A Beckwith–Wiedemann-Associated CDKN1C Mutation Allows the Identification of a Novel Nuclear Localization Signal in Human p57Kip2
Abstract
1. Introduction
2. Results
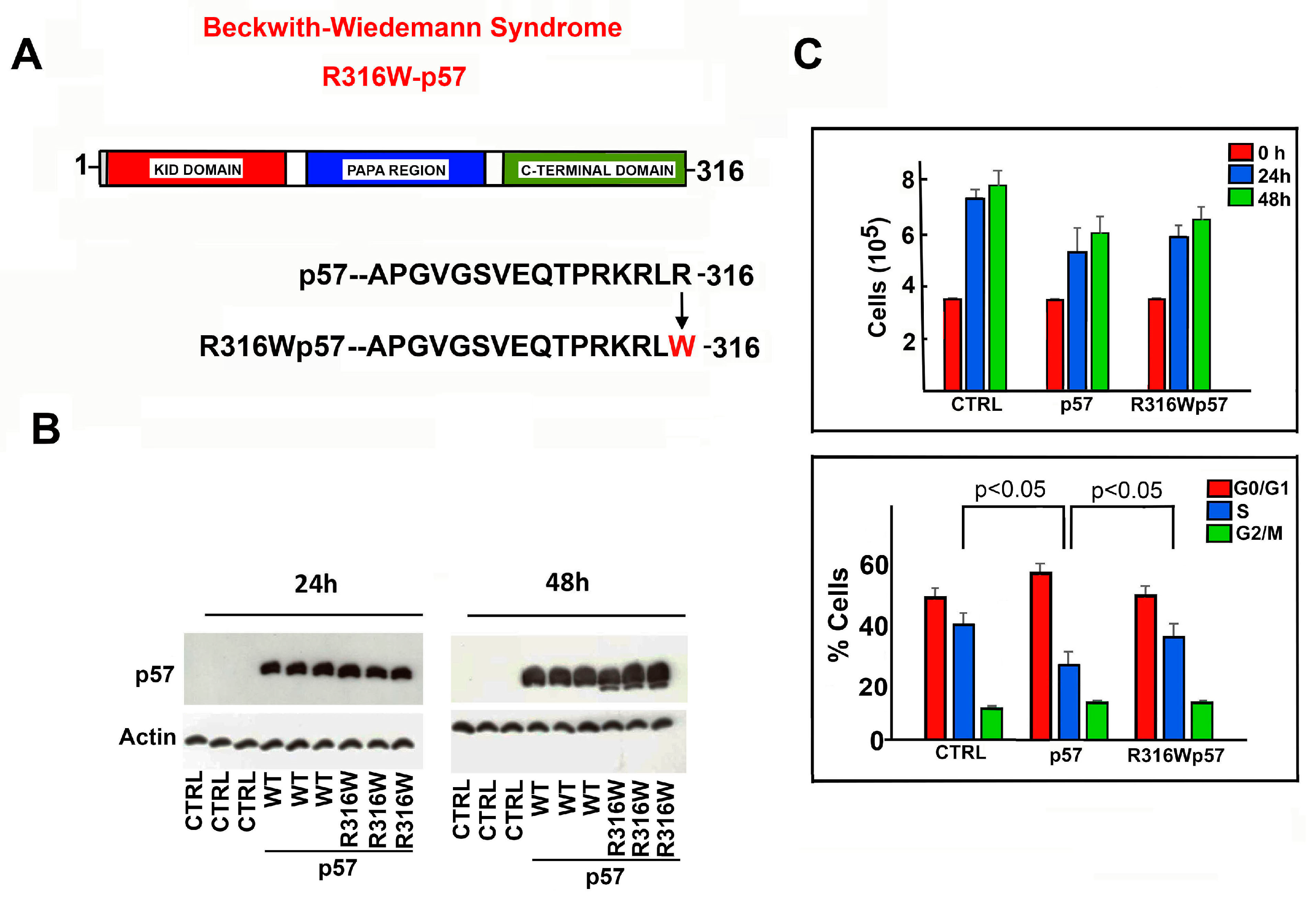
2.1. Effects of R316W-p57 Transfection on Cell Proliferation Rate and Cell Cycle Phase Distribution
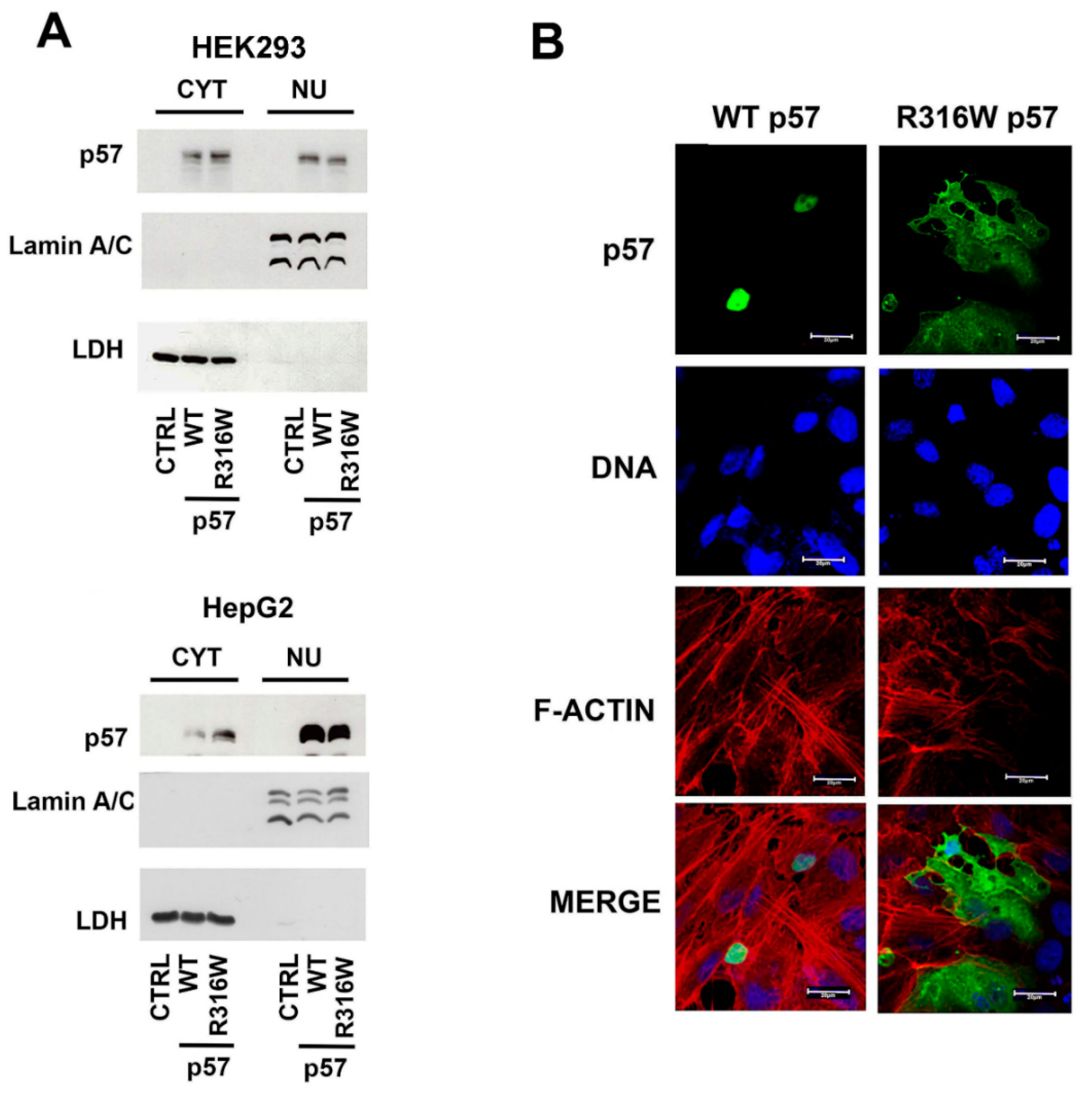
2.2. Subcellular Localization of R316W-p57
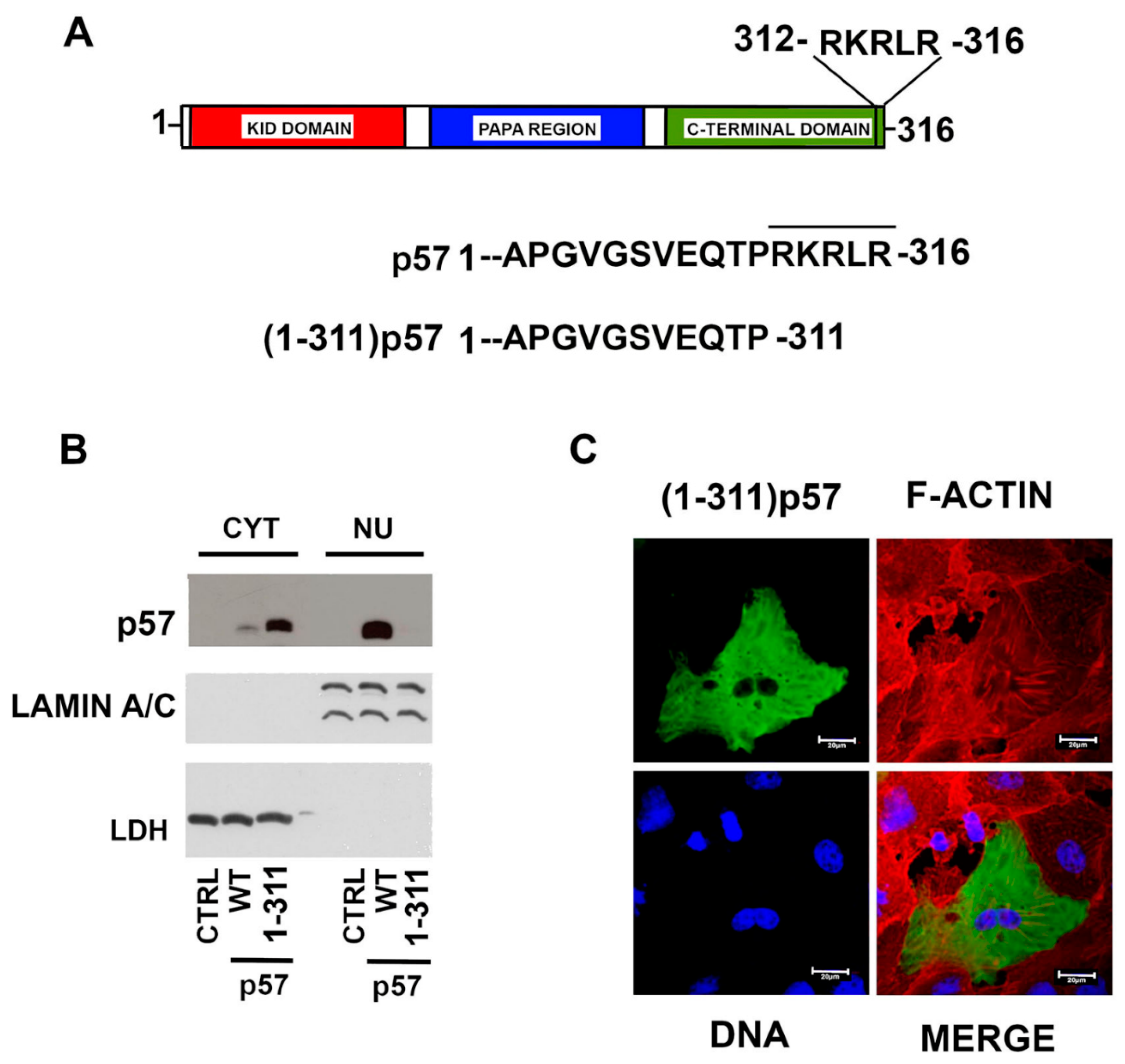
2.3. The Nuclear Localization of p57 Depends on the C-Terminal Domain
2.4. A Putative NLS Sequence Occurs at the End of the p57 C-Terminal Domain
2.5. Identification of p57 NLS
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Cell Extracts Preparation and Immunoblotting Analyses
4.2. Plasmid Preparation and Transfection
4.3. Analysis of Proliferation Rate and Cell Cycle Distribution
4.4. Immunofluorescence Microscopy
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, M.H.; Reynisdóttir, I.; Massagué, J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995, 9, 639–649. [Google Scholar] [CrossRef]
- Matsuoka, S.; Edwards, M.C.; Bai, C.; Parker, S.; Zhang, P.; Baldini, A.; Harper, J.W.; Elledge, S.J. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995, 9, 650–662. [Google Scholar] [CrossRef]
- Borriello, A.; Caldarelli, I.; Bencivenga, D.; Criscuolo, M.; Cucciolla, V.; Tramontano, A.; Oliva, A.; Perrotta, S.; Della Ragione, F. p57(Kip2) and cancer: Time for a critical appraisal. Mol. Cancer Res. 2011, 9, 1269–1284. [Google Scholar] [CrossRef]
- Eggermann, T.; Binder, G.; Brioude, F.; Maher, R.E.; Lapunzina, P.; Cubellis, M.V.; Bergadá, I.; Prawitt, D.; Begemann, M. CDKN1C mutations: Two sides of the same coin. Trends Mol. Med. 2014, 20, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.N.; Andresini, O.; Matteini, F.; Maione, R. Transcriptional regulation of p57kip2 expression during development, differentiation and disease. Front. Biosci. 2018, 23, 83–108. [Google Scholar] [CrossRef]
- Stampone, E.; Caldarelli, I.; Zullo, A.; Bencivenga, D.; Mancini, F.P.; Della Ragione, F.; Borriello, A. Genetic and Epigenetic Control of CDKN1C Expression: Importance in Cell Commitment and Differentiation, Tissue Homeostasis and Human Diseases. Int. J. Mol. Sci. 2018, 19, 1055. [Google Scholar] [CrossRef]
- Russo, G.L.; Stampone, E.; Cervellera, C.; Borriello, A. Regulation of p27Kip1 and p57Kip2 Functions by Natural Polyphenols. Biomolecules 2020, 10, 1316. [Google Scholar] [CrossRef] [PubMed]
- Creff, J.; Besson, A. Functional Versatility of the CDK Inhibitor p57Kip2. Front. Cell Dev. Biol. 2020, 8, 584590. [Google Scholar] [CrossRef]
- Russo, A.A.; Jeffrey, P.D.; Patten, A.K.; Massagué, J.; Pavletich, N.P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 1996, 382, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Yokoo, T.; Toyoshima, H.; Miura, M.; Miura, M.; Wang, Y.; Iida, K.T.; Suzuki, H.; Sone, H.; Shimano, H.; Gotoda, T.; et al. p57Kip2 regulates actin dynamics by binding and translocating LIM-kinase 1 to the nucleus. J. Biol. Chem. 2003, 278, 52919–52923. [Google Scholar] [CrossRef]
- Vlachos, P.; Joseph, B. The Cdk inhibitor p57(Kip2) controls LIM-kinase 1 activity and regulates actin cytoskeleton dynamics. Oncogene 2009, 28, 4175–4188. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S.; Kim, M.J.; Ryoo, K.; Park, J.; Eom, S.-J.; Shim, J.; Nakayama, K.I.; Nakayama, K.; Tomita, M.; Takahashi, K.; et al. p57KIP2 modulates stress-activated signaling by inhibiting c-Jun NH2-terminal kinase/stress-activated protein Kinase. J. Biol. Chem. 2003, 278, 48092–48098. [Google Scholar] [CrossRef]
- Luo, Y.; Hurwitz, J.; Massagué, J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature 1995, 375, 159–161. [Google Scholar] [CrossRef]
- Watanabe, H.; Pan, Z.Q.; Schreiber-Agus, N.; De Pinho, R.A.; Hurwitz, J.; Xiong, Y. Suppression of cell transformation by the cyclin-dependent kinase inhibitor p57KIP2 requires binding to proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA 1998, 95, 1392–1397. [Google Scholar] [CrossRef]
- Polyak, K.; Lee, M.H.; Erdjument-Bromage, H.; Koff, A.; Roberts, J.M.; Tempst, P.; Massagué, J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 1994, 78, 59–66. [Google Scholar] [CrossRef]
- Zeng, Y.; Hirano, K.; Hirano, M.; Nishimura, J.; Kanaide, H. Minimal requirements for the nuclear localization of p27(Kip1), a cyclin-dependent kinase inhibitor. Biochem. Biophys. Res. Commun. 2000, 274, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Frisén, J.; Lee, M.H.; Massagué, J.; Barbacid, M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997, 11, 973–983. [Google Scholar] [CrossRef]
- Zhang, P.; Liégeois, N.J.; Wong, C.; Finegold, M.; Hou, H.; Thompson, J.C.; Silverman, A.; Harper, J.W.; DePinho, R.A.; Elledge, S.J. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature 1997, 387, 151–158. [Google Scholar] [CrossRef]
- Arboleda, V.A.; Lee, H.; Parnaik, R.; Fleming, A.; Banerjee, A.; Ferraz-de-Souza, B.; Délot, E.C.; Rodriguez-Fernandez, I.A.; Braslavsky, D.; Bergadá, I. Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat. Genet. 2012, 44, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Eggermannm, T.; Brioude, F.; Russo, S.; Lombardi, M.P.; Bliek, J.; Maher, E.R.; Larizza, L.; Prawitt, D.; Netchine, I.; Gonzale, M. Prenatal molecular testing for Beckwith-Wiedemann and Silver-Russell syndromes: A challenge for molecular analysis and genetic counseling. Eur. J. Hum. Genet. 2016, 24, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Brioude, F.; Netchine, I.; Praz, F.; Le Jule, M.; Calmel, C.; Lacombe, D.; Edery, P.; Catala, M.; Odent, S.; Isidor, B.; et al. Mutations of the Imprinted CDKN1C Gene as a Cause of the Overgrowth Beckwith-Wiedemann Syndrome: Clinical Spectrum and Functional Characterization. Hum. Mutat. 2015, 36, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Hatada, I.; Inazawa, J.; Abe, T.; Nakayama, M.; Kaneko, Y.; Jinno, Y.; Niikawa, N.; Ohashi, H.; Fukushima, Y.; Iida, K. Genomic imprinting of human p57KIP2 and its reduced expression in Wilms’ tumors. Hum. Mol. Genet. 1996, 5, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.W.; Hatada, I.; Ohishi, S.; Mukai, T.; Joyce, J.A.; Cole, T.R.; Donnai, D.; Reik, W.; Schofield, P.N.; Maher, E.R. Analysis of germline CDKN1C (p57KIP2) mutations in familial and sporadic Beckwith-Wiedemann syndrome (BWS) provides a novel genotype-phenotype correlation. J. Med. Genet. 1999, 36, 518–523, PMID: 10424811, PMCID: PMC1734395. [Google Scholar] [PubMed]
- Boulikas, T. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 1993, 3, 193–227, PMID:8241603. [Google Scholar] [PubMed]
- Brioude, F.; Oliver-Petit, I.; Blaise, A.; Praz, F.; Rossignol, S.; Le Jule, M.; Thibaud, N.; Faussat, A.M.; Tauber, M.; Le Bouc, Y.; et al. CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell Silver syndrome. J. Med. Genet. 2013, 50, 823–830. [Google Scholar] [CrossRef]
- Heide, S.; Chantot-Bastaraud, S.; Keren, B.; Harbison, M.D.; Azzi, S.; Rossignol, S.; Michot, C.; Lackmy-Port Lys, M.; Demeer, B.; Heinrichs, C. Chromosomal rearrangements in the 11p15 imprinted region: 17 new 11p15.5 duplications with associated phenotypes and putative functional consequences. J. Med. Genet. 2018, 55, 205–213. [Google Scholar] [CrossRef]
- Brioude, F.; Kalish, J.M.; Mussa, A.; Foster, A.C.; Bliek, J.; Ferrero, G.B.; Boonen, S.E.; Cole, T.; Baker, R.; Bertoletti, M. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: An international consensus statement. Nat. Rev. Endocrinol. 2018, 14, 229–249. [Google Scholar] [CrossRef]
- Wakeling, E.L.; Brioude, F.; Lokulo-Sodipe, O.; O’Connell, S.M.; Salem, J.; Bliek, J.; Canton, A.P.M.; Chrzanowska, K.H.; Davies, J.H.; Dias, R.P.; et al. Diagnosis and management of Silver-Russell syndrome: First international consensus statement. Nat. Rev. Endocrinol. 2017, 13, 105–124. [Google Scholar] [CrossRef]
- Bennett, J.; Schrier Vergano, S.A.; Deardorff, M.A. IMAGe Syndrome. 2014 [updated 2016 September 8]. In GeneReviews® [Internet]; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Eds.; University of Washington: Seattle, WA, USA, 1993–2020; PMID: 24624461. [Google Scholar]
- Choufani, S.; Shuman, C.; Weksberg, R. Beckwith-Wiedemann syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2010, 154C, 343–354. [Google Scholar] [CrossRef]
- Cabrera-Salcedo, C.; Kumar, P.; Hwa, V.; Dauber, A. IMAGe and Related Undergrowth Syndromes: The Complex Spectrum of Gain-of-Function CDKN1C Mutations. Pediatric Endocrinol. Rev. 2017, 14, 289–297. [Google Scholar] [CrossRef]
- Hamajima, N.; Johmura, Y.; Suzuki, S.; Nakanishi, M.; Saitoh, S. Increased protein stability of CDKN1C causes a gain-of-function phenotype in patients with IMAGe syndrome. PLoS ONE 2013, 8, e75137. [Google Scholar] [CrossRef]
- Borges, K.S.; Arboleda, V.A.; Vilain, E. Mutations in the PCNA-binding site of CDKN1C inhibit cell proliferation by impairing the entry into S phase. Cell Div. 2015, 10, 1–6. [Google Scholar] [CrossRef]
- Bhuiyan, Z.A.; Yatsuki, H.; Sasaguri, T.; Joh, K.; Soejima, H.; Zhu, X.; Hatada, I.; Morisaki, H.; Morisaki, T.; Mukai, T. Functional analysis of the p57KIP2 gene mutation in Beckwith-Wiedemann syndrome. Hum. Genet. 1999, 104, 205–210. [Google Scholar] [CrossRef]
- Eggermann, T.; Algar, E.; Lapunzina, P.; Mackay, D.; Maher, E.R.; Mannens, M.; Netchine, I.; Prawitt, D.; Riccio, A. Clinical utility gene card for: Beckwith-Wiedemann Syndrome. Eur. J. Hum. Genet. 2014, 22. [Google Scholar] [CrossRef] [PubMed]
- Iolascon, A.; Giordani, L.; Moretti, A.; Basso, G.; Borriello, A.; Della Ragione, F. Analysis of CDKN2A, CDKN2B, CDKN2C, and cyclin Ds gene status in hepatoblastoma. Hepatology 1998, 27, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Cucciolla, V.; Borriello, A.; Criscuolo, M.; Sinisi, A.A.; Bencivenga, D.; Tramontano, A.; Scudieri, A.C.; Oliva, A.; Zappia, V.; Della Ragione, F. Histone deacetylase inhibitors upregulate p57Kip2 level by enhancing its expression through Sp1 transcription factor. Carcinogenesis 2008, 29, 560–567. [Google Scholar] [CrossRef][Green Version]
- Natalicchio, M.I.; Improta, G.; Zupa, A.; Cursio, O.E.; Stampone, E.; Possidente, L.; Gerardi, A.M.T.; Vita, G.; Martini, G.; Cassano, A. Pyrosequencing evaluation of low-frequency KRAS mutant alleles for EGF receptor therapy selection in metastatic colorectal carcinoma. Future Oncol. 2014, 10, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Borriello, A.; Naviglio, S.; Bencivenga, D.; Caldarelli, I.; Tramontano, A.; Speranza, M.C.; Stampone, E.; Sapio, L.; Negri, A.; Oliva, A. Histone Deacetylase Inhibitors Increase p27(Kip1) by Affecting Its Ubiquitin-Dependent Degradation through Skp2 Downregulation. Oxidative Med. Cell. Longev. 2016, 2016, 2481865. [Google Scholar] [CrossRef] [PubMed]
- Stampone, E.; Bencivenga, D.; Barone, C.; Aulitto, A.; Verace, F.; Della Ragione, F.; Borriello, A. High Dosage Lithium Treatment Induces DNA Damage and p57Kip2 Decrease. Int. J. Mol. Sci. 2020, 21, 1169. [Google Scholar] [CrossRef] [PubMed]
- Bencivenga, D.; Stampone, E.; Aulitto, A.; Tramontano, A.; Barone, C.; Negri, A.; Roberti, D.; Perrotta, S.; Della Ragione, F.; Borriello, A. A cancer-associated CDKN1B mutation induces p27 phosphorylation on a novel residue: A new mechanism for tumor suppressor loss-of-function. Mol. Oncol. 2021, 15, 915–941. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stampone, E.; Bencivenga, D.; Barone, C.; Di Finizio, M.; Della Ragione, F.; Borriello, A. A Beckwith–Wiedemann-Associated CDKN1C Mutation Allows the Identification of a Novel Nuclear Localization Signal in Human p57Kip2. Int. J. Mol. Sci. 2021, 22, 7428. https://doi.org/10.3390/ijms22147428
Stampone E, Bencivenga D, Barone C, Di Finizio M, Della Ragione F, Borriello A. A Beckwith–Wiedemann-Associated CDKN1C Mutation Allows the Identification of a Novel Nuclear Localization Signal in Human p57Kip2. International Journal of Molecular Sciences. 2021; 22(14):7428. https://doi.org/10.3390/ijms22147428
Chicago/Turabian StyleStampone, Emanuela, Debora Bencivenga, Clementina Barone, Marilena Di Finizio, Fulvio Della Ragione, and Adriana Borriello. 2021. "A Beckwith–Wiedemann-Associated CDKN1C Mutation Allows the Identification of a Novel Nuclear Localization Signal in Human p57Kip2" International Journal of Molecular Sciences 22, no. 14: 7428. https://doi.org/10.3390/ijms22147428
APA StyleStampone, E., Bencivenga, D., Barone, C., Di Finizio, M., Della Ragione, F., & Borriello, A. (2021). A Beckwith–Wiedemann-Associated CDKN1C Mutation Allows the Identification of a Novel Nuclear Localization Signal in Human p57Kip2. International Journal of Molecular Sciences, 22(14), 7428. https://doi.org/10.3390/ijms22147428








