Toward the Specificity of Bare Nanomaterial Surfaces for Protein Corona Formation
Abstract
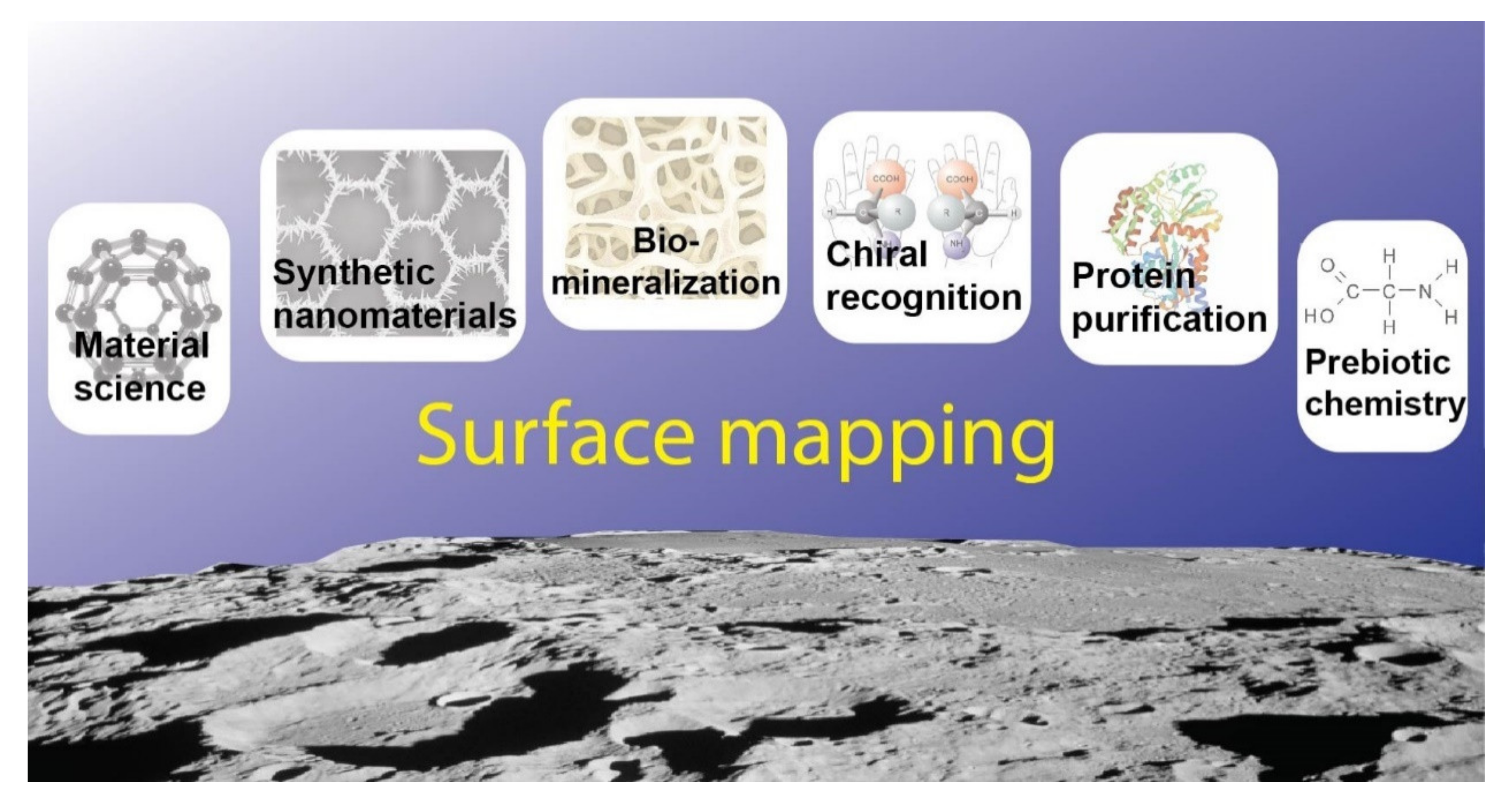
1. Introduction
2. The Role of Protein Selectivity in Biomedical Applications of Nanomaterials
3. Carbon-Based Nanomaterials
4. Molecular Biomimetics and Biomineralization
5. Prebiotic Chemistry and Chirality Selection
6. Protein and Peptide Purification by Bare Nanomaterials
7. Surface Energy and Protein Corona
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | Accelerated blood clearance |
| CNMs | carbon based nanomaterials |
| CNTs | carbon nanotubes |
| CQDs | carbon quantum dots |
| MWCNTs | carboxylated multiwalled CNTs |
| PEG | polyethylene glycol |
| AuNPs | gold nanoparticles |
| γ-Fe2O3 | maghemite |
| Fe3O4 | magnetite |
| IONPs | iron oxide nanoparticles |
| MSPs | mesoporous silica nanoparticles |
| MEAM | modified embedded atom method |
| GEPIs | genetically engineered proteins for inorganics |
| GO | graphene oxide |
| GFP | green fluorescent protein |
| EISM | effective implicit surface model |
| PEG | polyethylene glycol |
| SWCNTs | single-walled CNTs |
| TEG | tetra(ethylene glycol) |
References
- Aggarwal, P.; Hall, J.B.; McLeland, C.B.; Dobrovolskaia, M.A.; McNeil, S.E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 2009, 61, 428–437. [Google Scholar] [CrossRef]
- Kang, B.; Okwieka, P.; Schöttler, S.; Seifert, O.; Kontermann, R.E.; Pfizenmaier, K.; Musyanovych, A.; Meyer, R.; Diken, M.; Sahin, U.; et al. Tailoring the stealth properties of biocompatible polysaccharide nanocontainers. Biomaterials 2015, 49, 125–134. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Bertrand, N.; Zope, H.; Farokhzad, O. Emerging understanding of the protein corona at the nano-bio interfaces. Nano Today 2016, 11, 817–832. [Google Scholar] [CrossRef]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle-protein corona using methods to quntify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Lynch, I.; Salvati, A.; Dawson, K.A. Protein-nanoparticle interactions: What does the cell see? Nat. Nanotechnol. 2009, 4, 546–547. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, A.; Fenaroli, F.; Monopoli, M.P.; Åberg, C.; Dawson, K.A.; Salvati, A. Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano 2012, 6, 5845–5857. [Google Scholar] [CrossRef] [PubMed]
- Moyano, D.F.; Saha, K.; Prakash, G.; Yan, B.; Kong, H.; Yazdani, M.; Rotello, V.M. Fabrication of Corona-Free Nanoparticles with Tunable Hydrophobicity. ACS Nano 2014, 8, 6748–6755. [Google Scholar] [CrossRef]
- Saha, K.; Rahimi, M.; Yazdani, M.; Kim, S.T.; Moyano, D.F.; Hou, S.; Das, R.; Mout, R.; Rezaee, F.; Mahmoudi, M.; et al. Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona. ACS Nano 2016, 10, 4421–4430. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Lin, J.; Huang, P.; Chen, X. Development of endogenous enzyme-responsive nanomaterials for theranostics. Chem. Soc. Rev. 2018, 47, 5554–5573. [Google Scholar] [CrossRef] [PubMed]
- De la Rica, R.; Aili, D.; Stevens, M.M. Enzyme-responsive nanoparticles for drug release and diagnostics. Adv. Drug Deliv. Rev. 2012, 64, 967–978. [Google Scholar] [CrossRef]
- Hong, R.; Fischer, N.; Verma, A.; Goodman, C.; Emrick, T.; Rotello, V.M. Control of Protein Structure and Function through Surface Recognition by Tailored Nanoparticle Scaffolds. J. Am. Chem. Soc. 2004, 126, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Worrall, J.W.E.; Verma, A.; Yan, H.; Rotello, V.M. “Cleaning” of nanoparticle inhibitors via proteolysis of adsorbed proteins. Chem. Commun. 2006, 2338–2340, 2338–2340. [Google Scholar] [CrossRef] [PubMed]
- Jakschitz, T.A.E.; Rode, B.M. Chemical evolution from simple inorganic compounds to chiral peptides. Chem. Soc. Rev. 2012, 41, 5484–5489. [Google Scholar] [CrossRef]
- Dong, L.; Luo, Q.; Cheng, K.; Shi, H.; Wang, Q.; Weng, W.; Han, W.-Q. Facet-Specific Assembly of Proteins on SrTiO3 Polyhedral Nanocrystals. Sci. Rep. 2014, 4, 5084. [Google Scholar] [CrossRef]
- Lu, X.; Xu, P.; Ding, H.-M.; Yu, Y.-S.; Huo, D.; Ma, Y.-Q. Tailoring the component of protein corona via simple chemistry. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Piloni, A.; Wong, C.K.; Chen, F.; Lord, M.; Walther, A.; Stenzel, M.H. Surface roughness influences the protein corona formation of glycosylated nanoparticles and alter their cellular uptake. Nanoscale 2019, 11, 23259–23267. [Google Scholar] [CrossRef] [PubMed]
- Hirsh, S.L.; McKenzie, D.R.; Nosworthy, N.J.; Denman, J.A.; Sezerman, O.U.; Bilek, M. The Vroman effect: Competitive protein exchange with dynamic multilayer protein aggregates. Colloids Surf. B Biointerfaces 2013, 103, 395–404. [Google Scholar] [CrossRef]
- Walkey, C.D.; Chan, W.C.W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef]
- Schöttler, S.; Landfester, K.; Mailänder, V. Controlling the Stealth Effect of Nanocarriers through Understanding the Protein Corona. Angew. Chem. Int. Ed. 2016, 55, 8806–8815. [Google Scholar] [CrossRef]
- Berdon, A.L.B.; Pozzi, D.; Caracciolo, G.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Riccioli, A.; Palchetti, S.; Laganà, A. Time Evolution of Nanoparticle–Protein Corona in Human Plasma: Relevance for Targeted Drug Delivery. Langmuir 2013, 29, 6485–6494. [Google Scholar] [CrossRef]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef]
- Bellion, M.; Santen, L.; Mantz, H.; Hähl, H.; Quinn, A.; Nagel, A.; Gilow, C.; Weitenberg, C.; Schmitt, Y.; Jacobs, K. Protein adsorption on tailored substrates: Long-range forces and conformational changes. J. Physics: Condens. Matter 2008, 20, 404226–404237. [Google Scholar] [CrossRef]
- Secundo, F. Conformational changes of enzymes upon immobilisation. Chem. Soc. Rev. 2013, 42, 6250–6261. [Google Scholar] [CrossRef]
- Marruecos, D.F.; Schwartz, D.K.; Kaar, J.L. Impact of surface interactions on protein conformation. Curr. Opin. Colloid Interface Sci. 2018, 38, 45–55. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Nidetzky, B. On the relationship between structure and catalytic effectiveness in solid surface-immobilized enzymes: Advances in methodology and the quest for a single-molecule perspective. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2020, 1868, 140333. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, C. Comparing proteins by their internal dynamics: Exploring structure–function relationships beyond static structural alignments. Phys. Life Rev. 2013, 10, 1–26. [Google Scholar] [CrossRef]
- Orozco, M. A theoretical view of protein dynamics. Chem. Soc. Rev. 2014, 43, 5051–5066. [Google Scholar] [CrossRef] [PubMed]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.V.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Maiolo, D.; del Pino, P.; Metrangolo, P.; Parak, W.J.; Baldelli Bombelli, F. Nanomedicine delivery: Does protein corona route to the target or off road? Nanomedicine 2015, 10, 3231–3247. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Kim, H.S.; Palanikumar, L.; Go, E.M.; Jana, B.; Park, S.A.; Kim, H.Y.; Kim, K.; Seo, J.K.; Kwak, S.K.; et al. Cloaking nanoparticles with protein corona shield for targeted drug delivery. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Ishida, T.; Ichihara, M.; Wang, X.; Yamamoto, K.; Kimura, J.; Majima, E.; Kiwada, H. Injection of PEGylated liposomes in rats elicits PEG-specific IgM, which is responsible for rapid elimination of a second dose of PEGylated liposomes. J. Control. Release 2006, 112, 15–25. [Google Scholar] [CrossRef]
- Wang, X.; Ishida, T.; Kiwada, H. Anti-PEG IgM elicited by injection of liposomes is involved in the enhanced blood clearance of a subsequent dose of PEGylated liposomes. J. Control. Release 2007, 119, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kiwada, H. Accelerated blood clearance (ABC) phenomenon upon repeated injection of PEGylated liposomes. Int. J. Pharm. 2008, 354, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar] [PubMed]
- Hatakeyama, H.; Akita, H.; Harashima, H. The Polyethyleneglycol Dilemma: Advantage and Disadvantage of PEGylation of Liposomes for Systemic Genes and Nucleic Acids Delivery to Tumors. Biol. Pharm. Bull. 2013, 36, 892–899. [Google Scholar] [CrossRef]
- Schöttler, S.; Becker, G.; Winzen, S.; Steinbach, T.; Mohr, K.; Landfester, K.; Mailänder, V.; Wurm, F.R. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. [Google Scholar] [CrossRef]
- Ke, P.C.; Lin, S.; Parak, W.J.; Davis, T.P.; Caruso, F. A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. [Google Scholar] [CrossRef]
- Nienhaus, K.; Wang, H.; Nienhaus, G. Nanoparticles for biomedical applications: Exploring and exploiting molecular interactions at the nano-bio interface. Mater. Today Adv. 2020, 5, 100036. [Google Scholar] [CrossRef]
- Khanbeigi, R.A.; Abelha, T.F.; Woods, A.; Rastoin, O.; Harvey, R.D.; Jones, M.-C.; Forbes, B.; Green, M.A.; Collins, H.; Dailey, L.A. Surface Chemistry of Photoluminescent F8BT Conjugated Polymer Nanoparticles Determines Protein Corona Formation and Internalization by Phagocytic Cells. Biomacromolecules 2015, 16, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Limo, M.J.; Rabada, A.S.; Boix, E.; Thota, V.; Westcott, Z.C.; Puddu, V.; Perry, C.C. Interactions between Metal Oxides and Biomolecules: From Fundamental Understanding to Applications. Chem. Rev. 2018, 118, 11118–11193. [Google Scholar] [CrossRef]
- Lynch, I.; Cedervall, T.; Lundqvist, M.; Cabaleiro-Lago, C.; Linse, S.; Dawson, K.A. The nanoparticle–protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv. Colloid Interface Sci. 2007, 134–135, 167–174. [Google Scholar] [CrossRef]
- Sengupta, B.; Gregory, W.E.; Zhu, J.; Dasetty, S.; Karakaya, M.; Brown, J.M.; Rao, A.M.; Barrows, J.K.; Sarupria, S.; Podila, R. Influence of carbon nanomaterial defects on the formation of protein corona. RSC Adv. 2015, 5, 82395–82402. [Google Scholar] [CrossRef]
- Comparetti, E.J.; Pedrosa, V.; Kaneno, R. Carbon Nanotube as a Tool for Fighting Cancer. Bioconjug. Chem. 2018, 29, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Bai, W.; Raghavendra, A.; Podila, R. Defect density in multiwalled carbon nanotubes influences ovalbumin adsorption and promotes macrophage activation and CD4+ T-cell proliferation. Int. J. Nanomed. 2016, 11, 4357–4371. [Google Scholar] [CrossRef] [PubMed]
- Care, A.; Bergquist, P.L.; Sunna, A. Solid-binding peptides: Smart tools for nanobiotechnology. Trends Biotechnol. 2015, 33, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Humphreys, E.S.; Chung, S.-Y.; Delduco, D.F.; Lustig, S.R.; Wang, H.; Parker, K.N.; Rizzo, N.W.; Subramoney, S.; Chiang, Y.-M.; et al. Peptides with selective affinity for carbon nanotubes. Nat. Mater. 2003, 2, 196–200. [Google Scholar] [CrossRef]
- Calvaresi, M.; Zerbetto, F. The Devil and Holy Water: Protein and Carbon Nanotube Hybrids. Acc. Chem. Res. 2013, 46, 2454–2463. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.J.; Fritz, K.; Fu, S.; Brown, J.M.; Podila, R.; Shannahan, J.H. Variations in biocorona formation related to defects in the structure of single walled carbon nanotubes and the hyperlipidemic disease state. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Machova, I.; Hubalek, M.; Belinova, T.; Fucikova, A.; Stehlik, S.; Rezek, B.; Kalbacova, M.H. The bio-chemically selective interaction of hydrogenated and oxidized ultra-small nanodiamonds with proteins and cells. Carbon 2020, 162, 650–661. [Google Scholar] [CrossRef]
- Kumar, S.; Parekh, S.H. Linking graphene-based material physicochemical properties with molecular adsorption, structure and cell fate. Commun. Chem. 2020, 3, 1–11. [Google Scholar] [CrossRef]
- Kim, S.S.; Kuang, Z.; Ngo, Y.H.; Farmer, B.L.; Naik, R.R. Biotic–Abiotic Interactions: Factors that Influence Peptide–Graphene Interactions. ACS Appl. Mater. Interfaces 2015, 7, 20447–20453. [Google Scholar] [CrossRef]
- Henriques, P.C.; Pereira, A.T.; Pires, A.L.; Pereira, A.M.; Magalhães, F.D.; Gonçalves, I.C. Graphene Surfaces Interaction with Proteins, Bacteria, Mammalian Cells, and Blood Constituents: The Impact of Graphene Platelet Oxidation and Thickness. ACS Appl. Mater. Interfaces 2020, 12, 21020–21035. [Google Scholar] [CrossRef]
- Kim, S.N.; Kuang, Z.; Slocik, J.M.; Jones, S.E.; Cui, Y.; Farmer, B.L.; McAlpine, M.; Naik, R.R. Preferential Binding of Peptides to Graphene Edges and Planes. J. Am. Chem. Soc. 2011, 133, 14480–14483. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Jen, A.; Schulten, K.; Baneyx, F. Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar] [CrossRef]
- So, C.R.; Liu, J.; Fears, K.P.; Leary, D.H.; Golden, J.P.; Wahl, K.J. Self-Assembly of Protein Nanofibrils Orchestrates Calcite Step Movement through Selective Nonchiral Interactions. ACS Nano 2015, 9, 5782–5791. [Google Scholar] [CrossRef] [PubMed]
- Tamerler, C.; Sarikaya, M. Genetically Designed Peptide-Based Molecular Materials. ACS Nano 2009, 3, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, S.V.; Emami, F.S.; Berry, R.J.; Jones, S.E.; Naik, R.R.; Deschaume, O.; Heinz, H.; Perry, C.C. Chemistry of aqueous silica nanoparticle surfaces and the mechanism of selective peptide adsorption. J. Am. Chem. Soc. 2012, 134, 6244–6256. [Google Scholar] [CrossRef]
- Palafox-Hernandez, J.P.; Tang, Z.; Hughes, Z.E.; Li, Y.; Swihart, M.T.; Prasad, P.N.; Walsh, T.R.; Knecht, M.R. Comparative study of materials-binding peptide interactions with gold and silver surfaces and nanostructures: A thermodynamic basis for biological selectivity of inorganic materials. Chem. Mater. 2014, 26, 4960–4969. [Google Scholar] [CrossRef]
- Hughes, Z.E.; Kochandra, R.; Walsh, T.R. Facet-Specific Adsorption of Tripeptides at Aqueous Au Interfaces: Open Questions in Reconciling Experiment and Simulation. Langmuir 2017, 33, 3742–3754. [Google Scholar] [CrossRef] [PubMed]
- Hernández, B.; Tinacci, L.; Coïc, Y.-M.; Chenal, A.; Cohen, R.; Sanchez-Cortes, S.; Ghomi, M. Tryptophan Tight Binding to Gold Nanoparticles Induces Drastic Changes in Indole Ring Raman Markers. J. Phys. Chem. C 2018, 122, 13034–13046. [Google Scholar] [CrossRef]
- Hernández, B.; Coïc, Y.-M.; López-Tobar, E.; Sanchez-Cortes, S.; Baron, B.; Pflüger, F.; Kruglik, S.G.; Cohen, R.; Ghomi, M. Dynamical Behavior of Somatostatin-14 and Its Cyclic Analogues as Analyzed in Bulk and on Plasmonic Silver Nanoparticles. In Advances in Protein Chemistry and Structural Biology; Academic Press Inc.: Cambridge, MA, USA, 2018; Volume 112, pp. 81–121. [Google Scholar]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef] [PubMed]
- Linse, S. Monomer-dependent secondary nucleation in amyloid formation. Biophys. Rev. 2017, 9, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Törnquist, M.; Michaels, T.C.T.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.A.; Knowles, T.P.J.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [PubMed]
- Linse, S.; Cabaleiro-Lago, C.; Xue, W.-F.; Lynch, I.; Lindman, S.; Thulin, E.; Radford, S.; Dawson, K.A. Nucleation of protein fibrillation by nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 8691–8696. [Google Scholar] [CrossRef]
- Cabaleiro-Lago, C.; Quinlan-Pluck, F.; Lynch, I.; Lindman, S.; Minogue, A.M.; Thulin, E.; Walsh, D.M.; Dawson, K.A.; Linse, S. Inhibition of Amyloid β Protein Fibrillation by Polymeric Nanoparticles. J. Am. Chem. Soc. 2008, 130, 15437–15443. [Google Scholar] [CrossRef]
- Rocha, S.; Thünemann, A.F.; Pereira, M.D.C.; Coelho, M.; Moehwald, H.; Brezesinski, G. Influence of fluorinated and hydrogenated nanoparticles on the structure and fibrillogenesis of amyloid beta-peptide. Biophys. Chem. 2008, 137, 35–42. [Google Scholar] [CrossRef]
- Cabaleiro-Lago, C.; Szczepankiewicz, O.; Linse, S. The effect of nanoparticles on amyloid aggregation depends on the protein stability and intrinsic aggregation rate. Langmuir 2012, 28, 1852–1857. [Google Scholar] [CrossRef]
- Szczepankiewicz, O.; Cabaleiro-Lago, C.; Tartaglia, G.G.; Vendruscolo, M.; Hunter, T.; Hunter, G.J.; Nilsson, H.; Thulin, E.; Linse, S. Interactions in the native state of monellin, which play a protective role against aggregationw. J. C. R. Soc. Chem. 2011, 7, 521. [Google Scholar]
- Magro, M.; Baratella, D.; Bonaiuto, E.; Roger, J.D.A.; Chemello, G.; Pasquaroli, S.; Mancini, L.; Olivotto, I.; Zoppellaro, G.; Ugolotti, J.; et al. Stealth Iron Oxide Nanoparticles for Organotropic Drug Targeting. Biomacromolecules 2019, 20, 1375–1384. [Google Scholar] [CrossRef]
- Naidja, A.; Liu, C.; Huang, P. Formation of Protein–Birnessite Complex: XRD, FTIR, and AFM Analysis. J. Colloid Interface Sci. 2002, 251, 46–56. [Google Scholar] [CrossRef]
- Najafpour, M.M. Biomineralization: A proposed evolutionary origin for inorganic cofactors of enzymes. Theory Biosci. 2012, 131, 265–272. [Google Scholar] [CrossRef]
- Bernal, J.D. The physical basis of life. Proc. Phys. Soc. Sect. B 1949, 62, 597–618. [Google Scholar] [CrossRef]
- Rode, B.M.; Son, H.L.; Suwannachot, Y. The combination of salt induced peptide formation reaction and clay catalysis: A way to higher peptides under primitive earth conditions. Orig. Life Evol. Biosph. 1999, 29, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Orgel, L.E. Polymerization on the rocks: β-amino acids and arginine. Orig. Life Evol. Biosph. 1998, 28, 245–257. [Google Scholar] [CrossRef]
- Pollet, R.; Boehme, C.; Marx, D. AB INITIO Simulations of Desorption and Reactivity of Glycine at a Water-Pyrite Interface at “Iron-Sulfur World” Prebiotic Conditions. Orig. Life Evol. Biosph. 2006, 36, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.P.; Berndt, G.; de Junior, I.G.; di Mauro, E.; Paesano, A., Jr.; de Santana, H.; da Costa, A.C.S.; Zaia, C.T.B.V.; Zaia, D.A.M. Adsorption of cysteine on hematite, magnetite and ferrihydrite: FT-IR, Mössbauer, EPR spectroscopy and X-ray diffractometry studies. Amino Acids 2011, 40, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Zaia, D.A.M.; Zaia, C.T.B.V.; de Santana, H. Which Amino Acids Should Be Used in Prebiotic Chemistry Studies? Orig. Life Evol. Biosph. 2008, 38, 469–488. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.-F. Adsorption and Polymerization of Amino Acids on Mineral Surfaces: A Review. Orig. Life Evol. Biosph. 2008, 38, 211–242. [Google Scholar] [CrossRef] [PubMed]
- Cairns-Smith, A.G. Seven Clues to the Origin of Life; Cambridge University Press: Cambridge, UK, 1987. [Google Scholar]
- Russell, M.J.; Allen, J.; Milner-White, E.J. Inorganic Complexes Enabled the Onset of Life and Oxygenic Photosynthesis. In Photosynthesis. Energy from the Sun; Springer: Dordrecht, The Netherlands, 2008; pp. 1187–1192. [Google Scholar] [CrossRef]
- Kauffman, S.A. The Origins of Order: Self-Organization and Selection in Evolution; Springer: Dordrecht, The Netherlands, 1992; pp. 61–100. [Google Scholar] [CrossRef]
- Lundqvist, M.; Nygren, P.; Jonsson, B.-H.; Broo, K. Induction of Structure and Function in a Designed Peptide upon Adsorption on a Silica Nanoparticle. Angew. Chem. Int. Ed. 2006, 45, 8169–8173. [Google Scholar] [CrossRef] [PubMed]
- Nygren, P.; Lundqvist, M.; Broo, K.; Jonsson, B.-H. Fundamental Design Principles That Guide Induction of Helix upon Formation of Stable Peptide−Nanoparticle Complexes. Nano Lett. 2008, 8, 1844–1852. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.; Yeo, K.B.; Ki, M.-R.; Pack, S.P. Self-encapsulation and controlled release of recombinant proteins using novel silica-forming peptides as fusion linkers. Int. J. Biol. Macromol. 2019, 125, 1175–1183. [Google Scholar] [CrossRef]
- Crick, F. Life Itself: Its Origin and Nature; Simon and Schuster: New York, NY, USA, 1982. [Google Scholar]
- Hazen, R.M.; Sholl, D.S. Chiral selection on inorganic crystalline surfaces. Nat. Mater. 2003, 2, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Zaera, F. Chirality in adsorption on solid surfaces. Chem. Soc. Rev. 2017, 46, 7374–7398. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.G.; Fitz, D.; Jakschitz, T.; Rode, B.M. Selective adsorption and chiral amplification of amino acids in vermiculite clay-implications for the origin of biochirality. Phys. Chem. Chem. Phys. 2011, 13, 831–838. [Google Scholar] [CrossRef]
- Ferris, J.P. Montmorillonite-catalysed formation of RNA oligomers: The possible role of catalysis in the origins of life. Philos. Trans. R. Soc. B: Biol. Sci. 2006, 361, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Szostak, J.W.; Bartel, D.P.; Luisi, P.L. Synthesizing life. Nat. Cell Biol. 2001, 409, 387–390. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Wang, Y.; Chen, H. Emerging chirality in nanoscience. Chem. Soc. Rev. 2013, 42, 2930–2962. [Google Scholar] [CrossRef]
- Magro, M.; Molinari, S.; Venerando, A.; Baratella, D.; Zoppellaro, G.; Salviulo, G.; Zboril, R.; Vianello, F. Colloidal maghemite nanoparticles with oxyhydroxide-like interface and chiroptical properties. Appl. Surf. Sci. 2020, 534, 147567. [Google Scholar] [CrossRef]
- Stensballe, A.; Andersen, S.; Jensen, O.N. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics 2001, 1, 207–222. [Google Scholar] [CrossRef]
- Stensballe, A.; Jensen, O.N. Phosphoric acid enhances the performance of Fe(III) affinity chromatography and matrix-assisted laser desorption/ionization tandem mass spectrometry for recovery, detection and sequencing of phosphopeptides. Rapid Commun. Mass Spectrom. 2004, 18, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Raska, C.S.; Parker, C.E.; Dominski, Z.; Marzluff, W.F.; Glish, G.; Pope, R.M.; Borchers, C.H. Direct MALDI-MS/MS of Phosphopeptides Affinity-Bound to Immobilized Metal Ion Affinity Chromatography Beads. Anal. Chem. 2002, 74, 3429–3433. [Google Scholar] [CrossRef]
- Zhou, H.; Ye, M.; Dong, J.; Han, G.; Jiang, X.; Wu, R.; Zou, H. Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J. Proteome Res. 2008, 7, 3957–3967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, J.; Jensen, O.N.; Roepstorff, P. Highly efficient phosphopeptide enrichment by calcium phosphate precipitation combined with subsequent IMAC enrichment. Mol. Cell. Proteom. 2007, 6, 2032–2042. [Google Scholar] [CrossRef]
- Wang, Z.G.; Lv, N.; Bi, W.Z.; Zhang, J.L.; Ni, J.Z. Development of the affinity materials for phosphorylated proteins/peptides enrichment in phosphoproteomics analysis. ACS Appl. Mater. Interfaces 2015, 7, 8377–8392. [Google Scholar] [CrossRef]
- Wolschin, F.; Wienkoop, S.; Weckwerth, W. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC). Proteomics 2005, 5, 4389–4397. [Google Scholar] [CrossRef]
- Ikeguchi, Y.; Nakamura, H. Selective Enrichment of Phospholipids by Titania. Anal. Sci. 2000, 16, 541–543. [Google Scholar] [CrossRef]
- Jiang, Z.-T.; Zuo, Y.-M. Synthesis of porous titania microspheres for HPLC packings by polymerization-induced colloid aggregation (PICA). Anal. Chem. 2000, 73, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Nakamura, H.; Nakajima, T. Titania and zirconia: Possible new ceramic microparticulates for high-performance liquid chromatography. J. Chromatogr. A 1990, 515, 149–158. [Google Scholar] [CrossRef]
- Sano, A.; Nakamura, H. Chemo-affinity of Titania for the Column-switching HPLC Analysis of Phosphopeptides. Anal. Sci. 2004, 20, 565–566. [Google Scholar] [CrossRef] [PubMed]
- Pinkse, M.W.H.; Uitto, P.M.; Hilhorst, M.J.; Ooms, B.; Heck, A.J.R. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 2004, 76, 3935–3943. [Google Scholar] [CrossRef]
- Pinkse, M.W.H.; Mohammed, S.; Gouw, J.W.; van Breukelen, B.; Vos, H.R.; Heck, A.J.R. Highly Robust, Automated, and Sensitive Online TiO2-Based Phosphoproteomics Applied To Study Endogenous Phosphorylation in Drosophila melanogaster. J. Proteome Res. 2008, 7, 687–697. [Google Scholar] [CrossRef]
- Park, S.-S.; Maudsley, S. Discontinuous pH gradient-mediated separation of TiO2-enriched phosphopeptides. Anal. Biochem. 2011, 409, 81–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, R.; Hu, J.; Cai, Z.; Ju, H. Highly selective enrichment of phosphopeptides with high-index facets exposed octahedral tin dioxide nanoparticles for mass spectrometric analysis. Talanta 2014, 119, 452–457. [Google Scholar] [CrossRef]
- Magro, M.; Venerando, A.; Macone, A.; Canettieri, G.; Agostinelli, E.; Vianello, F. Nanotechnology-Based Strategies to Develop New Anticancer Therapies. Biomolecules 2020, 10, 735. [Google Scholar] [CrossRef]
- Schubert, J.; Chanana, M. Coating Matters: Review on Colloidal Stability of Nanoparticles with Biocompatible Coatings in Biological Media, Living Cells and Organisms. Curr. Med. Chem. 2018, 25, 4553–4586. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.M.; Tavallaie, R.; Sandiford, L.; Tilley, R.; Gooding, J.J. Gold coated magnetic nanoparticles: From preparation to surface modification for analytical and biomedical applications. Chem. Commun. 2016, 52, 7528–7540. [Google Scholar] [CrossRef] [PubMed]
- Kudr, J.; Haddad, Y.A.E.; Richtera, L.; Heger, Z.; Cernak, M.; Adam, V.; Zitka, O. Magnetic Nanoparticles: From Design and Synthesis to Real World Applications. Nanomaterials 2017, 7, 243. [Google Scholar] [CrossRef]
- Blank-Shim, S.A.; Schwaminger, S.P.; Borkowska-Panek, M.; Anand, P.; Yamin, P.; Fraga-García, P.; Fink, K.; Wenzel, W.; Berensmeier, S. Binding patterns of homo-peptides on bare magnetic nanoparticles: Insights into environmental dependence. Sci. Rep. 2017, 7, 14047. [Google Scholar] [CrossRef]
- Schwaminger, S.P.; Anand, P.; Borkowska-Panek, M.; Blank-Shim, S.A.; Fraga-Garcia, P.; Fink, K.; Berensmeier, S.; Wenzel, W. Rational Design of Iron Oxide Binding Peptide Tags. Langmuir 2019, 35, 8472–8481. [Google Scholar] [CrossRef] [PubMed]
- Schwaminger, S.P.; Fraga-García, P.; Blank-Shim, S.A.; Straub, T.; Haslbeck, M.; Muraca, F.; Dawson, K.A.; Berensmeier, S. Magnetic One-Step Purification of His-Tagged Protein by Bare Iron Oxide Nanoparticles. ACS Omega 2019, 4, 3790–3799. [Google Scholar] [CrossRef]
- Magro, M.; Vianello, F. Bare Iron Oxide Nanoparticles: Surface Tunability for Biomedical, Sensing and Environmental Applications. Nanomaterials 2019, 9, 1608. [Google Scholar] [CrossRef]
- Magro, M.; Cozza, G.; Molinari, S.; Venerando, A.; Baratella, D.; Miotto, G.; Zennaro, L.; Rossetto, M.; Frömmel, J.; Kopečná, M.; et al. Role of carboxylic group pattern on protein surface in the recognition of iron oxide nanoparticles: A key for protein corona formation. Int. J. Biol. Macromol. 2020, 164, 1715–1728. [Google Scholar] [CrossRef] [PubMed]
- Hadjidemetriou, M.; Al-Ahmady, Z.; Buggio, M.; Swift, J.; Kostarelos, K. A novel scavenging tool for cancer biomarker discovery based on the blood-circulating nanoparticle protein corona. Biomater. 2019, 188, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Safavi-Sohi, R.; Malekzadeh, R.; Poustchi, H.; Vasighi, M.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Hajipour, M.; di Domenico, M.; et al. Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horiz. 2019, 4, 1063–1076. [Google Scholar] [CrossRef]
- Magro, M.; Zaccarin, M.; Miotto, G.; da Dalt, L.; Baratella, D.; Fariselli, P.; Gabai, G.; Vianello, F. Analysis of hard protein corona composition on selective iron oxide nanoparticles by MALDI-TOF mass spectrometry: Identification and amplification of a hidden mastitis biomarker in milk proteome. Anal. Bioanal. Chem. 2018, 410, 2949–2959. [Google Scholar] [CrossRef] [PubMed]
- Somorjai, G.A.; Li, Y. Introduction to Surface Chemistry and Catalysis; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Vollath, D.; Fischer, F.D.; Holec, D. Surface energy of nanoparticles—influence of particle size and structure. Beilstein J. Nanotechnol. 2018, 9, 2265–2276. [Google Scholar] [CrossRef]
- Foroozandeh, P.; Aziz, A.A. Merging Worlds of Nanomaterials and Biological Environment: Factors Governing Protein Corona Formation on Nanoparticles and Its Biological Consequences. Nanoscale Res. Lett. 2015, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, A.; Konduru, N.V. The role of natural processes and surface energy of inhaled engineered nanoparticles on aggregation and corona formation. NanoImpact 2016, 2, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Norde, W. Colloids and Interfaces in Life Sciences; Marcel Dekker: New York, USA, 2003. [Google Scholar]
- Wohlleben, W.; Driessen, M.D.; Raesch, S.S.; Schaefer, U.F.; Schulze, C.; von Vacano, B.; Vennemann, A.; Wiemann, M.; Ruge, C.A.; Platsch, H.; et al. Influence of agglomeration and specific lung lining lipid/protein interaction on short-term inhalation toxicity. Nanotoxicology 2016, 10, 970–980. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.-Y.; Tian, N.; Li, J.-T.; Broadwell, I.; Sun, S.-G. Nanomaterials of high surface energy with exceptional properties in catalysis and energy storage. Chem. Soc. Rev. 2011, 40, 4167–4185. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.Z.; Jiang, H.B.; Qiao, S.Z.; Yang, H.G.; Lu, G.Q. Synthesis of high-reactive facets dominated anatase TiO. J. Mater. Chem. 2011, 21, 7052–7061. [Google Scholar] [CrossRef]
- Wang, C.; Daimon, H.; Onodera, T.; Koda, T.; Sun, S. A General Approach to the Size- and Shape-Controlled Synthesis of Platinum Nanoparticles and Their Catalytic Reduction of Oxygen. Angew. Chem. Int. Ed. 2008, 47, 3588–3591. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, S.; Wu, W. Shape control of inorganic nanoparticles from solution. Nanoscale 2016, 8, 1237–1259. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, F.; Gao, K.; Wu, J.; Xue, D. Recent developments in the chemical synthesis of inorganic porous capsules. J. Mater. Chem. 2009, 19, 6073–6084. [Google Scholar] [CrossRef]
- Jun, Y.-W.; Choi, J.-S.; Cheon, J. Shape Control of Semiconductor and Metal Oxide Nanocrystals through Nonhydrolytic Colloidal Routes. Angew. Chem. Int. Ed. 2006, 45, 3414–3439. [Google Scholar] [CrossRef] [PubMed]
- Grzelczak, M.; Pérez-Juste, J.; Mulvaney, P.; Liz-Marzán, L.M. Shape control in gold nanoparticle synthesis. Chem. Soc. Rev. 2008, 37, 1783–1791. [Google Scholar] [CrossRef]
- Baskes, M.I. Modified embedded-atom potentials for cubic materials and impurities. Phys. Rev. B 1992, 46, 2727–2742. [Google Scholar] [CrossRef]
- Ma, F.; Xu, K.-W. Using dangling bond density to characterize the surface energy of nanomaterials. Surf. Interface Anal. 2007, 39, 611–614. [Google Scholar] [CrossRef]
- Holec, D.; Dumitraschkewitz, P.; Vollath, D.; Fischer, F.D. Surface Energy of Au Nanoparticles Depending on Their Size and Shape. Nanomaterials 2020, 10, 484. [Google Scholar] [CrossRef]
- Ferrer, M.; Fabris, G.D.S.L.; de Faria, B.V.; Martins, J.B.; Moreira, M.L.; Sambrano, J.R. Quantitative evaluation of the surface stability and morphological changes of Cu2O particles. Heliyon 2019, 5, e02500. [Google Scholar] [CrossRef]
- Madathiparambil Visalakshan, R.; García, L.E.G.; Benzigar, M.R.; Ghazaryan, A.; Simon, J.; Mierczynska-Vasilev, A.; Michl, T.D.; Vinu, A.; Mailänder, V.; Morsbach, S.; et al. The Influence of Nanoparticle Shape on Protein Corona Formation. Small 2020, 16, 2000285. [Google Scholar] [CrossRef]
- Schulman, R.D.; Trejo, M.; Salez, T.; Raphaël, E.; Dalnoki-Veress, K. Surface energy of strained amorphous solids. Nat. Commun. 2018, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tran, R.; Xu, Z.; Radhakrishnan, B.; Winston, D.; Sun, W.; Persson, K.A.; Ong, S.P. Data Descriptor: Surface energies of elemental crystals. Sci. Data 2016, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Qin, R.; Fu, G.; Zheng, N. Surface Coordination Chemistry of Metal Nanomaterials. J. Am. Chem. Soc. 2017, 139, 2122–2131. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Veracoechea, F.J.; Frenkel, D. Designing super selectivity in multivalent nano-particle binding. Proc. Natl. Acad. Sci. USA 2011, 108, 10963–10968. [Google Scholar] [CrossRef]
- Luo, L.; Liu, Y.; Gao, T.; Wang, X.; Chen, J.; Wang, H.; Liu, Y.; Cao, A. Characterization of the specific interactions between nanoparticles and proteins at residue-resolution by alanine scanning mutagenesis. ACS Appl. Mater. Interfaces 2020, 12, 34514–34523. [Google Scholar] [CrossRef]
- Cai, K.; Bossert, J.; Jandt, K.D. Does the nanometre scale topography of titanium influence protein adsorption and cell proliferation? Colloids Surf. B Biointerfaces 2006, 49, 136–144. [Google Scholar] [CrossRef]
- Heinz, H.; Ramezani-Dakhel, H. Simulations of inorganic–bioorganic interfaces to discover new materials: Insights, comparisons to experiment, challenges, and opportunities. Chem. Soc. Rev. 2016, 45, 412–448. [Google Scholar] [CrossRef] [PubMed]
- Penna, M.; Yarovsky, I. Nanoscale in silico classification of ligand functionalised surfaces for protein adsorption resistance. Nanoscale 2020, 12, 7240–7255. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Weikl, T.; Ma, Y.-Q. Theoretical modeling of interactions at the bio-nano interface. Nanoscale 2020, 12, 10426–10429. [Google Scholar] [CrossRef]
- Bourassin, N.; Baaden, M.; Lojou, E.; Sacquin-Mora, S. Implicit Modeling of the Impact of Adsorption on Solid Surfaces for Protein Mechanics and Activity with a Coarse-Grained Representation. J. Phys. Chem. B 2020, 124, 8516–8523. [Google Scholar] [CrossRef]
- Ozboyaci, M.; Kokh, D.B.; Corni, S.; Wade, R.C. Modeling and simulation of protein-surface interactions: Achievements and challenges. Q. Rev. Biophys. 2016, 49, e4. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, I.; Fubini, B.; Ghibaudi, E.M.; Turci, F. Multiple aspects of the interaction of biomacromolecules with inorganic surfaces. Adv. Drug Deliv. Rev. 2011, 63, 1186–1209. [Google Scholar] [CrossRef] [PubMed]
- Lynch, I.; Afantitis, A.; Exner, T.; Himly, M.; Lobaskin, V.; Doganis, P.; Maier, D.; Sanabria, N.; Papadiamantis, A.; Rybinska-Fryca, A.; et al. Can an InChI for Nano Address the Need for a Simplified Representation of Complex Nanomaterials across Experimental and Nanoinformatics Studies? Nanomaterials 2020, 10, 2493. [Google Scholar] [CrossRef] [PubMed]
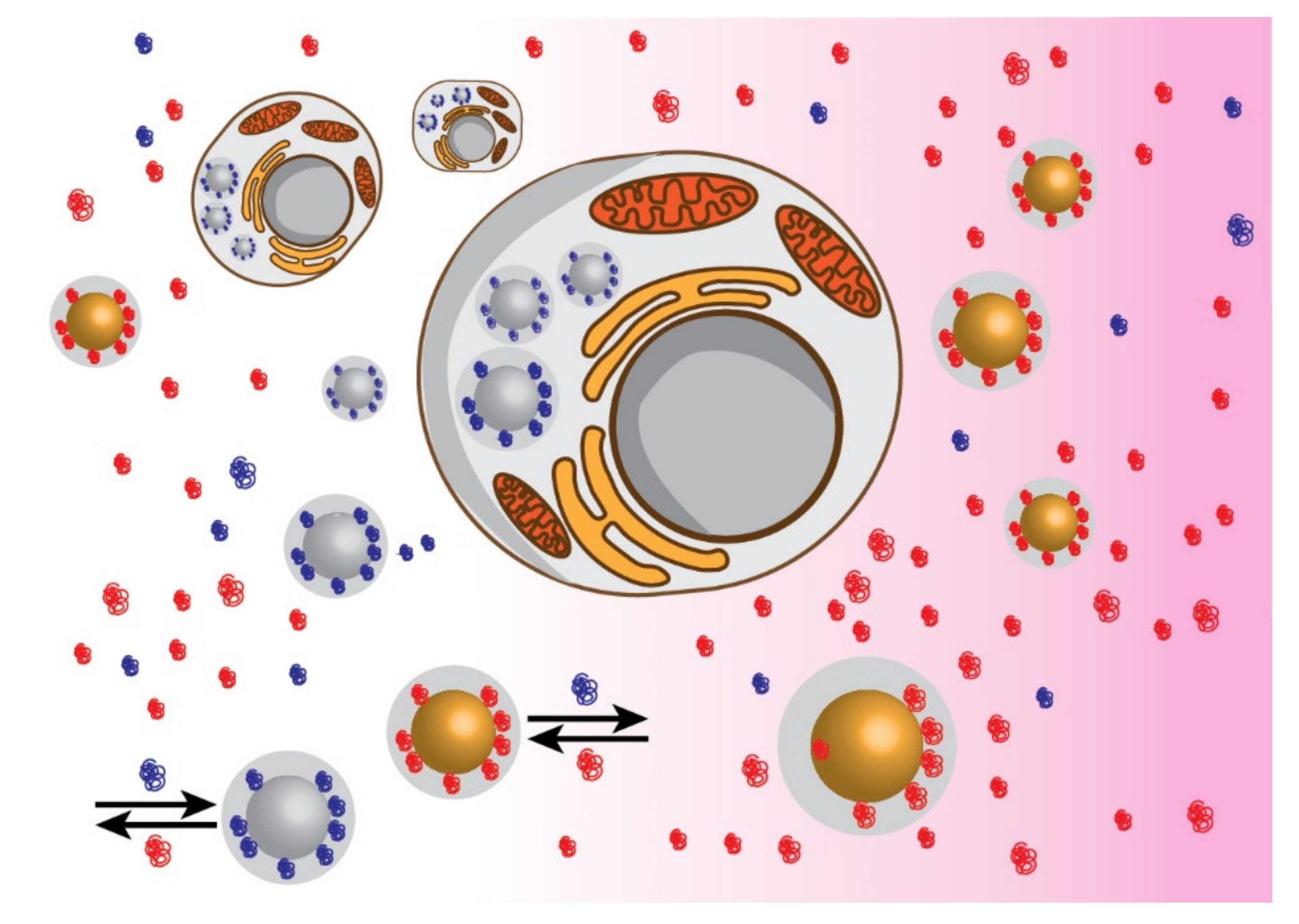
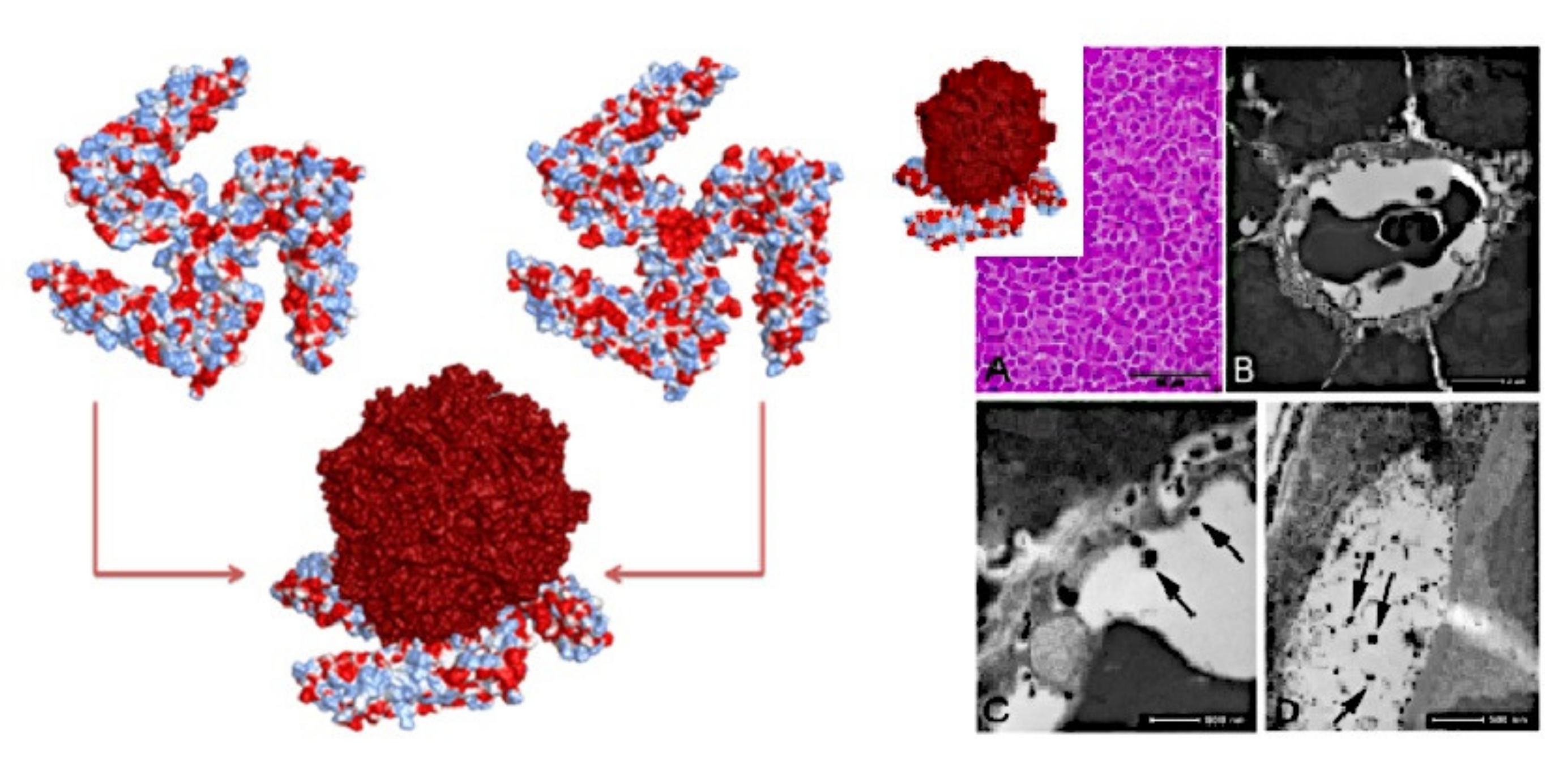
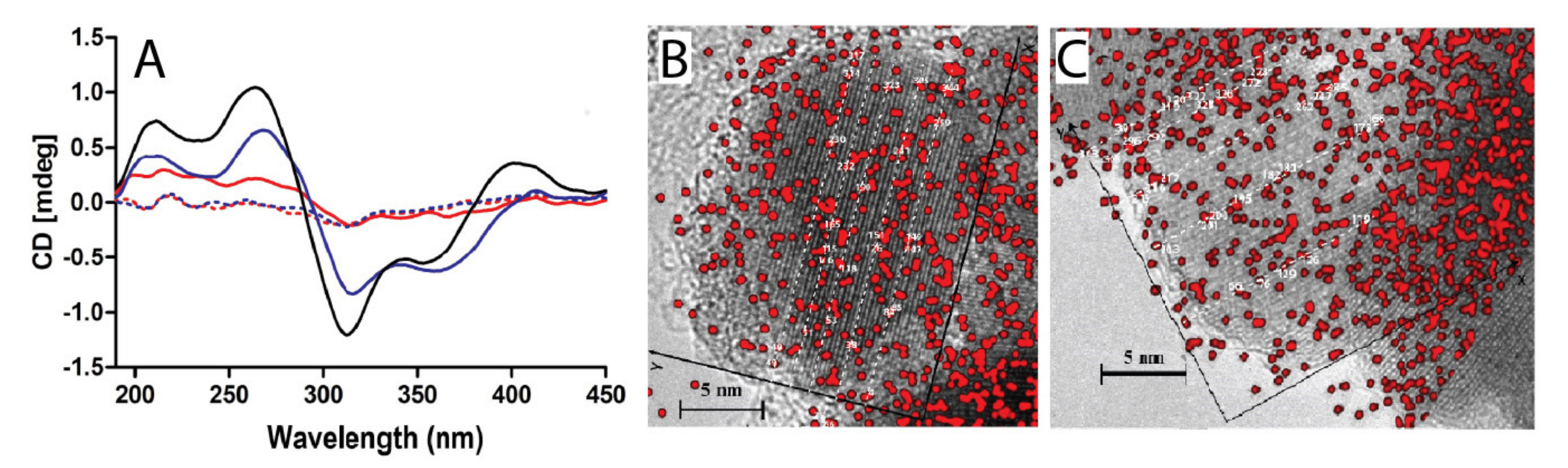
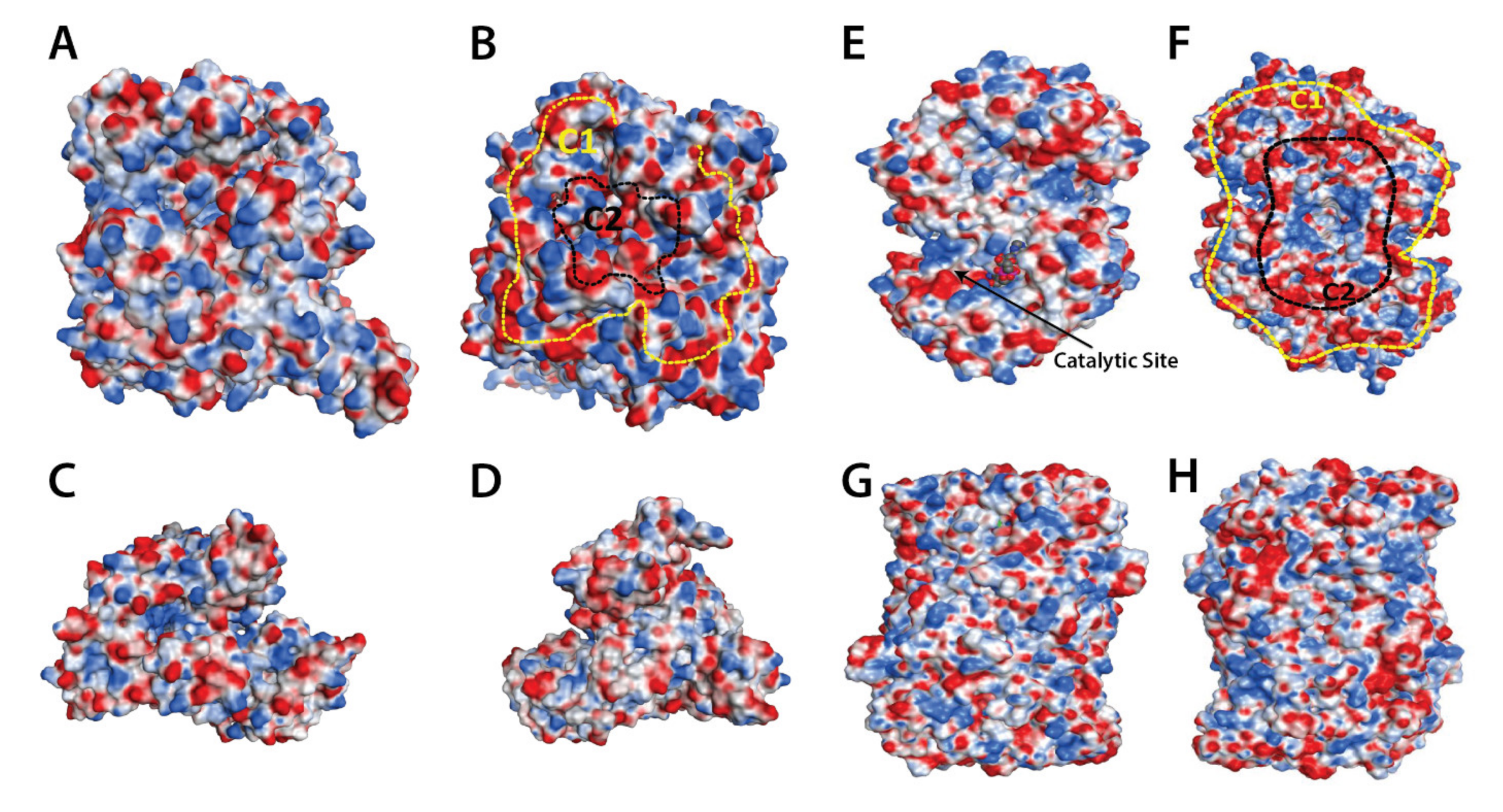
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vianello, F.; Cecconello, A.; Magro, M. Toward the Specificity of Bare Nanomaterial Surfaces for Protein Corona Formation. Int. J. Mol. Sci. 2021, 22, 7625. https://doi.org/10.3390/ijms22147625
Vianello F, Cecconello A, Magro M. Toward the Specificity of Bare Nanomaterial Surfaces for Protein Corona Formation. International Journal of Molecular Sciences. 2021; 22(14):7625. https://doi.org/10.3390/ijms22147625
Chicago/Turabian StyleVianello, Fabio, Alessandro Cecconello, and Massimiliano Magro. 2021. "Toward the Specificity of Bare Nanomaterial Surfaces for Protein Corona Formation" International Journal of Molecular Sciences 22, no. 14: 7625. https://doi.org/10.3390/ijms22147625
APA StyleVianello, F., Cecconello, A., & Magro, M. (2021). Toward the Specificity of Bare Nanomaterial Surfaces for Protein Corona Formation. International Journal of Molecular Sciences, 22(14), 7625. https://doi.org/10.3390/ijms22147625







