Applying Proteomics and Integrative “Omics” Strategies to Decipher the Chronic Kidney Disease-Related Atherosclerosis
Abstract
1. Introduction
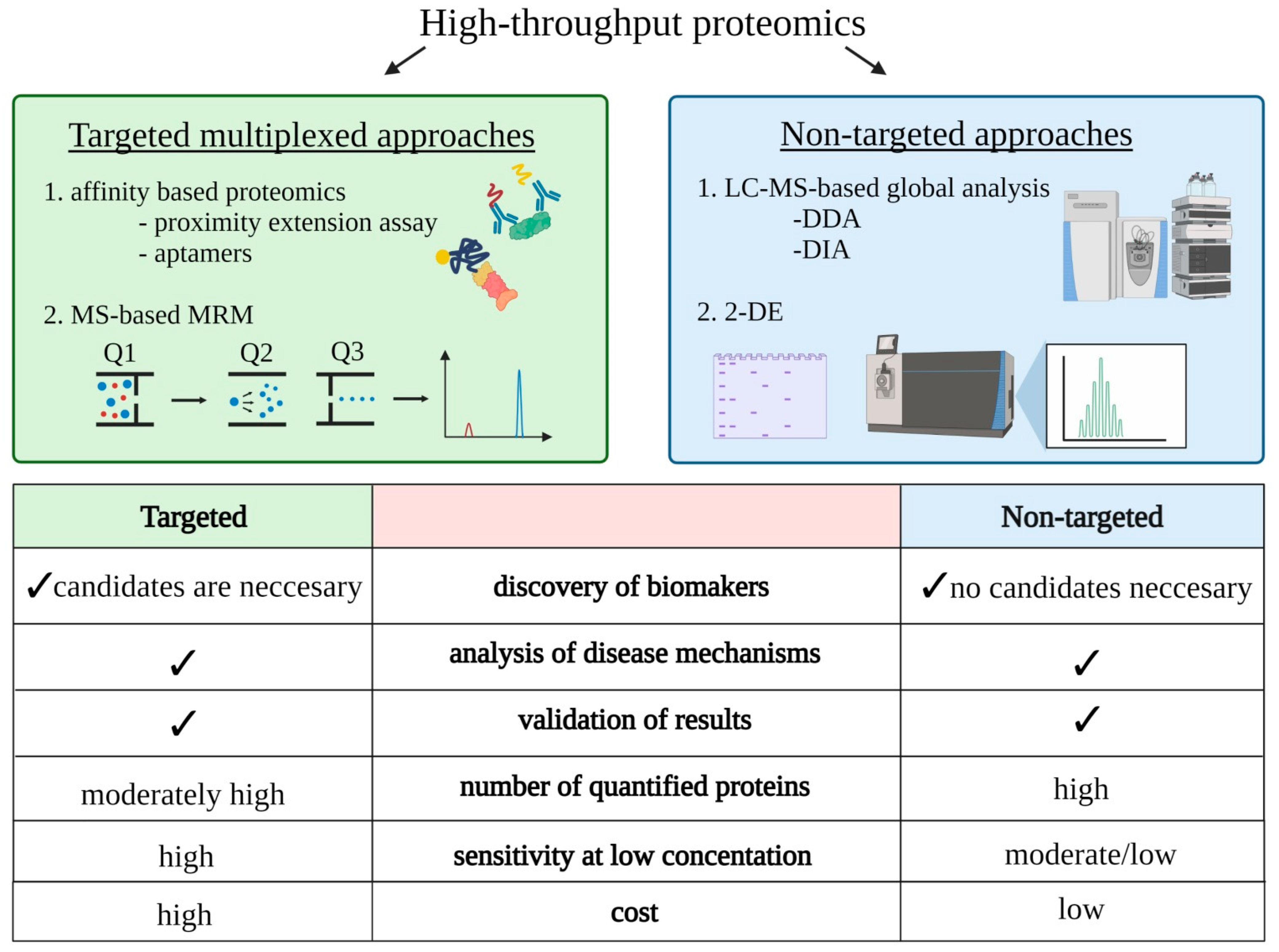
2. Key Concepts of Proteomics in Disease Analysis
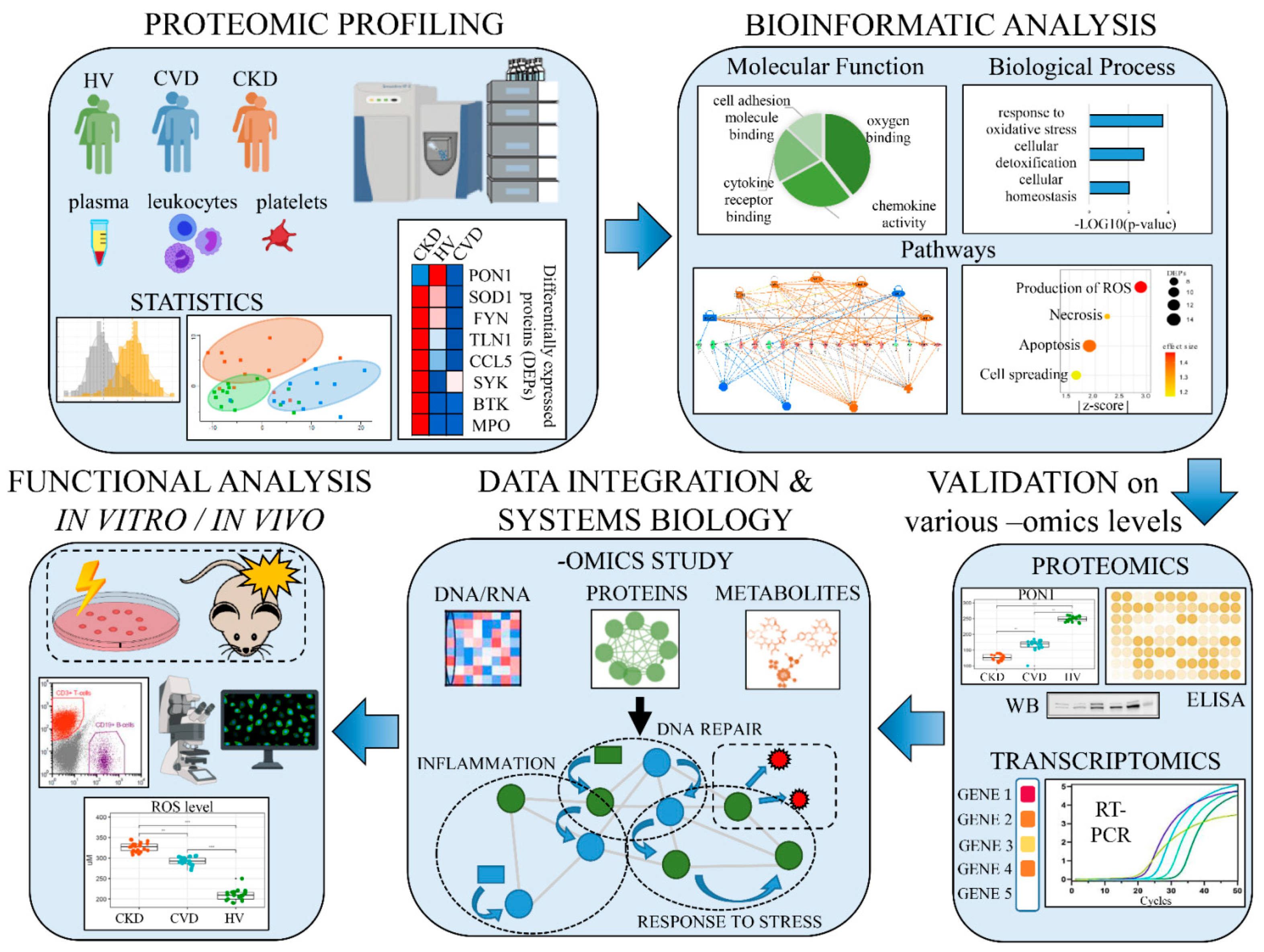
3. Application of Proteomics to Understanding Molecular Mechanism of CKD-A
3.1. Proteomic and Lipidomic Analyses on Lipid Metabolism in CKD-A
3.2. “Omics” Strategies to Decipher the Vascular Calcification Mechanism in CKD-A
3.3. Proteomics and Metabolomics in Analysis of Inflammation and Endothelial Dysfunction in CKD-A
3.4. The Potential and Future Perspectives of “Omics” Approaches in Understanding Molecular Mechanism of CKD-A
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levin, A.; Stevens, P.E.; Bilous, R.W.; Coresh, J.; De Francisco, A.L.M.; De Jong, P.E.; Griffith, K.E.; Hemmelgarn, B.R.; Iseki, K.; Lamb, E.J.; et al. Kidney disease: Improving global outcomes (KDIGO) CKD work group. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int. Suppl. 2013, 3, 1–150. [Google Scholar] [CrossRef]
- UK, National Clinical Guideline Centre. Chronic Kidney Disease (Partial Update): Early Identification and Management of Chronic Kidney Disease in Adults in Primary and Secondary Care; National Institute for Health and Care Excellence: London, UK, 2014. [Google Scholar]
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic Kidney Disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef]
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.K.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef]
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Naylor, K.L.; Kim, S.J.; McArthur, E.; Garg, A.X.; McCallum, M.K.; Knoll, G.A. Mortality in Incident Maintenance Dialysis Patients Versus Incident Solid Organ Cancer Patients: A Population-Based Cohort. Am. J. Kidney Dis. 2019, 73, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Cockwell, P.; Fisher, L.-A. The global burden of chronic kidney disease. Lancet 2020, 395, 662–664. [Google Scholar] [CrossRef]
- Vallianou, N.G.; Mitesh, S.; Gkogkou, A.; Geladari, E. Chronic Kidney Disease and Cardiovascular Disease: Is there Any Relationship? Curr. Cardiol. Rev. 2019, 15, 55–63. [Google Scholar] [CrossRef]
- Gansevoort, R.T.; Correa-Rotter, R.; Hemmelgarn, B.R.; Jafar, T.H.; Heerspink, H.J.L.; Mann, J.F.; Matsushita, K.; Wen, C.P. Chronic kidney disease and cardiovascular risk: Epidemiology, mechanisms, and prevention. Lancet 2013, 382, 339–352. [Google Scholar] [CrossRef]
- Wanner, C.; Amann, K.; Shoji, T. The heart and vascular system in dialysis. Lancet 2016, 388, 276–284. [Google Scholar] [CrossRef]
- Wen, C.P.; Cheng, T.Y.D.; Tsai, M.K.; Chang, Y.C.; Chan, H.T.; Tsai, S.P.; Chiang, P.H.; Hsu, C.C.; Sung, P.K.; Hsu, Y.H.; et al. All-cause mortality attributable to chronic kidney disease: A prospective cohort study based on 462 293 adults in Taiwan. Lancet 2008, 371, 2173–2182. [Google Scholar] [CrossRef]
- Hemmelgarn, B.R.; Manns, B.J.; Lloyd, A.; James, M.T.; Klarenbach, S.; Quinn, R.R.; Wiebe, N.; Tonelli, M. Alberta Kidney Disease Network Relation between kidney function, proteinuria, and adverse outcomes. JAMA 2010, 303, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Schiffrin, E.L.; Lipman, M.L.; Mann, J.F.E. Chronic kidney disease: Effects on the cardiovascular system. Circulation 2007, 116, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.C.; Lu, L.; Cao, Y.; Sun, R.R.; Chen, S.; Zhang, P.Y. Cardiovascular disease and its relationship with chronic kidney disease. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 2918–2926. [Google Scholar]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Mohler, E.R.; Xie, D.; Shlipak, M.; Townsend, R.R.; Appel, L.J.; Ojo, A.; Schreiber, M.; Nessel, L.; Zhang, X.; et al. Traditional and non-traditional risk factors for incident peripheral arterial disease among patients with chronic kidney disease. Nephrol. Dial. Transplant. 2016, 31, 1145–1151. [Google Scholar] [CrossRef]
- Weiner, D.E.; Tighiouart, H.; Elsayed, E.F.; Griffith, J.L.; Salem, D.N.; Levey, A.S.; Sarnak, M.J. The relationship between nontraditional risk factors and outcomes in individuals with stage 3 to 4 CKD. Am. J. Kidney Dis. 2008, 51, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Major, R.W.; Cheng, M.R.I.; Grant, R.A.; Shantikumar, S.; Xu, G.; Oozeerally, I.; Brunskill, N.J.; Gray, L.J. Cardiovascular disease risk factors in chronic kidney disease: A systematic review and meta-analysis. PLoS ONE 2018, 13, e0192895. [Google Scholar] [CrossRef]
- Kendrick, J.; Chonchol, M.B. Nontraditional risk factors for cardiovascular disease in patients with chronic kidney disease. Nat. Clin. Pract. Nephrol. 2008, 4, 672–681. [Google Scholar] [CrossRef]
- Kalantar-Zadeh, K.; Block, G.; Humphreys, M.H.; Kopple, J.D. Reverse epidemiology of cardiovascular risk factors in maintenance dialysis patients. Kidney Int. 2003, 63, 793–808. [Google Scholar] [CrossRef]
- Maini, R.; Wong, D.B.; Addison, D.; Chiang, E.; Weisbord, S.D.; Jneid, H. Persistent underrepresentation of kidney disease in randomized, controlled trials of cardiovascular disease in the contemporary era. J. Am. Soc. Nephrol. 2018, 29, 2782–2786. [Google Scholar] [CrossRef]
- Romagnani, P.; Remuzzi, G.; Glassock, R.; Levin, A.; Jager, K.J.; Tonelli, M.; Massy, Z.; Wanner, C.; Anders, H.-J. Chronic kidney disease. Nat. Rev. Dis. Prim. 2017, 3, 17088. [Google Scholar] [CrossRef] [PubMed]
- Daenen, K.; Andries, A.; Mekahli, D.; Schepdael, A.V.; Jouret, F.; Bammens, B. Oxidative stress in chronic kidney disease. Iran. J. Kidney Dis. 2015, 9, 165. [Google Scholar] [CrossRef]
- Valdivielso, J.M.; Rodríguez-Puyol, D.; Pascual, J.; Barrios, C.; Bermúdez-López, M.; Sánchez-Niño, M.D.; Pérez-Fernández, M.; Ortiz, A. Atherosclerosis in Chronic Kidney Disease. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1938–1966. [Google Scholar] [CrossRef]
- Tummalapalli, L.; Nadkarni, G.N.; Coca, S.G. Biomarkers for predicting outcomes in chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2016, 25, 480–486. [Google Scholar] [CrossRef]
- Saucedo, A.L.; Perales-Quintana, M.M.; Paniagua-Vega, D.; Sanchez-Martinez, C.; Cordero-Perez, P.; Minsky, N.W. Chronic Kidney Disease and the Search for New Biomarkers for Early Diagnosis. Curr. Med. Chem. 2018, 25, 3719–3747. [Google Scholar] [CrossRef]
- Taherkhani, A.; Farrokhi Yekta, R.; Mohseni, M.; Saidijam, M.; Arefi Oskouie, A. Chronic kidney disease: A review of proteomic and metabolomic approaches to membranous glomerulonephritis, focal segmental glomerulosclerosis, and IgA nephropathy biomarkers. Proteome Sci. 2019, 17, 7. [Google Scholar] [CrossRef]
- Mayeux, R. Biomarkers: Potential Uses and Limitations. NeuroRx 2004, 1, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Parikh, C.R.; Thiessen-Philbrook, H. Key concepts and limitations of statistical methods for evaluating biomarkers of kidney disease. J. Am. Soc. Nephrol. 2014, 25, 1621–1629. [Google Scholar] [CrossRef]
- Drucker, E.; Krapfenbauer, K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-Y.; Ravassa, S.; Pejchinovski, M.; Yang, W.-Y.; Zürbig, P.; López, B.; Wei, F.-F.; Thijs, L.; Jacobs, L.; González, A.; et al. A Urinary Fragment of Mucin-1 Subunit α is a Novel Biomarker Associated with Renal Dysfunction in the General Population. Kidney Int. Rep. 2017, 2, 811–820. [Google Scholar] [CrossRef]
- Siwy, J.; Zürbig, P.; Argiles, A.; Beige, J.; Haubitz, M.; Jankowski, J.; Julian, B.A.; Linde, P.G.; Marx, D.; Mischak, H.; et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol. Dial. Transplant. 2017, 32, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Knoflach, M.; Kiechl, S.; Mantovani, A.; Cuccovillo, I.; Bottazzi, B.; Xu, Q.; Xiao, Q.; Gasperi, A.; Mayr, A.; Kehrer, M.; et al. Pentraxin-3 as a marker of advanced atherosclerosis results from the Bruneck, ARMY and ARFY Studies. PLoS ONE 2012, 7, e31474. [Google Scholar] [CrossRef]
- Martínez, P.J.; Baldán-Martín, M.; López, J.A.; Martín-Lorenzo, M.; Santiago-Hernández, A.; Agudiez, M.; Cabrera, M.; Calvo, E.; Vázquez, J.; Ruiz-Hurtado, G.; et al. Identification of six cardiovascular risk biomarkers in the young population: A promising tool for early prevention. Atherosclerosis 2019, 282, 67–74. [Google Scholar] [CrossRef]
- Han, W.K.; Bailly, V.; Abichandani, R.; Thadhani, R.; Bonventre, J.V. Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury. Kidney Int. 2002, 62, 237–244. [Google Scholar] [CrossRef]
- Mishra, J.; Qing, M.A.; Prada, A.; Mitsnefes, M.; Zahedi, K.; Yang, J.; Barasch, J.; Devarajan, P. Identification of neutrophil gelatinase-associated lipocalin as a novel early urinary biomarker for ischemic renal injury. J. Am. Soc. Nephrol. 2003, 14, 2534–2543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.R.; Parikh, C.R. Biomarkers of Acute and Chronic Kidney Disease. Annu. Rev. Physiol. 2019, 81, 309–333. [Google Scholar] [CrossRef]
- Good, D.M.; Zürbig, P.; Argilés, À.; Bauer, H.W.; Behrens, G.; Coon, J.J.; Dakna, M.; Decramer, S.; Delles, C.; Dominiczak, A.F.; et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol. Cell. Proteom. 2010, 9, 2424–2437. [Google Scholar] [CrossRef] [PubMed]
- Persson, F.; Rossing, P. Urinary Proteomics and Precision Medicine for Chronic Kidney Disease: Current Status and Future Perspectives. Proteom. Clin. Appl. 2019, 13, 1800176. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.A.; Hamza, E.; Guerrero-Hue, M.; Rayego-Mateos, S.; García-Caballero, C.; Vallejo-Mudarra, M.; Metzinger, L.; Metzinger-Le Meuth, V. Non-Coding RNAs in Kidney Diseases: The Long and Short of Them. Int. J. Mol. Sci. 2021, 22, 6077. [Google Scholar] [CrossRef]
- Mischak, H.; Allmaier, G.; Apweiler, R.; Attwood, T.; Baumann, M.; Benigni, A.; Bennett, S.E.; Bischoff, R.; Bongcam-Rudloff, E.; Capasso, G.; et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef]
- Gromova, M.; Vaggelas, A.; Dallmann, G.; Seimetz, D. Biomarkers: Opportunities and Challenges for Drug Development in the Current Regulatory Landscape. Biomark. Insights 2020, 15, 117727192097465. [Google Scholar] [CrossRef]
- Gupta, S.; Venkatesh, A.; Ray, S.; Srivastava, S. Challenges and prospects for biomarker research: A current perspective from the developing world. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 899–908. [Google Scholar] [CrossRef]
- Lundberg, M.; Eriksson, A.; Tran, B.; Assarsson, E.; Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011, 39, e102. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.; Carter, J.; Cunningham, V.; Dalby, A.; Eaton, B.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. Nat. Preced. 2010. [Google Scholar] [CrossRef]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gústafsdóttir, S.M.; Ostman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef] [PubMed]
- Percy, A.J.; Chambers, A.G.; Smith, D.S.; Borchers, C.H. Standardized Protocols for Quality Control of MRM-based Plasma Proteomic Workflows. J. Proteome Res. 2013, 12, 222–233. [Google Scholar] [CrossRef]
- Rezeli, M.; Sjödin, K.; Lindberg, H.; Gidlöf, O.; Lindahl, B.; Jernberg, T.; Spaak, J.; Erlinge, D.; Marko-Varga, G. Quantitation of 87 Proteins by nLC-MRM/MS in Human Plasma: Workflow for Large-Scale Analysis of Biobank Samples. J. Proteome Res. 2017, 16, 3242–3254. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Forne, I.; Imhof, A. Bioinformatic analysis of proteomics data. BMC Syst. Biol. 2014, 8, S3. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the panther classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB—A database for integrating human functional interaction networks. Nucleic Acids Res. 2009, 37, D623–D628. [Google Scholar] [CrossRef] [PubMed]
- Schwanhüusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Edfors, F.; Danielsson, F.; Hallström, B.M.; Käll, L.; Lundberg, E.; Pontén, F.; Forsström, B.; Uhlén, M. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 2016, 12, 883. [Google Scholar] [CrossRef]
- Venturini, G.; Malagrino, P.A.; Padilha, K.; Tanaka, L.Y.; Laurindo, F.R.; Dariolli, R.; Carvalho, V.M.; Cardozo, K.H.M.; Krieger, J.E.; Pereira, A.d.C. Integrated proteomics and metabolomics analysis reveals differential lipid metabolism in human umbilical vein endothelial cells under high and low shear stress. Am. J. Physiol. Cell Physiol. 2019, 317, C326–C338. [Google Scholar] [CrossRef]
- Marczak, L.; Idkowiak, J.; Tracz, J.; Stobiecki, M.; Perek, B.; Kostka-Jeziorny, K.; Tykarski, A.; Wanic-Kossowska, M.; Borowski, M.; Osuch, M.; et al. Mass spectrometry-based lipidomics reveals differential changes in the accumulated lipid classes in chronic kidney disease. Metabolites 2021, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Rubinow, K.B.; Henderson, C.M.; Robinson-Cohen, C.; Himmelfarb, J.; de Boer, I.H.; Vaisar, T.; Kestenbaum, B.; Hoofnagle, A.N. Kidney function is associated with an altered protein composition of high-density lipoprotein. Kidney Int. 2017, 92, 1526–1535. [Google Scholar] [CrossRef]
- Holzer, M.; Birner-Gruenberger, R.; Stojakovic, T.; El-Gamal, D.; Binder, V.; Wadsack, C.; Heinemann, A.; Marsche, G. Uremia alters HDL composition and function. J. Am. Soc. Nephrol. 2011, 22, 1631–1641. [Google Scholar] [CrossRef]
- Weichhart, T.; Kopecky, C.; Kubicek, M.; Haidinger, M.; Döller, D.; Katholnig, K.; Suarna, C.; Eller, P.; Tölle, M.; Gerner, C.; et al. Serum amyloid A in uremic HDL promotes inflammation. J. Am. Soc. Nephrol. 2012, 23, 934–947. [Google Scholar] [CrossRef]
- Shao, B.; De Boer, I.; Tang, C.; Mayer, P.S.; Zelnick, L.; Afkarian, M.; Heinecke, J.W.; Himmelfarb, J. A Cluster of Proteins Implicated in Kidney Disease Is Increased in High-Density Lipoprotein Isolated from Hemodialysis Subjects. J. Proteome Res. 2015, 14, 2792–2806. [Google Scholar] [CrossRef]
- Mangé, A.; Goux, A.; Badiou, S.; Patrier, L.; Canaud, B.; Maudelonde, T.; Cristol, J.-P.; Solassol, J. HDL proteome in hemodialysis patients: A quantitative nanoflow liquid chromatography-tandem mass spectrometry approach. PLoS ONE 2012, 7, e34107. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.M.; Chung, J.H.; Playford, M.P.; Dey, A.K.; Sviridov, D.; Seifuddin, F.; Chen, Y.C.; Pirooznia, M.; Chen, M.Y.; Mehta, N.N.; et al. High density lipoprotein proteome is associated with cardiovascular risk factors and atherosclerosis burden as evaluated by coronary CT angiography. Atherosclerosis 2018, 278, 278–285. [Google Scholar] [CrossRef]
- Luczak, M.; Formanowicz, D.; Marczak, Ł.; Suszyńska-Zajczyk, J.; Pawliczak, E.; Wanic-Kossowska, M.; Stobiecki, M. ITRAQ-based proteomic analysis of plasma reveals abnormalities in lipid metabolism proteins in chronic kidney disease-related atherosclerosis. Sci. Rep. 2016, 6, 32511. [Google Scholar] [CrossRef] [PubMed]
- Braun, F.; Rinschen, M.M.; Bartels, V.; Frommolt, P.; Habermann, B.; Hoeijmakers, J.H.J.; Schumacher, B.; Dollé, M.E.T.; Müller, R.-U.; Benzing, T.; et al. Altered lipid metabolism in the aging kidney identified by three layered omic analysis. Aging 2016, 8, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Seckler, H.D.S.; Fornelli, L.; Mutharasan, R.K.; Thaxton, C.S.; Fellers, R.; Daviglus, M.; Sniderman, A.; Rader, D.; Kelleher, N.L.; Lloyd-Jones, D.M.; et al. A Targeted, Differential Top-Down Proteomic Methodology for Comparison of ApoA-I Proteoforms in Individuals with High and Low HDL Efflux Capacity. J. Proteome Res. 2018, 17, 2156–2164. [Google Scholar] [CrossRef] [PubMed]
- Luczak, M.; Formanowicz, D.; Marczak, Ł.; Pawliczak, E.; Wanic-Kossowska, M.; Figlerowicz, M.; Stobiecki, M. Deeper insight into chronic kidney disease-related atherosclerosis: Comparative proteomic studies of blood plasma using 2DE and mass spectrometry. J. Transl. Med. 2015, 13, 20. [Google Scholar] [CrossRef]
- Luczak, M.; Suszynska-Zajczyk, J.; Marczak, L.; Formanowicz, D.; Pawliczak, E.; Wanic-Kossowska, M.; Stobiecki, M. Label-free quantitative proteomics reveals differences in molecular mechanism of atherosclerosis related and non-related to chronic kidney disease. Int. J. Mol. Sci. 2016, 17, 631. [Google Scholar] [CrossRef]
- Simic, P.; Kim, W.; Zhou, W.; Pierce, K.A.; Chang, W.; Sykes, D.B.; Aziz, N.B.; Elmariah, S.; Ngo, D.; Pajevic, P.D.; et al. Glycerol-3-phosphate is an FGF23 regulator derived from the injured kidney. J. Clin. Investig. 2020, 130, 1513–1526. [Google Scholar] [CrossRef]
- Mihai, S.; Codrici, E.; Popescu, I.D.; Enciu, A.-M.; Rusu, E.; Zilisteanu, D.; Albulescu, R.; Anton, G.; Tanase, C. Proteomic Biomarkers Panel: New Insights in Chronic Kidney Disease. Dis. Markers 2016, 2016, 3185232. [Google Scholar] [CrossRef]
- Oliveira, E.; Araújo, J.E.; Gómez-Meire, S.; Lodeiro, C.; Perez-Melon, C.; Iglesias-Lamas, E.; Otero-Glez, A.; Capelo, J.L.; Santos, H.M. Proteomics analysis of the peritoneal dialysate effluent reveals the presence of calciumregulation proteins and acute inflammatory response. Clin. Proteom. 2014, 11. [Google Scholar] [CrossRef]
- Wang, C.; Tang, Y.; Wang, Y.; Li, G.; Wang, L.; Li, Y. Label-free quantitative proteomics identifies Smarca4 is involved in vascular calcification. Ren. Fail. 2019, 41, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Opdebeeck, B.; Maudsley, S.; Azmi, A.; De Maré, A.; De Leger, W.; Meijers, B.; Verhulst, A.; Evenepoel, P.; D’Haese, P.C.; Neven, E. Indoxyl Sulfate and p-Cresyl Sulfate Promote Vascular Calcification and Associate with Glucose Intolerance. J. Am. Soc. Nephrol. 2019, 30, 751–766. [Google Scholar] [CrossRef]
- Nakano, T.; Katsuki, S.; Chen, M.; Decano, J.L.; Halu, A.; Lee, L.H.; Pestana, D.V.S.; Kum, A.S.T.; Kuromoto, R.K.; Golden, W.S.; et al. Uremic Toxin Indoxyl Sulfate Promotes Proinflammatory Macrophage Activation Via the Interplay of OATP2B1 and Dll4-Notch Signaling: Potential Mechanism for Accelerated Atherogenesis in Chronic Kidney Disease. Circulation 2019, 139, 78–96. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-H.; Lin, Y.-T.; Chiu, Y.-W.; Baldanzi, G.; Huang, J.-C.; Liang, S.-S.; Lee, S.-C.; Chen, S.-C.; Hsu, Y.-L.; Kuo, M.-C.; et al. The relationship of indoxyl sulfate and p-cresyl sulfate with target cardiovascular proteins in hemodialysis patients. Sci. Rep. 2021, 11, 3786. [Google Scholar] [CrossRef]
- Lin, Y.H.; Platt, M.P.; Fu, H.; Gui, Y.; Wang, Y.; Gonzalez-Juarbe, N.; Zhou, D.; Yu, Y. Global Proteome and Phosphoproteome Characterization of Sepsis-induced Kidney Injury. Mol. Cell. Proteom. 2020, 19, 2030–2046. [Google Scholar] [CrossRef]
- Yang, J.; Brody, E.N.; Murthy, A.C.; Mehler, R.E.; Weiss, S.J.; DeLisle, R.K.; Ostroff, R.; Williams, S.A.; Ganz, P. Impact of Kidney Function on the Blood Proteome and on Protein Cardiovascular Risk Biomarkers in Patients With Stable Coronary Heart Disease. J. Am. Heart Assoc. 2020, 9, e016463. [Google Scholar] [CrossRef]
- Feldreich, T.; Nowak, C.; Fall, T.; Carlsson, A.C.; Carrero, J.J.; Ripsweden, J.; Qureshi, A.R.; Heimbürger, O.; Barany, P.; Stenvinkel, P.; et al. Circulating proteins as predictors of cardiovascular mortality in end-stage renal disease. J. Nephrol. 2019, 32, 111–119. [Google Scholar] [CrossRef]
- Tracz, J.; Handschuh, L.; Lalowski, M.; Marczak, Ł.; Kostka-Jeziorny, K.; Perek, B.; Wanic-Kossowska, M.; Podkowińska, A.; Tykarski, A.; Formanowicz, D.; et al. Proteomic Profiling of Leukocytes Reveals Dysregulation of Adhesion and Integrin Proteins in Chronic Kidney Disease-Related Atherosclerosis. J. Proteome Res. 2021, 20, 3067. [Google Scholar] [CrossRef]
- Kapustin, A.N.; Chatrou, M.L.L.; Drozdov, I.; Zheng, Y.; Davidson, S.M.; Soong, D.; Furmanik, M.; Sanchis, P.; De Rosales, R.T.M.; Alvarez-Hernandez, D.; et al. Vascular Smooth Muscle Cell Calcification Is Mediated by Regulated Exosome Secretion. Circ. Res. 2015, 116, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, G.; Mullen, W.; Duranton, F.; Filip, S.; Gayrard, N.; Husi, H.; Schepers, E.; Neirynck, N.; Schanstra, J.P.; Jankowski, J.; et al. New insights in molecular mechanisms involved in chronic kidney disease using high-resolution plasma proteome analysis. Nephrol. Dial. Transplant. 2015, 30, 1842–1852. [Google Scholar] [CrossRef]
- Luczak, M.; Formanowicz, D.; Pawliczak, E.; Wanic-Kossowska, M.; Wykretowicz, A.; Figlerowicz, M. Chronic kidney disease-related atherosclerosis—Proteomic studies of blood plasma. Proteome Sci. 2011, 9, 25. [Google Scholar] [CrossRef]
- Feng, Y.L.; Cao, G.; Chen, D.Q.; Vaziri, N.D.; Chen, L.; Zhang, J.; Wang, M.; Guo, Y.; Zhao, Y.Y. Microbiome–metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cell. Mol. Life Sci. 2019, 76, 4961–4978. [Google Scholar] [CrossRef]
- Kim, J.E.; Han, D.; Jeong, J.S.; Moon, J.J.; Moon, H.K.; Lee, S.; Kim, Y.C.; Yoo, K.D.; Lee, J.W.; Kim, D.K.; et al. Multisample Mass Spectrometry-Based Approach for Discovering Injury Markers in Chronic Kidney Disease. Mol. Cell. Proteom. 2021, 20, 100037. [Google Scholar] [CrossRef]
- Carbó, C.; Arderiu, G.; Escolar, G.; Fusté, B.; Cases, A.; Carrascal, M.; Abián, J.; Díaz-Ricart, M. Differential Expression of Proteins From Cultured Endothelial Cells Exposed to Uremic Versus Normal Serum. Am. J. Kidney Dis. 2008, 51, 603–612. [Google Scholar] [CrossRef]
- Palomo, M.; Vera, M.; Martin, S.; Torramadé-Moix, S.; Martinez-Sanchez, J.; Belen Moreno, A.; Carreras, E.; Escolar, G.; Cases, A.; Díaz-Ricart, M. Up-regulation of HDACs, a harbinger of uraemic endothelial dysfunction, is prevented by defibrotide. J. Cell. Mol. Med. 2020, 24, 1713–1723. [Google Scholar] [CrossRef]
- Nkuipou-Kenfack, E.; Duranton, F.; Gayrard, N.; Argilés, À.; Lundin, U.; Weinberger, K.M.; Dakna, M.; Delles, C.; Mullen, W.; Husi, H.; et al. Assessment of metabolomic and proteomic biomarkers in detection and prognosis of progression of renal function in chronic kidney disease. PLoS ONE 2014, 9, e96955. [Google Scholar] [CrossRef]
- Hao, W.; Friedman, A. The LDL-HDL Profile Determines the Risk of Atherosclerosis: A Mathematical Model. PLoS ONE 2014, 9, e90497. [Google Scholar] [CrossRef] [PubMed]
- Bandeali, S.; Farmer, J. High-Density Lipoprotein and Atherosclerosis: The Role of Antioxidant Activity. Curr. Atheroscler. Rep. 2012, 14, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Fellström, B.C.; Jardine, A.G.; Schmieder, R.E.; Holdaas, H.; Bannister, K.; Beutler, J.; Chae, D.-W.; Chevaile, A.; Cobbe, S.M.; Grönhagen-Riska, C.; et al. Rosuvastatin and Cardiovascular Events in Patients Undergoing Hemodialysis. N. Engl. J. Med. 2009, 360, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.O.; Bangalore, S.; Lavelle, M.P.; Pellikka, P.A.; Sidhu, M.S.; Boden, W.E.; Asif, A. Diagnosis and management of atherosclerotic cardiovascular disease in chronic kidney disease: A review. Kidney Int. 2017, 91, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, A.; Covic, A.; Fliser, D.; Fouque, D.; Goldsmith, D.; Kanbay, M.; Mallamaci, F.; Massy, Z.A.; Rossignol, P.; Vanholder, R.; et al. Epidemiology, contributors to, and clinical trials of mortality risk in chronic kidney failure. Lancet 2014, 383, 1831–1843. [Google Scholar] [CrossRef]
- Fleischmann, E.H.; Bower, J.D.; Salahudeen, A.K. Risk factor paradox in hemodialysis: Better nutrition as a partial explanation. ASAIO J. 2001, 47. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaysen, G.A. New insights into lipid metabolism in chronic kidney disease. J. Ren. Nutr. 2011, 21, 120–123. [Google Scholar] [CrossRef] [PubMed]
- van der Stoep, M.; Korporaal, S.J.A.; Van Eck, M. High-density lipoprotein as a modulator of platelet and coagulation responses. Cardiovasc. Res. 2014, 103, 362–371. [Google Scholar] [CrossRef]
- Navab, M.; Ananthramaiah, G.M.; Reddy, S.T.; Van Lenten, B.J.; Ansell, B.J.; Fonarow, G.C.; Vahabzadeh, K.; Hama, S.; Hough, G.; Kamranpour, N.; et al. The oxidation hypothesis of atherogenesis: The role of oxidized phospholipids and HDL. J. Lipid. Res. 2004, 45, 993–1007. [Google Scholar] [CrossRef] [PubMed]
- Marsche, G.; Furtmüller, P.G.; Obinger, C.; Sattler, W.; Malle, E. Hypochlorite-modified high-density lipoprotein acts as a sink for myeloperoxidase in vitro. Cardiovasc. Res. 2008, 79, 187–194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, Y.; Wu, Z.; Riwanto, M.; Gao, S.; Levison, B.S.; Gu, X.; Fu, X.; Wagner, M.A.; Besler, C.; Gerstenecker, G.; et al. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J. Clin. Investig. 2013, 123, 3815–3828. [Google Scholar] [CrossRef]
- Kratzer, A.; Giral, H.; Landmesser, U. High-density lipoproteins as modulators of endothelial cell functions: Alterations in patients with coronary artery disease. Cardiovasc. Res. 2014, 103, 350–361. [Google Scholar] [CrossRef]
- Kronenberg, F.; Kuen, E.; Ritz, E.; Konig, P.; Kraatz, G.; Lhotta, K.; Mann, J.F.; Muller, G.A.; Neyer, U.; Riegel, W.; et al. Apolipoprotein A-IV serum concentrations are elevated in patients with mild and moderate renal failure. J. Am. Soc. Nephrol. 2002, 13, 461–469. [Google Scholar] [CrossRef]
- Karohl, C.; D’Marco Gascón, L.; Raggi, P. Noninvasive imaging for assessment of calcification in chronic kidney disease. Nat. Rev. Nephrol. 2011, 7, 567–577. [Google Scholar] [CrossRef]
- Otsuka, F.; Sakakura, K.; Yahagi, K.; Joner, M.; Virmani, R. Has our understanding of calcification in human coronary atherosclerosis progressed? Arterioscler. Thromb. Vasc. Biol. 2014, 34, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Garland, J.S.; Holden, R.M.; Groome, P.A.; Lam, M.; Nolan, R.L.; Morton, A.R.; Pickett, W. Prevalence and Associations of Coronary Artery Calcification in Patients With Stages 3 to 5 CKD Without Cardiovascular Disease. Am. J. Kidney Dis. 2008, 52, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Ishibashi-Ueda, H.; Niizuma, S.; Yoshihara, F.; Horio, T.; Kawano, Y. Coronary calcification in patients with chronic kidney disease and coronary artery disease. Clin. J. Am. Soc. Nephrol. 2009, 4, 1892–1900. [Google Scholar] [CrossRef]
- Matsubara, K.; Stenvinkel, P.; Qureshi, A.R.; Carrero, J.J.; Axelsson, J.; Heimbürger, O.; Bárány, P.; Alvestrand, A.; Lindholm, B.; Suliman, M.E. Inflammation modifies the association of osteoprotegerin with mortality in chronic kidney disease. J. Nephrol. 2009, 22, 774–782. [Google Scholar] [PubMed]
- Stockler-Pinto, M.B.; Saldanha, J.F.; Yi, D.; Mafra, D.; Fouque, D.; Soulage, C.O. The uremic toxin indoxyl sulfate exacerbates reactive oxygen species production and inflammation in 3T3-L1 adipose cells. Free Radic. Res. 2016, 50, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-J.; Wu, V.; Wu, P.-C.; Wu, C.-J. Meta-Analysis of the Associations of p-Cresyl Sulfate (PCS) and Indoxyl Sulfate (IS) with Cardiovascular Events and All-Cause Mortality in Patients with Chronic Renal Failure. PLoS ONE 2015, 10, e0132589. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Osaka, M.; Edamatsu, T.; Itoh, Y.; Yoshida, M. Crucial Role of the Aryl Hydrocarbon Receptor (AhR) in Indoxyl Sulfate-Induced Vascular Inflammation. J. Atheroscler. Thromb. 2016, 23, 960–975. [Google Scholar] [CrossRef] [PubMed]
- Serradell, M.; Díaz-Ricart, M.; Cases, A.; Zurbano, M.; López-Pedret, J.; Arranz, O.; Ordinas, A.; Escolar, G. Uremic medium causes expression, redistribution and shedding of adhesion molecules in cultured endothelial cells. Haematologica 2002, 87, 1053–1061. [Google Scholar]
- Barreto, F.C.; Barreto, D.V.; Liabeuf, S.; Meert, N.; Glorieux, G.; Temmar, M.; Choukroun, G.; Vanholder, R.; Massy, Z.A.; European Uremic Toxin Work Group (EUTox), on behalf of the E.U.T.W.G. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin. J. Am. Soc. Nephrol. 2009, 4, 1551–1558. [Google Scholar] [CrossRef]
- Liabeuf, S.; Barreto, D.V.; Barreto, F.C.; Meert, N.; Glorieux, G.; Schepers, E.; Temmar, M.; Choukroun, G.; Vanholder, R.; Massy, Z.A.; et al. Free p-cresylsulphate is a predictor of mortality in patients at different stages of chronic kidney disease. Nephrol. Dial. Transplant. 2010, 25, 1183–1191. [Google Scholar] [CrossRef]
- Meijers, B.K.I.; Van kerckhoven, S.; Verbeke, K.; Dehaen, W.; Vanrenterghem, Y.; Hoylaerts, M.F.; Evenepoel, P. The Uremic Retention Solute p-Cresyl Sulfate and Markers of Endothelial Damage. Am. J. Kidney Dis. 2009, 54, 891–901. [Google Scholar] [CrossRef]
- Watanabe, H.; Miyamoto, Y.; Honda, D.; Tanaka, H.; Wu, Q.; Endo, M.; Noguchi, T.; Kadowaki, D.; Ishima, Y.; Kotani, S.; et al. P-Cresyl sulfate causes renal tubular cell damage by inducing oxidative stress by activation of NADPH oxidase. Kidney Int. 2013, 83, 582–592. [Google Scholar] [CrossRef]
- Cozzolino, M.; Ciceri, P.; Galassi, A.; Mangano, M.; Carugo, S.; Capelli, I.; Cianciolo, G. The Key Role of Phosphate on Vascular Calcification. Toxins 2019, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Xu, J.; Zhang, S.; Jin, J. Meta-Analysis of the Association between Fibroblast Growth Factor 23 and Mortality and Cardiovascular Events in Hemodialysis Patients. Blood Purif. 2019, 47, 24–30. [Google Scholar] [CrossRef]
- Reynolds, J.L.; Joannides, A.J.; Skepper, J.N.; McNair, R.; Schurgers, L.J.; Proudfoot, D.; Jahnen-Dechent, W.; Weissberg, P.L.; Shanahan, C.M. Human vascular smooth muscle cells undergo vesicle-mediated calcification in response to changes in extracellular calcium and phosphate concentrations: A potential mechanism for accelerated vascular calcification in ESRD. J. Am. Soc. Nephrol. 2004, 15, 2857–2867. [Google Scholar] [CrossRef]
- M’baya-Moutoula, E.; Louvet, L.; Molinié, R.; Guerrera, I.C.; Cerutti, C.; Fourdinier, O.; Nourry, V.; Gutierrez, L.; Morlière, P.; Mesnard, F.; et al. A multi-omics analysis of the regulatory changes induced by miR-223 in a monocyte/macrophage cell line. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2664–2678. [Google Scholar] [CrossRef] [PubMed]
- Gimbrone, M.A.; García-Cardeña, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef]
- Mohindra, R.; Agrawal, D.K.; Thankam, F.G. Altered Vascular Extracellular Matrix in the Pathogenesis of Atherosclerosis. J. Cardiovasc. Transl. Res. 2021, 1–14. [Google Scholar] [CrossRef]
- Trojanowicz, B.; Ulrich, C.; Kohler, F.; Bode, V.; Seibert, E.; Fiedler, R.; Girndt, M. Monocytic angiotensin-converting enzyme 2 relates to atherosclerosis in patients with chronic kidney disease. Nephrol. Dial. Transplant. 2016, 32, gfw206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, C.-Y.; Tarng, D.-C. Diet, gut microbiome and indoxyl sulphate in chronic kidney disease patients. Nephrology 2018, 23, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Fowler, M.J. Microvascular and Macrovascular Complications of Diabetes. Clin. Diabetes 2008, 26, 77–82. [Google Scholar] [CrossRef]
- Abe, M.; Kalantar-Zadeh, K. Haemodialysis-induced hypoglycaemia and glycaemic disarrays. Nat. Rev. Nephrol. 2015, 11, 302–313. [Google Scholar] [CrossRef]
- Pecoits-Filho, R.; Abensur, H.; Betônico, C.C.R.; Machado, A.D.; Parente, E.B.; Queiroz, M.; Salles, J.E.N.; Titan, S.; Vencio, S. Interactions between kidney disease and diabetes: Dangerous liaisons. Diabetol. Metab. Syndr. 2016, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- La Sala, L.; Prattichizzo, F.; Ceriello, A. The link between diabetes and atherosclerosis. Eur. J. Prev. Cardiol. 2019, 26, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Caubet, C.; Camus, M.; Makridakis, M.; Denis, C.; Gilet, M.; Feuillet, G.; Rascalou, S.; Neau, E.; Garrigues, L.; et al. Connectivity mapping of glomerular proteins identifies dimethylaminoparthenolide as a new inhibitor of diabetic kidney disease. Sci. Rep. 2020, 10, 14898. [Google Scholar] [CrossRef]
- Santana, M.F.M.; Lira, A.L.A.; Pinto, R.S.; Minanni, C.A.; Silva, A.R.M.; Sawada, M.I.B.A.C.; Nakandakare, E.R.; Correa-Giannella, M.L.C.; Queiroz, M.S.; Ronsein, G.E.; et al. Enrichment of apolipoprotein A-IV and apolipoprotein D in the HDL proteome is associated with HDL functions in diabetic kidney disease without dialysis. Lipids Health Dis. 2020, 19, 205. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Ma, Z.; Livingston, M.J.; Zhang, W.; Yuan, Y.; Guo, C.; Liu, Y.; Fu, P.; Dong, Z. Decreased secretion and profibrotic activity of tubular exosomes in diabetic kidney disease. Am. J. Physiol. Ren. Physiol. 2020, 319, F664–F673. [Google Scholar] [CrossRef]
- Gajjala, P.R.; Bruck, H.; Noels, H.; Heinze, G.; Ceccarelli, F.; Kribben, A.; Saez-Rodriguez, J.; Marx, N.; Zidek, W.; Jankowski, J.; et al. Novel plasma peptide markers involved in the pathology of CKD identified using mass spectrometric approach. J. Mol. Med. 2019, 97, 1451–1463. [Google Scholar] [CrossRef]
- Romanova, Y.; Laikov, A.; Markelova, M.; Khadiullina, R.; Makseev, A.; Hasanova, M.; Rizvanov, A.; Khaiboullina, S.; Salafutdinov, I. Proteomic analysis of human serum from patients with chronic kidney disease. Biomolecules 2020, 10, 257. [Google Scholar] [CrossRef]
- Konvalinka, A.; Batruch, I.; Tokar, T.; Dimitromanolakis, A.; Reid, S.; Song, X.; Pei, Y.; Drabovich, A.P.; Diamandis, E.P.; Jurisica, I.; et al. Quantification of angiotensin II-regulated proteins in urine of patients with polycystic and other chronic kidney diseases by selected reaction monitoring. Clin. Proteom. 2016, 13, 16. [Google Scholar] [CrossRef]
- Øvrehus, M.A.; Zürbig, P.; Vikse, B.E.; Hallan, S.I. Urinary proteomics in chronic kidney disease: Diagnosis and risk of progression beyond albuminuria. Clin. Proteom. 2015, 12. [Google Scholar] [CrossRef][Green Version]
- Jiang, S.; He, H.; Tan, L.; Wang, L.; Su, Z.; Liu, Y.; Zhu, H.; Zhang, M.; Hou, F.F.; Li, A. Proteomic and phosphoproteomic analysis of renal cortex in a salt-load rat model of advanced kidney damage. Sci. Rep. 2016, 6, 35906. [Google Scholar] [CrossRef] [PubMed]
- Herrington, D.M.; Mao, C.; Parker, S.J.; Fu, Z.; Yu, G.; Chen, L.; Venkatraman, V.; Fu, Y.; Wang, Y.; Howard, T.D.; et al. Proteomic Architecture of Human Coronary and Aortic Atherosclerosis. Circulation 2018, 137, 2741–2756. [Google Scholar] [CrossRef] [PubMed]
- Olson, F.J.; Sihlbom, C.; Davidsson, P.; Hulthe, J.; Fagerberg, B.; Bergström, G. Consistent differences in protein distribution along the longitudinal axis in symptomatic carotid atherosclerotic plaques. Biochem. Biophys. Res. Commun. 2010, 401, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Wallentin, L.; Eriksson, N.; Olszowka, M.; Grammer, T.B.; Hagström, E.; Held, C.; Kleber, M.E.; Koenig, W.; März, W.; Stewart, R.A.H.; et al. Plasma proteins associated with cardiovascular death in patients with chronic coronary heart disease: A retrospective study. PLoS Med. 2021, 18, e1003513. [Google Scholar] [CrossRef]
- Yong, K.; Ooi, E.M.; Dogra, G.; Mannion, M.; Boudville, N.; Chan, D.; Lim, E.M.; Lim, W.H. Cytokine Elevated interleukin-12 and interleukin-18 in chronic kidney disease are not associated with arterial stiffness. Cytokine 2013, 64, 39–42. [Google Scholar] [CrossRef]
- Scholze, A.; Krueger, K.; Diedrich, M.; Räth, C.; Torges, A.; Jankowski, V.; Maier, A.; Thilo, F.; Zidek, W.; Tepel, M. Superoxide dismutase type 1 in monocytes of chronic kidney disease patients. Amino Acids 2011, 41, 427–438. [Google Scholar] [CrossRef]
- Marques, M.; Sacristán, D.; Mateos-Cáceres, P.J.; Herrero, J.; Arribas, M.J.; González-Armengol, J.J.; Villegas, A.; Macaya, C.; Barrientos, A.; López-Farré, A.J. Different protein expression in normal and dysfunctional platelets from uremic patients. J. Nephrol. 2010, 23, 90–101. [Google Scholar] [PubMed]
- Carlsson, A.C.; Ingelsson, E.; Sundström, J.; Carrero, J.J.; Gustafsson, S.; Feldreich, T.; Stenemo, M.; Larsson, A.; Lind, L.; Ärnlöv, J. Use of proteomics to investigate kidney function decline over 5 years. Clin. J. Am. Soc. Nephrol. 2017, 12, 1226–1235. [Google Scholar] [CrossRef]
- Feldreich, T.; Nowak, C.; Carlsson, A.C.; Östgren, C.J.; Nyström, F.H.; Sundström, J.; Carrero-Roig, J.J.; Leppert, J.; Hedberg, P.; Giedraitis, V.; et al. The association between plasma proteomics and incident cardiovascular disease identifies MMP-12 as a promising cardiovascular risk marker in patients with chronic kidney disease. Atherosclerosis 2020, 307, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Beck, S. Making multi-omics data accessible to researchers. Sci. Data 2019, 6, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.Y.; Ballard, S.; Batlle, D.; Bonventre, J.V.; Böttinger, E.P.; Feldman, H.I.; Klein, J.B.; Coresh, J.; Eckfeldt, J.H.; Inker, L.A.; et al. Cross-disciplinary biomarkers research: Lessons learned by the CKD biomarkers consortium. Clin. J. Am. Soc. Nephrol. 2015, 10, 894–902. [Google Scholar] [CrossRef] [PubMed]
| Studied Mechanism/Process in CKD-A | Applied “Omics” Strategy | References |
|---|---|---|
| Unique dyslipidemia | Lipidomics | [57] |
| Alterations in lipid transport- and metabolism-related molecules | MS-based proteomics Transcriptomics Metabolomics | [58,59,60,61,62,63,64,65] |
| Disturbances in chemical composition, structure and functionality of HDL | MS-based proteomics Lipidomics | [58,59,60,61,62,63,64,66] |
| Association of vascular calcification with accelerated inflammation and the progression of CKD | MS-based proteomics | [67,68,69,70,71,72] |
| Relationships between uremic toxins and development of vascular calcification | MS-based proteomics Aptamer-based proteomics Metabolomics | [69,73,74] |
| The mechanism of inflammation and atherogenesis development in renal and non-renal conditions | MS-based proteomics Transcriptomics Aptamer-based proteomics | [75,76,77,78,79] |
| The mechanism of VSMC calcification | MS-based proteomics | [80] |
| Enhanced inflammatory state or inflammation imbalance | MS-based proteomics | [67,68,76,79,81,82,83,84] |
| Disturbances in ECM turnover, endothelial adhesion and transmigration | MS-based proteomics Metabolomics Transcriptomics | [79,84,85,86,87] |
| Dysregulation of hemostasis and coagulation process | MS-based proteomics | [67,68,75,79,81,82,86,87] |
| The mechanism of hypoxia and oxidative stress | MS-based proteomics Transcriptomics | [84] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tracz, J.; Luczak, M. Applying Proteomics and Integrative “Omics” Strategies to Decipher the Chronic Kidney Disease-Related Atherosclerosis. Int. J. Mol. Sci. 2021, 22, 7492. https://doi.org/10.3390/ijms22147492
Tracz J, Luczak M. Applying Proteomics and Integrative “Omics” Strategies to Decipher the Chronic Kidney Disease-Related Atherosclerosis. International Journal of Molecular Sciences. 2021; 22(14):7492. https://doi.org/10.3390/ijms22147492
Chicago/Turabian StyleTracz, Joanna, and Magdalena Luczak. 2021. "Applying Proteomics and Integrative “Omics” Strategies to Decipher the Chronic Kidney Disease-Related Atherosclerosis" International Journal of Molecular Sciences 22, no. 14: 7492. https://doi.org/10.3390/ijms22147492
APA StyleTracz, J., & Luczak, M. (2021). Applying Proteomics and Integrative “Omics” Strategies to Decipher the Chronic Kidney Disease-Related Atherosclerosis. International Journal of Molecular Sciences, 22(14), 7492. https://doi.org/10.3390/ijms22147492






