Effects of Lipidation on a Proline-Rich Antibacterial Peptide
Abstract
:1. Introduction
2. Results
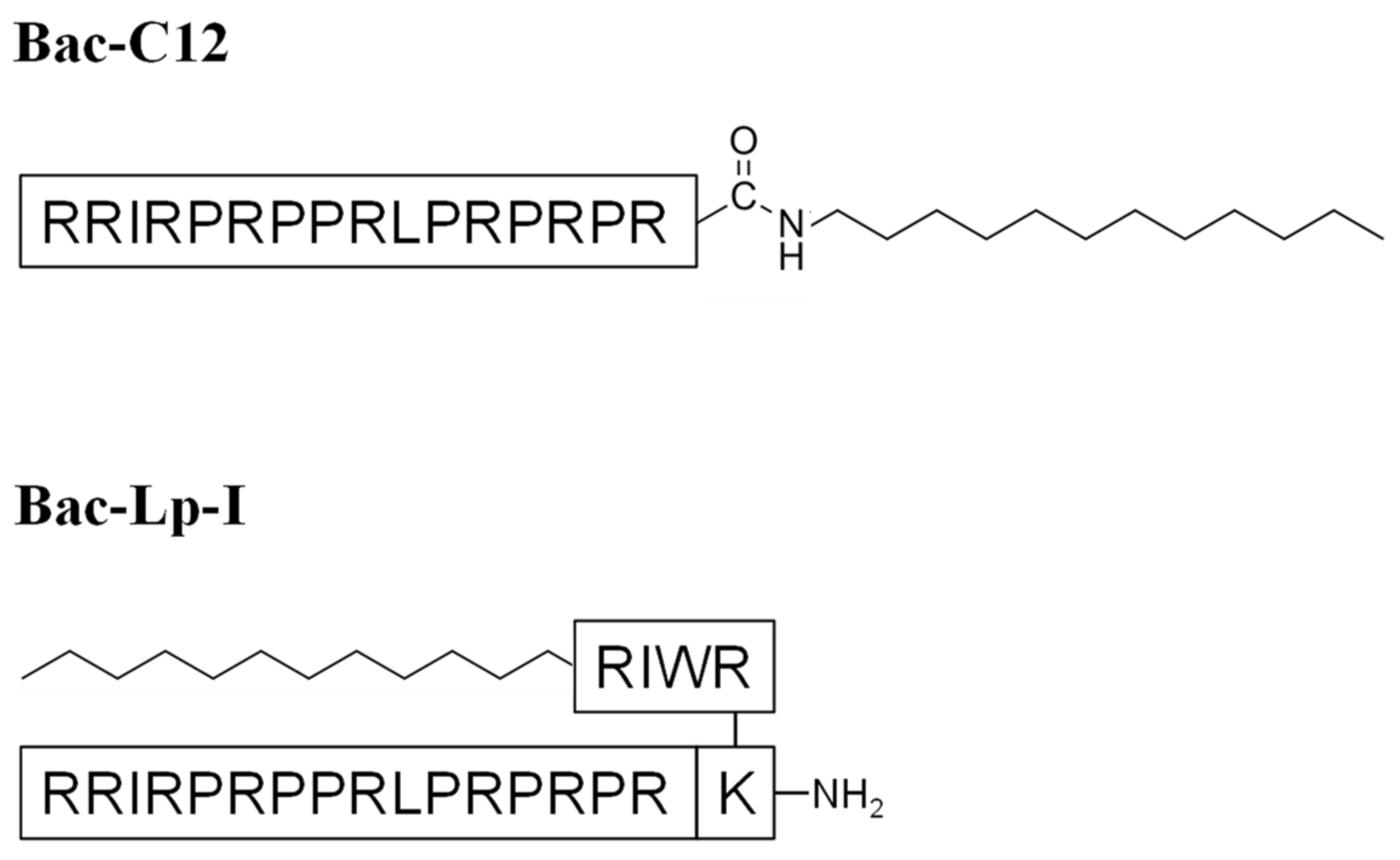
2.1. Bac-C12 and Bac-Lp-I Design
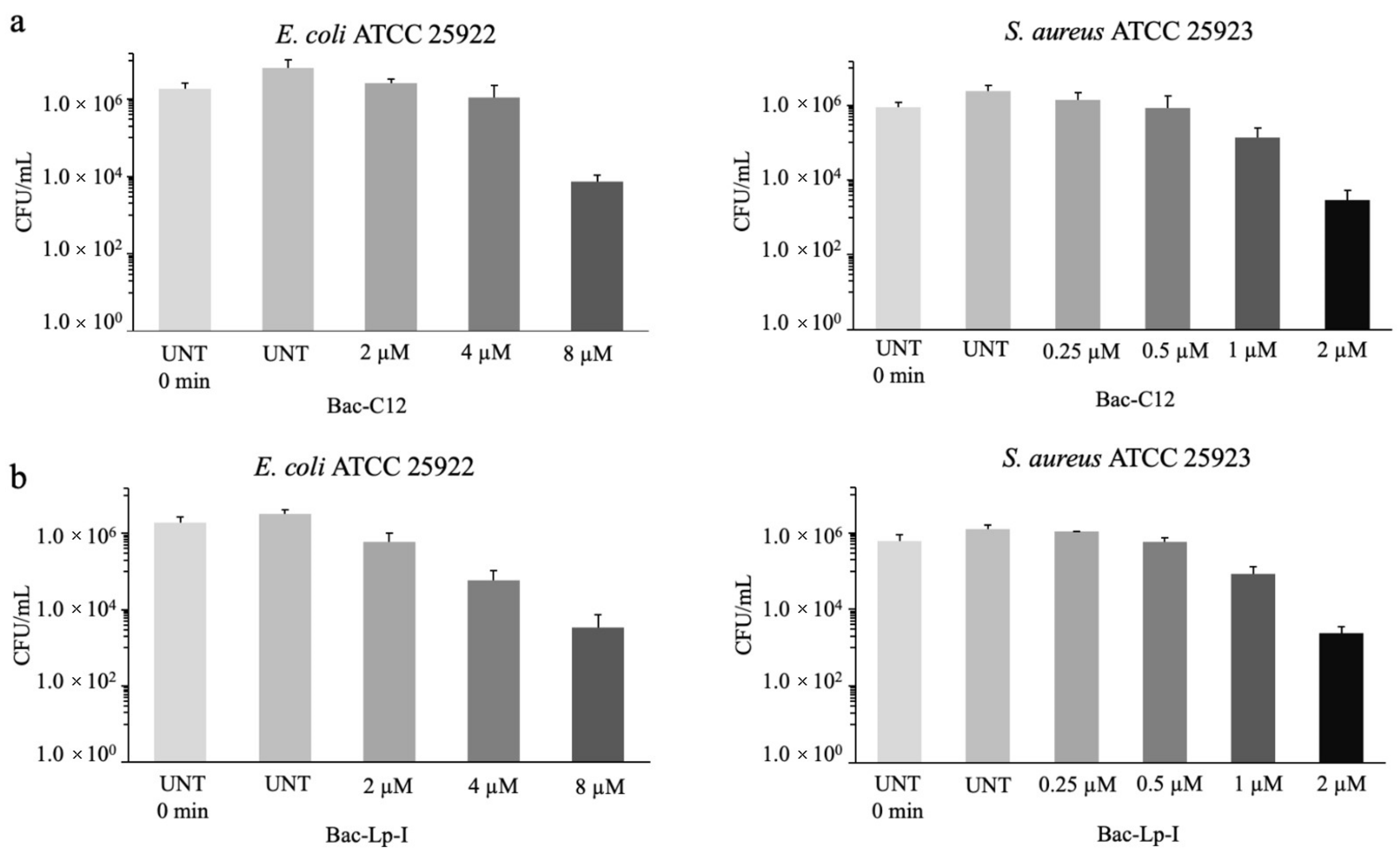
2.2. Antimicrobial Activity of Bac-C12 and Bac-Lp-I
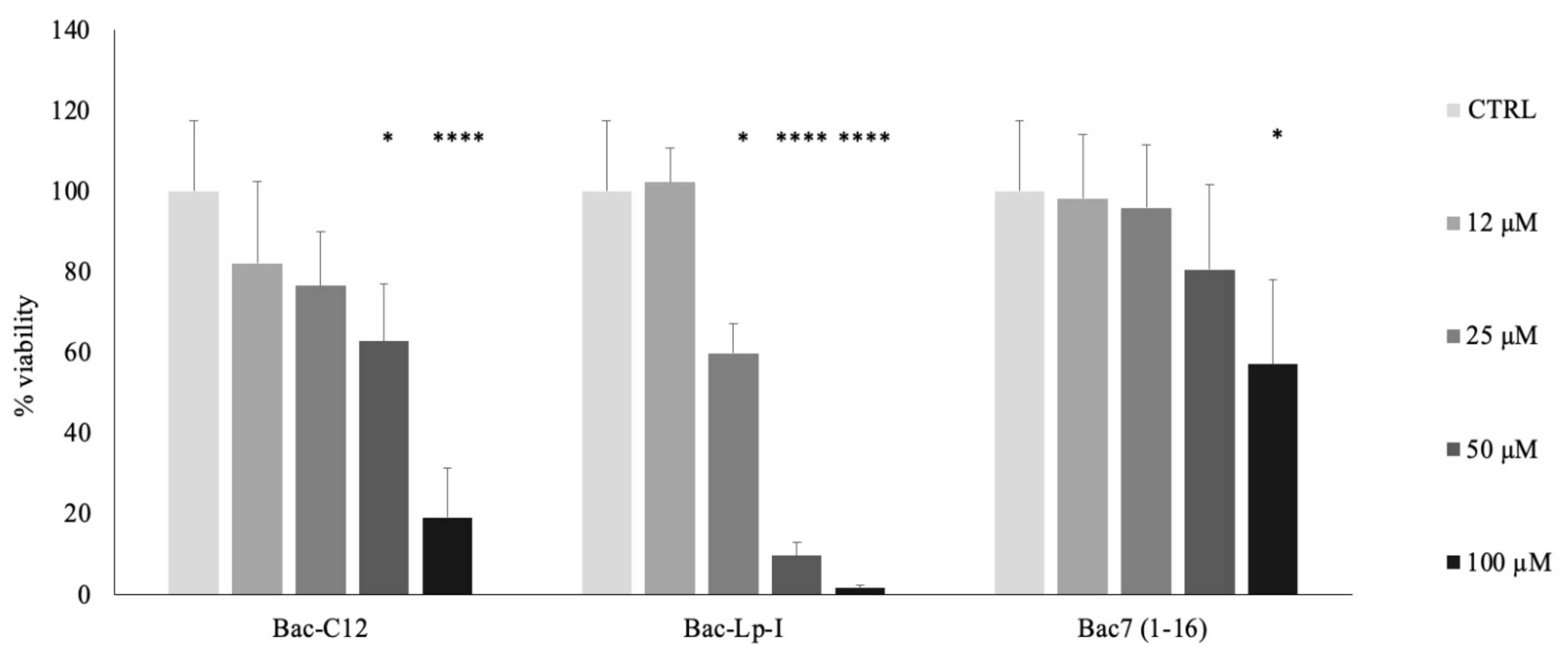
2.3. Evaluation of Cytotoxicity of Bac-C12 and Bac-Lp-I
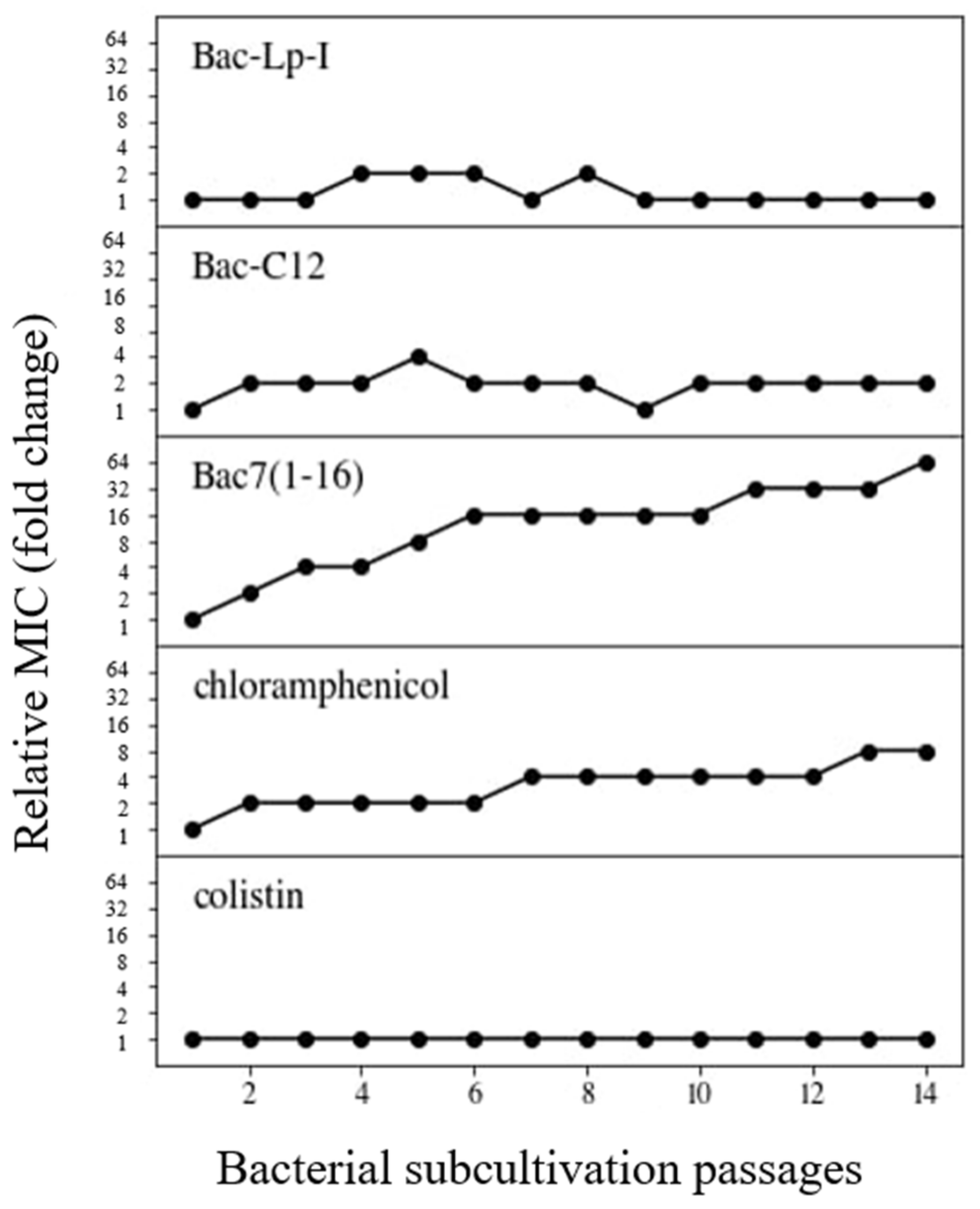
2.4. Resistance Selection in E. coli by Bac-C12 and Bac-Lp-I
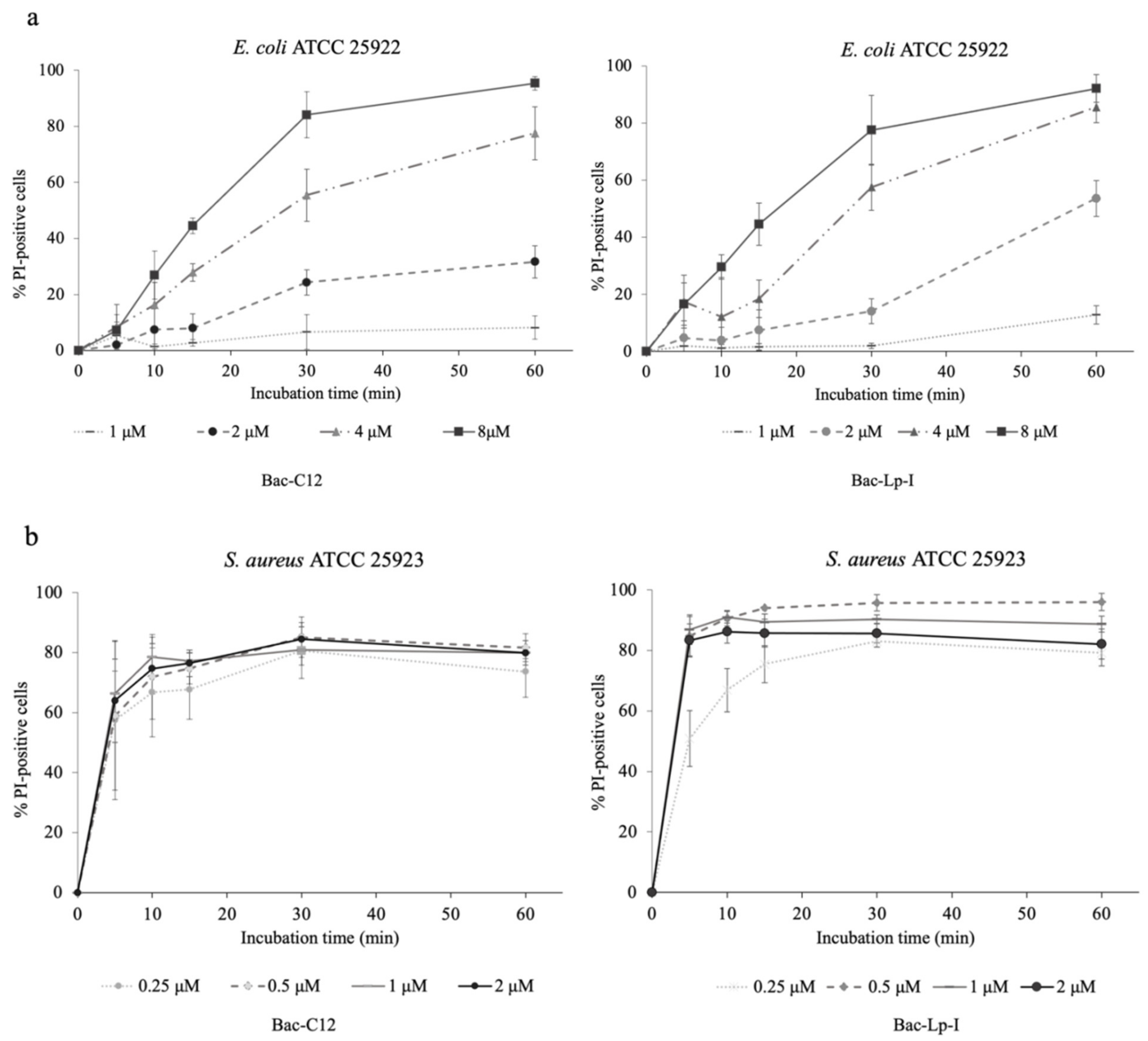
2.5. Assessment of the Integrity of the Bacterial Cell Membrane
3. Discussion
4. Materials and Methods
4.1. Peptides Synthesis
4.2. Bacterial Culture
4.3. Evaluation of the Antimicrobial Activity
4.4. In Vitro Toxicity Assays
4.5. Membrane Integrity Analysis
4.6. Resistance Selection by Serial Passages
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPs | Antimicrobial peptides |
| CFU | Colony-forming unit |
| CLSI | Clinical & Laboratory Standards Institute |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DMSO | Dimethyl sulfoxide |
| FBS | Fetal bovine serum |
| hRBCs | Human red blood cells |
| MBC | Minimal bactericidal concentration |
| MEC-1 | Human chronic lymphocytic leukemic B line |
| MHB | Mueller–Hinton broth |
| MIC | Minimum inhibitory concentration |
| MTT | Tetrazolium salts test |
| PBS | Phosphate-buffered saline |
| PI | Propidium iodide |
| PrAMPs | Proline-rich antimicrobial peptides |
| USCLs | Ultrashort cationic lipopeptides |
References
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miano, T.A.; Lautenbach, E.; Wilson, F.P.; Guo, W.; Borovskiy, Y.; Hennessy, S. Attributable Risk and Time Course of Colistin-Associated Acute Kidney Injury. Clin. J. Am. Soc. Nephrol. 2018, 13, 542–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in Development of Antimicrobial Peptidomimetics as Potential Drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahar, A.A.; Ren, D. Antimicrobial Peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Boto, A.; De La Lastra, J.M.P.; González, C.C. The Road from Host-Defense Peptides to a New Generation of Antimicrobial Drugs. Molecules 2018, 23, 311. [Google Scholar] [CrossRef] [Green Version]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Shai, Y. Mode of Action of Membrane Active Antimicrobial Peptides. Peptide Sci. 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2009, 276, 6483–6496. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, P. Multifunctional Host Defense Peptides: Intracellular-Targeting Antimicrobial Peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef]
- Li, W.; Tailhades, J.; O′Brien-Simpson, N.M.; Separovic, F.; Otvos, L.; Hossain, M.A.; Wade, J.D. Proline-rich antimicrobial peptides: Potential therapeutics against antibiotic-resistant bacteria. Amino Acids 2014, 46, 2287–2294. [Google Scholar] [CrossRef]
- Otvos, J.L. The short proline-rich antibacterial peptide family. Cell. Mol. Life Sci. 2002, 59, 1138–1150. [Google Scholar] [CrossRef]
- Scocchi, M.; Tossi, A.; Gennaro, R. Proline-rich antimicrobial peptides: Converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. 2011, 68, 2317–2330. [Google Scholar] [CrossRef]
- Berthold, N.; Hoffmann, R. Cellular uptake of apidaecin 1b and related analogs in Gram-negative bacteria reveals novel antibacterial mechanism for proline-rich antimicrobial peptides. Protein Pept. Lett. 2014, 21, 391–398. [Google Scholar] [CrossRef]
- Krizsan, A.; Knappe, D.; Hoffmann, R. Influence of theyjiL-mdtMGene Cluster on the Antibacterial Activity of Proline-Rich Antimicrobial Peptides Overcoming Escherichia coli Resistance Induced by the Missing SbmA Transporter System. Antimicrob. Agents Chemother. 2015, 59, 5992–5998. [Google Scholar] [CrossRef] [Green Version]
- Mattiuzzo, M.; Bandiera, A.; Gennaro, R.; Benincasa, M.; Pacor, S.; Antcheva, N.; Scocchi, M. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 2007, 66, 151–163. [Google Scholar] [CrossRef]
- Graf, M.; Mardirossian, M.; Nguyen, F.; Seefeldt, A.C.; Guichard, G.; Scocchi, M.; Innis, C.A.; Wilson, D.N. Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep. 2017, 34, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Grzela, R.; Giglione, C.; Meinnel, T.; Gennaro, R.; Mergaert, P.; Scocchi, M. The Host Antimicrobial Peptide Bac71-35 Binds to Bacterial Ribosomal Proteins and Inhibits Protein Synthesis. Chem. Biol. 2014, 21, 1639–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.N.; Lomakin, I.B.; Gagnon, M.G.; Steitz, T.A. The Mechanism of Inhibition of Protein Synthesis by the Proline-Rich Peptide Oncocin. Nat. Struct. Mol. Biol. 2015, 22, 466–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seefeldt, A.C.; Nguyen, F.; Antunes, S.; Pérébaskine, N.; Graf, M.; Arenz, S.; Inampudi, K.K.; Douat, C.; Guichard, G.; Wilson, D.N.; et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 2015, 22, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, A.C.; Graf, M.; Pérébaskine, N.; Nguyen, F.; Arenz, S.; Mardirossian, M.; Scocchi, M.; Wilson, D.N.; Innis, C.A. Structure of the mammalian antimicrobial peptide Bac7(1–16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016, 44, 2429–2438. [Google Scholar] [CrossRef] [Green Version]
- Zahn, M.; Berthold, N.; Kieslich, B.; Knappe, D.; Hoffmann, R.; Sträter, N. Structural Studies on the Forward and Reverse Binding Modes of Peptides to the Chaperone DnaK. J. Mol. Biol. 2013, 425, 2463–2479. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, M.; Scocchi, M.; Podda, E.; Skerlavaj, B.; Dolzani, L.; Gennaro, R. Antimicrobial Activity of Bac7 Fragments against Drug-Resistant Clinical Isolates. Peptides 2004, 25, 2055–2061. [Google Scholar] [CrossRef]
- Benincasa, M.; Pelillo, C.; Zorzet, S.; Garrovo, C.; Biffi, S.; Gennaro, R.; Scocchi, M. The Proline-Rich Peptide Bac7(1-35) Reduces Mortality from Salmonella Typhimurium in a Mouse Model of Infection. BMC Microbiol. 2010, 10, 178. [Google Scholar] [CrossRef] [Green Version]
- Dolzani, L.; Milan, A.; Scocchi, M.; Lagatolla, C.; Bressan, R.; Benincasa, M. Sub-MIC effects of a proline-rich antibacterial peptide on clinical isolates of Acinetobacter baumannii. J. Med. Microbiol. 2019, 68, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Sola, R.; Beckert, B.; Valencic, E.; Collis, D.W.P.; Borišek, J.; Armas, F.; Di Stasi, A.; Buchmann, J.; Syroegin, E.A.; et al. Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens. J. Med. Chem. 2020, 63, 9590–9602. [Google Scholar] [CrossRef] [PubMed]
- Melander, R.J.; Zurawski, D.V.; Melander, C. Narrow-spectrum antibacterial agents. MedChemComm 2018, 9, 12–21. [Google Scholar] [CrossRef]
- Kollef, M.H. Broad-Spectrum Antimicrobials and the Treatment of Serious Bacterial Infections: Getting It Right Up Front. Clin. Infect. Dis. 2008, 47, S3–S13. [Google Scholar] [CrossRef] [Green Version]
- Ting, D.S.J.; Beuerman, R.W.; Dua, H.S.; Lakshminarayanan, R.; Mohammed, I. Strategies in Translating the Therapeutic Potentials of Host Defense Peptides. Front. Immunol. 2020, 11, 983. [Google Scholar] [CrossRef]
- Laverty, G.; McLaughlin, M.; Shaw, C.; Gorman, S.P.; Gilmore, B.F. Antimicrobial Activity of Short, Synthetic Cationic Lipopeptides. Chem. Biol. Drug Des. 2010, 75, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Chu-Kung, A.F.; Bozzelli, K.N.; Lockwood, N.A.; Haseman, J.R.; Mayo, K.H.; Tirrell, M.V. Promotion of Peptide Antimicrobial Activity by Fatty Acid Conjugation. Bioconjugate Chem. 2004, 15, 530–535. [Google Scholar] [CrossRef]
- Li, W.; Separovic, F.; O′Brien-Simpson, N.M.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef] [PubMed]
- Serrano, G.N.; Zhanel, G.G.; Schweizer, F. Antibacterial Activity of Ultrashort Cationic Lipo-β-Peptides. Antimicrob. Agents Chemother. 2009, 53, 2215–2217. [Google Scholar] [CrossRef] [Green Version]
- Armas, F.; Pacor, S.; Ferrari, E.; Guida, F.; Pertinhez, T.A.; Romani, A.A.; Scocchi, M.; Benincasa, M. Design, Antimicrobial Activity and Mechanism of Action of Arg-Rich Ultra-Short Cationic Lipopeptides. PLoS ONE 2019, 14, e0212447. [Google Scholar] [CrossRef] [Green Version]
- Domalaon, R.; Sanchak, Y.; Koskei, L.C.; Lyu, Y.; Zhanel, G.G.; Arthur, G.; Schweizer, F. Short Proline-Rich Lipopeptide Potentiates Minocycline and Rifampin against Multidrug- and Extensively Drug-Resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [Green Version]
- Hong, H.A.; Huang, J.-M.; Khaneja, R.; Hiep, L.V.; Urdaci, M.C.; Cutting, S.M. The safety ofBacillus subtilisandBacillus indicusas food probiotics. J. Appl. Microbiol. 2008, 105, 510–520. [Google Scholar] [CrossRef]
- Bluhm, M.E.; Knappe, D.; Hoffmann, R. Structure-activity relationship study using peptide arrays to optimize Api137 for an increased antimicrobial activity against Pseudomonas aeruginosa. Eur. J. Med. Chem. 2015, 103, 574–582. [Google Scholar] [CrossRef]
- Knappe, D.; Adermann, K.; Hoffmann, R. Oncocin Onc72 is efficacious against antibiotic-susceptible Klebsiella pneumoniaeATCC 43816 in a murine thigh infection model. Peptide Sci. 2015, 104, 707–711. [Google Scholar] [CrossRef]
- Kopeikin, P.M.; Zharkova, M.S.; Kolobov, A.A.; Smirnova, M.P.; Sukhareva, M.S.; Umnyakova, E.S.; Kokryakov, V.N.; Orlov, D.S.; Milman, B.L.; Balandin, S.V.; et al. Caprine Bactenecins as Promising Tools for Developing New Antimicrobial and Antitumor Drugs. Front. Cell. Infect. Microbiol. 2020, 10, 552905. [Google Scholar] [CrossRef] [PubMed]
- Mardirossian, M.; Sola, R.; Beckert, B.; Collis, D.W.P.; Di Stasi, A.; Armas, F.; Hilpert, K.; Wilson, D.N.; Scocchi, M. Proline-Rich Peptides with Improved Antimicrobial Activity against E. coli, K. pneumoniae, and A. baumannii. ChemMedChem 2019, 14, 2025–2033. [Google Scholar] [CrossRef] [Green Version]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Pränting, M.; Negrea, A.; Rhen, M.; Andersson, D.I. Mechanism and Fitness Costs of PR-39 Resistance in Salmonella enterica Serovar Typhimurium LT2. Antimicrob. Agents Chemother. 2008, 52, 2734–2741. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, M.G.; Roy, R.N.; Lomakin, I.B.; Florin, T.; Mankin, A.S.; Steitz, T.A. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016, 44, 2439–2450. [Google Scholar] [CrossRef] [Green Version]
- Mardirossian, M.; Barrière, Q.; Timchenko, T.; Müller, C.; Pacor, S.; Mergaert, P.; Scocchi, M.; Wilsona, D.N. Fragments of the Nonlytic Proline-Rich Antimicrobial Peptide Bac5 Kill Escherichia coli Cells by Inhibiting Protein Synthesis. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 2016, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Podda, E.; Benincasa, M.; Pacor, S.; Micali, F.; Mattiuzzo, M.; Gennaro, R.; Scocchi, M. Dual Mode of Action of Bac7, a Proline-Rich Antibacterial Peptide. Biochim. Biophys. Acta Gen. Subj. 2006, 1760, 1732–1740. [Google Scholar] [CrossRef]
- Rounds, T.; Straus, S.K. Lipidation of Antimicrobial Peptides as a Design Strategy for Future Alternatives to Antibiotics. Int. J. Mol. Sci. 2020, 21, 9692. [Google Scholar] [CrossRef]
- Grimsey, E.; Collis, D.W.; Mikut, R.; Hilpert, K. The effect of lipidation and glycosylation on short cationic antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183195. [Google Scholar] [CrossRef]
- Albada, H.B.; Prochnow, P.; Bobersky, S.; Langklotz, S.; Schriek, P.; Bandow, J.E.; Metzler-Nolte, N. Tuning the Activity of a Short Arg-Trp Antimicrobial Peptide by Lipidation of a C- or N-Terminal Lysine Side-Chain. ACS Med. Chem. Lett. 2012, 3, 980–984. [Google Scholar] [CrossRef] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardirossian, M.; Pompilio, A.; Degasperi, M.; Runti, G.; Pacor, S.; Di Bonaventura, G.; Scocchi, M. D-BMAP18 Antimicrobial Peptide Is Active In vitro, Resists to Pulmonary Proteases but Loses Its Activity in a Murine Model of Pseudomonas aeruginosa Lung Infection. Front. Chem. 2017, 5, 40. [Google Scholar] [CrossRef] [PubMed]
| Bacteria Strain | MIC (μM) | MBC (μM) | ||||
|---|---|---|---|---|---|---|
| Bac7(1-16) | Lp-I | Bac-C12 | Bac-Lp-I | Bac-C12 | Bac-Lp-I | |
| B. subtilis DSMZ 4181 | 1 | 4 | 4 | 2 | 4 | 2 |
| E. faecalis ATCC 29212 | >64 | 8 | 8 | 4 | 8 | 8 |
| S. aureus ATCC 25923 | >64 | 4 | 4 | 2 | 8 | 4 |
| S. aureus ATCC 29213 | >64 | 4 | 2 | 4 | 4 | 4 |
| S. epidermidis ATCC 12228 | >64 | 8 | 2 | 2 | 2–4 | 2 |
| A. baumannii ATCC 17978 | 16 | 32 | 8 | 2 | 16 | 4 |
| A. baumannii ATCC 19606 | 64 | 32 | 8 | 4 | 8 | 4 |
| B. cepacia J2315 | >64 | 16 | 2 | 8 | n.d. | 8 |
| E. coli ATCC 25922 | 2 | 8 | 8 | 4 | 8 | 8 |
| E. coli BW25113 | 4 | 8 | 4 | 4 | 4 | 16 |
| E. coli BW25113ΔsbmA | 32 | n.d. | 4 | 4 | 4 | 4 |
| E. coli O18K1H7 # | >64 | 16 | 8 | 8 | 8 | 16 |
| K. pneumoniae ATCC 700603 | 8 | 64 | 8 | 4 | 8 | 4 |
| K. pneumoniae ATCC 13883 | 4 | >64 | 4 | 4 | 32 | 8 |
| P. aeruginosa ATCC 27853 | >64 | 16 | 8 | 8 | 16 | 16 |
| P. aeruginosa PAO1 | >64 | 16 | 8 | 8 | >64 | 16 |
| S. typhimurium ATCC 14028 | 2 | 32 | 4 | 2 | 8–16 | 4 |
| S. maltophilia ATCC 13637 | >64 | 8 | 2 | 8 | 4 | 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armas, F.; Di Stasi, A.; Mardirossian, M.; Romani, A.A.; Benincasa, M.; Scocchi, M. Effects of Lipidation on a Proline-Rich Antibacterial Peptide. Int. J. Mol. Sci. 2021, 22, 7959. https://doi.org/10.3390/ijms22157959
Armas F, Di Stasi A, Mardirossian M, Romani AA, Benincasa M, Scocchi M. Effects of Lipidation on a Proline-Rich Antibacterial Peptide. International Journal of Molecular Sciences. 2021; 22(15):7959. https://doi.org/10.3390/ijms22157959
Chicago/Turabian StyleArmas, Federica, Adriana Di Stasi, Mario Mardirossian, Antonello A. Romani, Monica Benincasa, and Marco Scocchi. 2021. "Effects of Lipidation on a Proline-Rich Antibacterial Peptide" International Journal of Molecular Sciences 22, no. 15: 7959. https://doi.org/10.3390/ijms22157959
APA StyleArmas, F., Di Stasi, A., Mardirossian, M., Romani, A. A., Benincasa, M., & Scocchi, M. (2021). Effects of Lipidation on a Proline-Rich Antibacterial Peptide. International Journal of Molecular Sciences, 22(15), 7959. https://doi.org/10.3390/ijms22157959






