in ovarian cancer. Front. Oncol. 2020, 10, 524128. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, K.; Wang, X.; Wang, H.; Wu, Q.; Wu, X.; Peng, J. Overexpression of RECQL4 is associated with poor prognosis in patients with gastric cancer. Oncol. Lett. 2018, 16, 5419–5425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Jin, J.; Liao, M.; Dang, W.; Chen, X.; Wu, Y.; Liao, W. Upregulation of RECQL4 expression predicts poor prognosis in hepatocellular carcinoma. Oncol. Lett. 2018, 15, 4248–4254. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Agarwal, D.; Abdel-Fatah, T.M.; Lu, H.; Croteau, D.L.; Moseley, P.; Aleskandarany, M.A.; Green, A.R.; Ball, G.; Rakha, E.A.; et al. RECQL4 helicase has oncogenic potential in sporadic breast cancers. J. Pathol. 2016, 238, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Upadhaya, S.; Yu, J.X.; Shah, M.; Correa, D.; Partridge, T.; Campbell, J. The clinical pipeline for cancer cell therapies. Nat. Rev. Drug Discov. 2021, 20, 503–504. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Lee, J.; Wang, H.; Miller, J.; Reik, A.; Gregory, P.D.; Dean, A.; Blobel, G.A. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012, 149, 1233–1244. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Rupon, J.W.; Krivega, I.; Breda, L.; Motta, I.; Jahn, K.S.; Reik, A.; Gregory, P.D.; Rivella, S.; Dean, A.; et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 2014, 158, 849–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, S.L.; Mariano, N.C.; Bermudez, A.; Arruda, N.L.; Wu, F.; Luo, Y.; Shankar, G.; Jia, L.; Chen, H.; Hu, J.F.; et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 2017, 8, 15993. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Pauklin, S. Revisiting 3D chromatin architecture in cancer development and progression. Nucleic Acids Res. 2020, 48, 10632–10647. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Servant, N.; Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.J.; Vert, J.P.; Heard, E.; Dekker, J.; Barillot, E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015, 16, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lareau, C.A.; Aryee, M.J. Hichipper: A preprocessing pipeline for calling DNA loops from HiChIP data. Nat. Methods 2018, 15, 155–156. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Sample Class | NO. | Sample | Data Source |
|---|---|---|---|
| Normal Colon | 1 | E075-H3K27ac.broadPeak | Roadmap |
| 2 | E101-H3K27ac.broadPeak | Roadmap | |
| 3 | E102-H3K27ac.broadPeak | Roadmap | |
| 4 | E106-H3K27ac.broadPeak | Roadmap | |
| 5 | ENCFF116PPS | ENCODE | |
| 6 | ENCFF154YYF | ENCODE | |
| 7 | ENCFF193RGE | ENCODE | |
| 8 | ENCFF345LME | ENCODE | |
| 9 | ENCFF389IUW | ENCODE | |
| 10 | ENCFF779JEH | ENCODE | |
| Colorectal Cancer | 1 | V400_H3K27ac | GSE36204 |
| 2 | V429_H3K27ac | GSE36204 | |
| 3 | V503_H3K27ac | GSE36204 | |
| 4 | V9M_H3K27ac | GSE36204 | |
| 5 | ENCFF436VPF | ENCODE | |
| 6 | ENCFF570UFJ | ENCODE | |
| 7 | ENCFF720SPY | ENCODE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Ma, Y.; Bi, J.; Wang, W.; He, A.; Su, G.; Zhao, Z.; Shi, J.; Zhang, L. Regulation Network of Colorectal-Cancer-Specific Enhancers in the Progression of Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 8337. https://doi.org/10.3390/ijms22158337
Chen B, Ma Y, Bi J, Wang W, He A, Su G, Zhao Z, Shi J, Zhang L. Regulation Network of Colorectal-Cancer-Specific Enhancers in the Progression of Colorectal Cancer. International Journal of Molecular Sciences. 2021; 22(15):8337. https://doi.org/10.3390/ijms22158337
Chicago/Turabian StyleChen, Bohan, Yiping Ma, Jinfang Bi, Wenbin Wang, Anshun He, Guangsong Su, Zhongfang Zhao, Jiandang Shi, and Lei Zhang. 2021. "Regulation Network of Colorectal-Cancer-Specific Enhancers in the Progression of Colorectal Cancer" International Journal of Molecular Sciences 22, no. 15: 8337. https://doi.org/10.3390/ijms22158337
APA StyleChen, B., Ma, Y., Bi, J., Wang, W., He, A., Su, G., Zhao, Z., Shi, J., & Zhang, L. (2021). Regulation Network of Colorectal-Cancer-Specific Enhancers in the Progression of Colorectal Cancer. International Journal of Molecular Sciences, 22(15), 8337. https://doi.org/10.3390/ijms22158337





