Probing the Energetic Metabolism of Resting Cysts under Different Conditions from Molecular and Physiological Perspectives in the Harmful Algal Blooms-Forming Dinoflagellate Scrippsiella trochoidea
Abstract
1. Introduction
2. Results
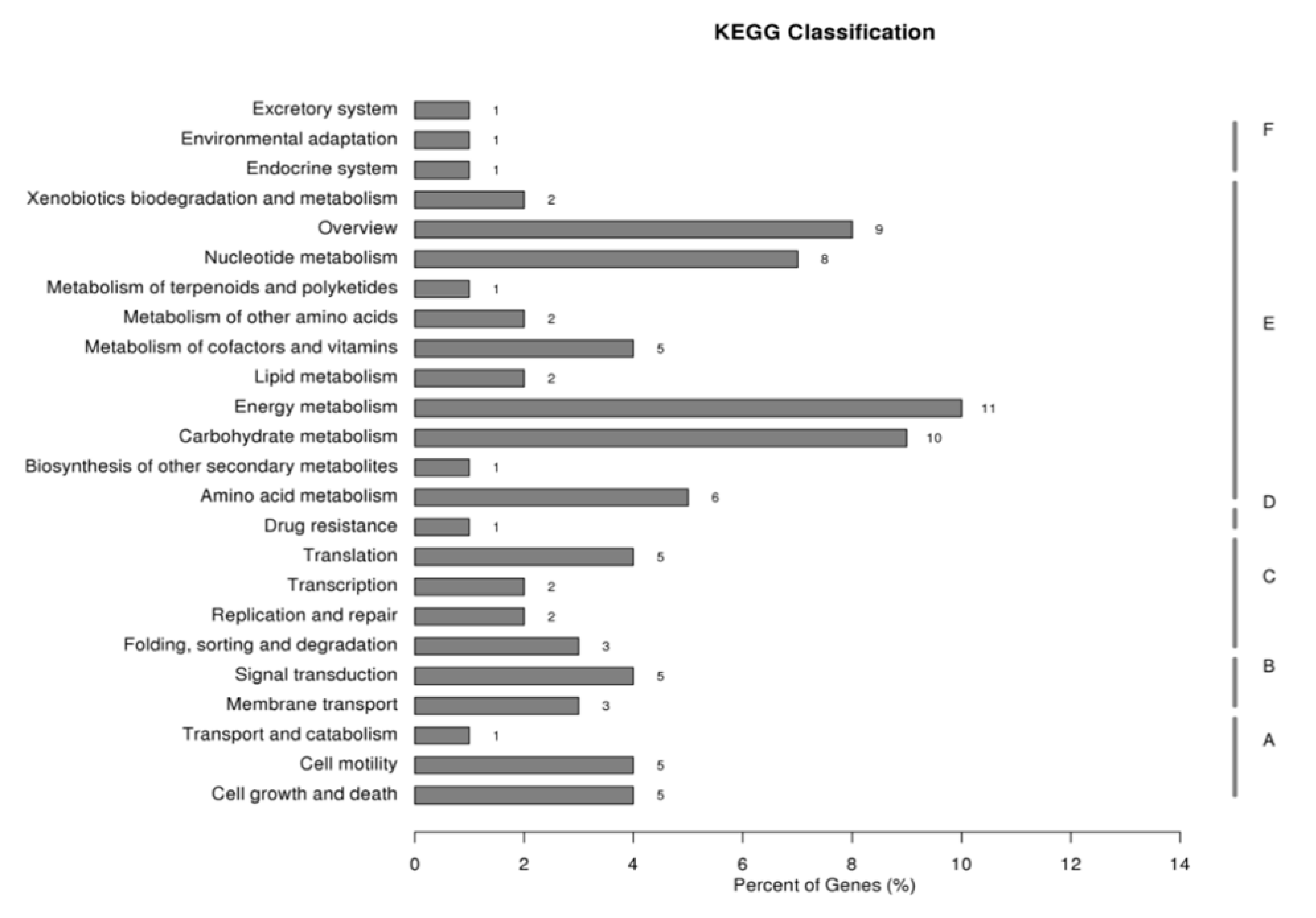
2.1. Gene Set Obtained from SSH Libraries
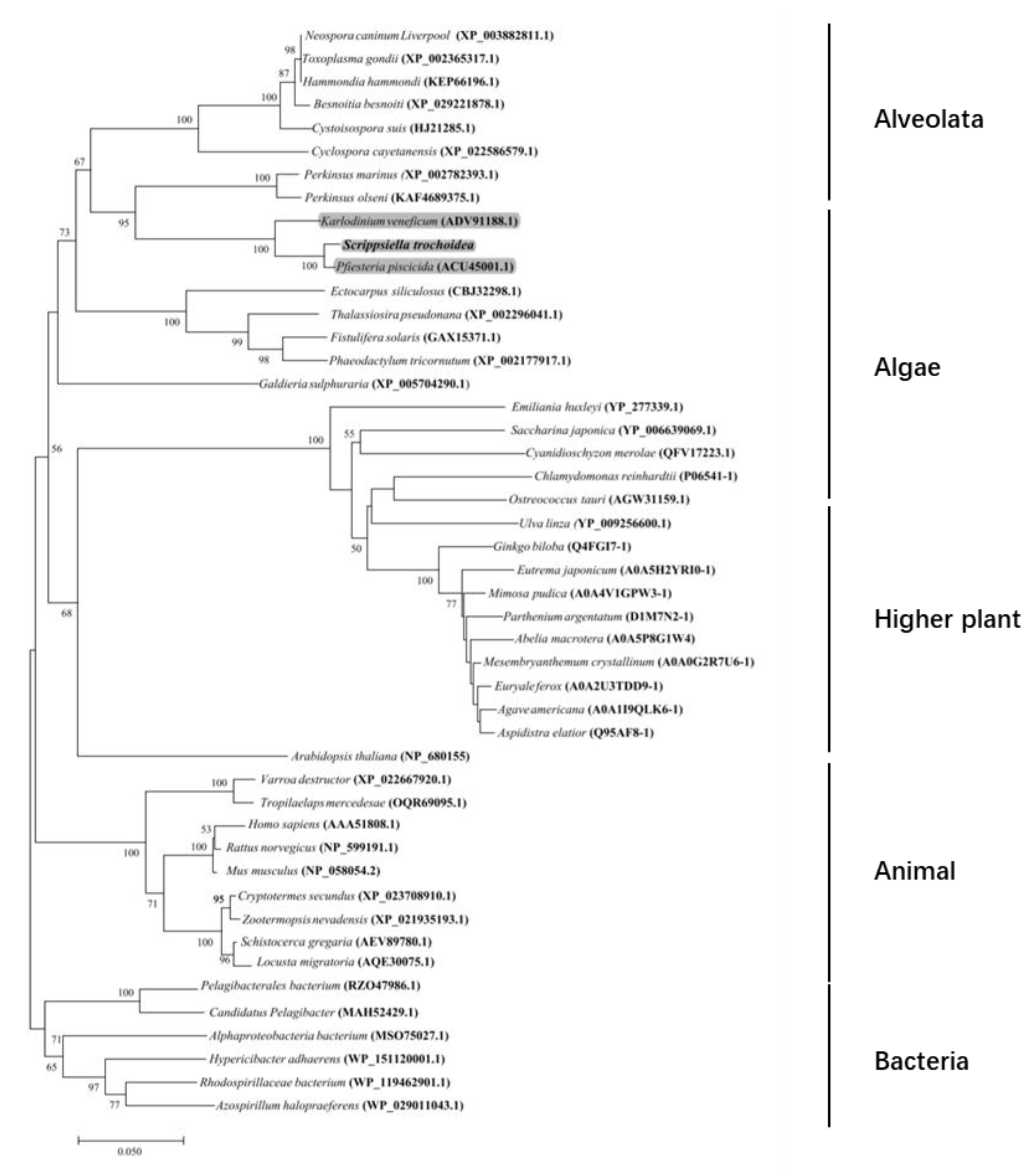
2.2. Characterization of the Full-Length cDNA Sequence of Stβ-F1-ATPase
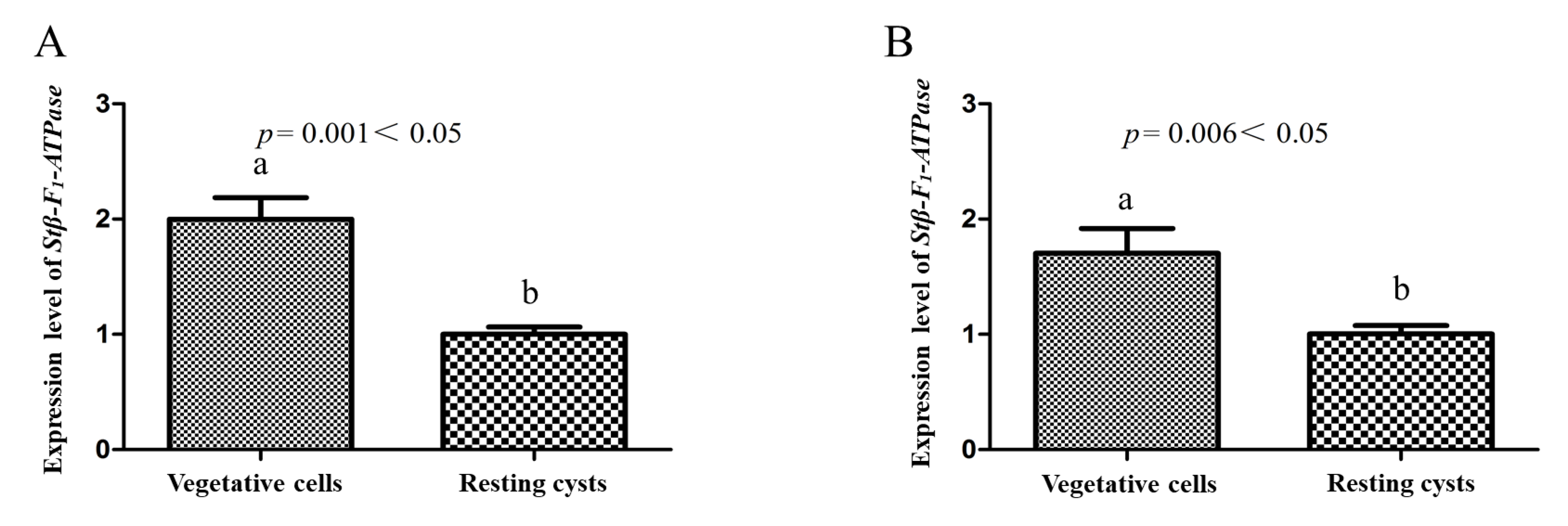
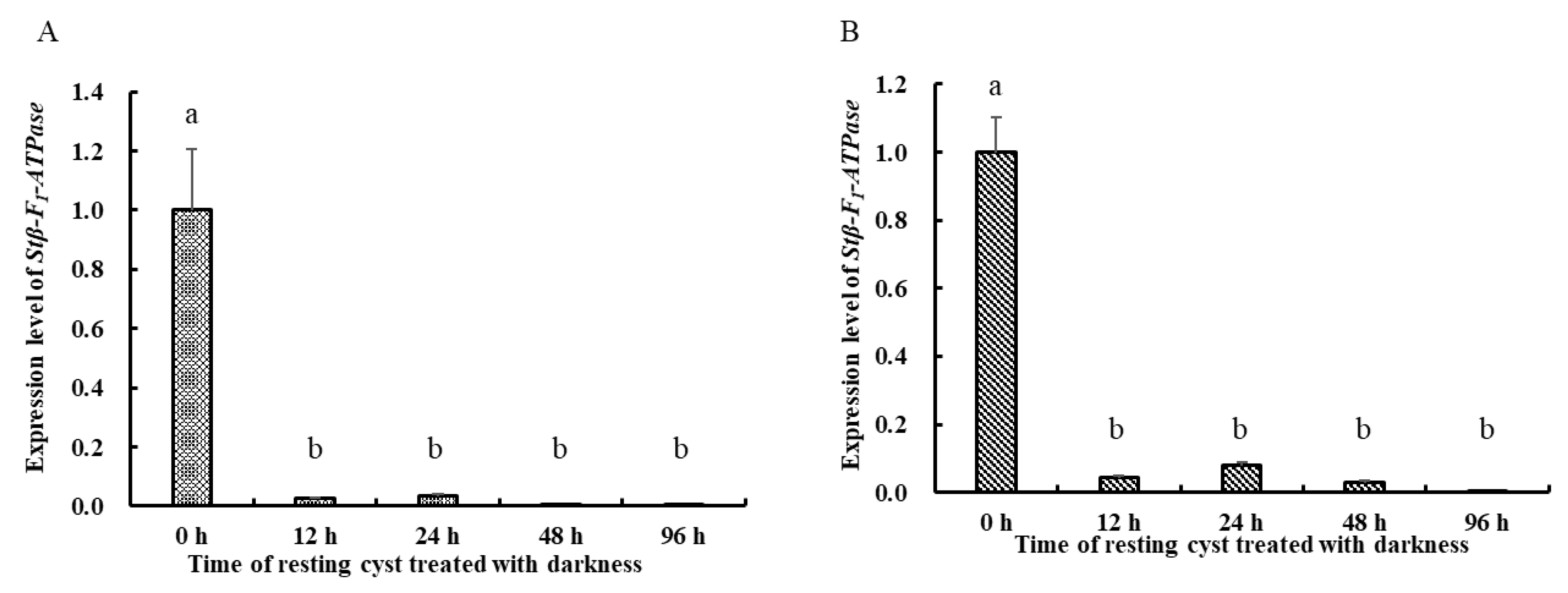
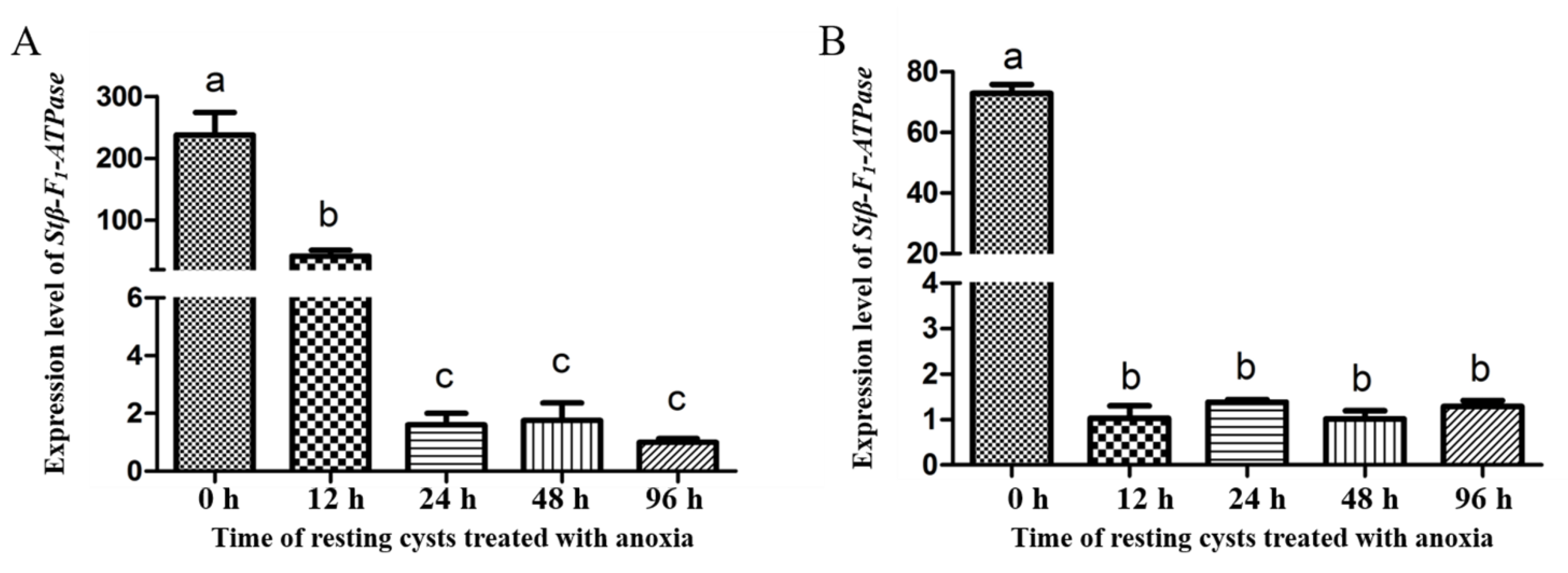
2.3. Transcriptional Profiles of Stβ-F1-ATPase
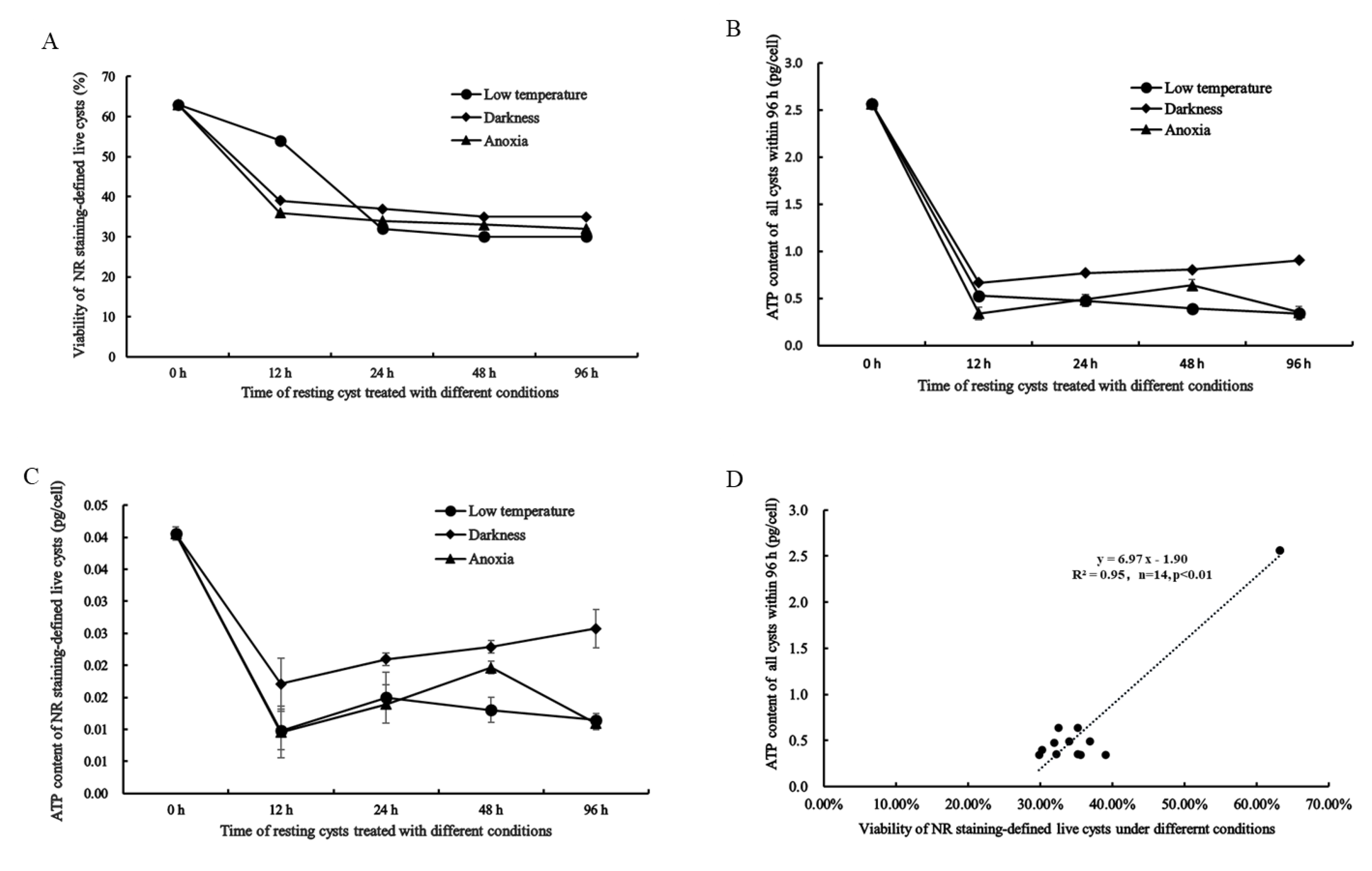
2.4. NR Staining-Defined Viability of Resting Cysts during Dormancy
2.5. Cellular ATP Content in All and Live Resting Cysts and Its Relation to NR Staining-Defined Viability
3. Discussion
3.1. Comments on the Results and Limitations of SSH
3.2. The Activity of Energetic Metabolism in Resting Cysts as Reflected in the Expression of Stβ-F1-ATPase, Viability and ATP Measurements
3.3. Further Discussion and Conclusions
4. Materials and Methods
4.1. Algal Cultures and Resting Cysts Harvesting
4.2. Treatments of Resting Cysts: Darkness, Lowered Temperature, and Anoxia
4.3. SSH Library Construction and Reverse Northern Blot Hybridization
4.4. Sequencing and Alignment Analysis
4.5. Full-Length cDNA Cloning of Stβ-F1-ATPase
4.6. Characteristics Analyses of Stβ-F1-ATPase
4.7. Validation of Real-Time Quantitative PCR (qPCR) Reference Genes Applicable for S. trochoidea Strain STBDH1 at Different Life Stages
4.8. Transcriptional Profiles of Stβ-F1-ATPase with qPCR Detection
4.9. Measurements for the Viability of S. trochoidea Cysts via Neutral Red (NR) Staining
4.10. Measurements of the Cellular ATP Content in Cysts
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, D.M.; Cembella, A.D.; Hallegraeff, G.M. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Annu. Rev. Mar. Sci. 2012, 4, 143–176. [Google Scholar] [CrossRef]
- Smayda, T.J. Harmful algal blooms: Their ecophysiology and general relevance to phytoplankton blooms in the sea. Limnol. Oceanogr. 1997, 42, 1137–1153. [Google Scholar] [CrossRef]
- Tang, Y.Z.; Gobler, C.J. Allelopathic effects of Cochlodinium polykrikoides isolates and blooms from the estuaries of Long Island, New York, on co-occurring phytoplankton. Mar. Ecol. Prog. Ser. 2010, 406, 19–31. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Hu, Z.X.; Deng, Y.Y.; Tang, Y.Z. Evidence for production of sexual resting cysts by the toxic dinoflagellate Karenia mikimotoi in clonal cultures and marine sediments. J. Phycol. 2020, 56, 121–134. [Google Scholar] [CrossRef]
- Song, X.; Hu, Z.; Shang, L.; Leaw, C.P.; Lim, P.T.; Tang, Y.Z. Contact micropredation may play a more important role than exotoxicity does in the lethal effects of Karlodinium australe blooms: Evidence from laboratory bioassays. Harmful Algae 2020, 99, 101926. [Google Scholar] [CrossRef]
- Tang, Y.; Hu, Z.; Deng, Y. Characteristical life history (resting cyst) provides a mechanism for recurrence and geographic expansion of harmful algal blooms of dinoflagellates: A review. Stud. Mar. Sin. 2016, 51, 132–154. [Google Scholar]
- Tang, Y.Z.; Gu, H.; Wang, Z.; Liu, D.; Wang, Y.; Lu, D.; Hu, Z.; Deng, Y.; Shang, L.; Qi, Y. Exploration of resting cysts (stages) and their relevance for possibly HABs-causing species in China. Harmful Algae 2021. [Google Scholar] [CrossRef]
- Matsuoka, K.; Fukuyo, Y. Technical Guide for Modern Dinoflagellate Cyst Study; IOC/WESTPAC-HAB; The University of Tokyo: Tokyo, Japan, 2000; 29p. [Google Scholar]
- Bravo, I.; Figueroa, R.I. Towards an ecological understanding of dinoflagellate cyst functions. Microorganisms 2014, 2, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Wall, D. Potential importance of benthic cysts of Gonyaulax tamarensis and G. excavata in initiating toxic dinoflagellate blooms 1, 2, 3. J. Phycol. 1978, 14, 224–234. [Google Scholar] [CrossRef]
- Anderson, D.M.; Morel, F.M. The seeding of two red tide blooms by the germination of benthic Gonyaulax tamarensis hypnocysts. Estuar. Coast. Mar. Sci. 1979, 8, 279–293. [Google Scholar] [CrossRef]
- Anderson, D.; Chisholm, S.T.; Watras, C. Importance of life cycle events in the population dynamics of Gonyaulax tamarensis. Mar. Biol. 1983, 76, 179–189. [Google Scholar] [CrossRef]
- Anderson, D.M. Cysts as factors in Pyrodinium bahamense ecology. In Biology, Epidemiology and Management of Pyrodinium Red Tides; Hallegraeff, G.M., Maclean, J.L., Eds.; Fisheries Department, Ministry of Development, Brunei Darussalam, and International Center for Living Aquatic Resources Management: Manila, Philippines, 1989; Volume 21, pp. 81–88. [Google Scholar]
- Hallegraeff, G.M.; Bolch, C.J. Transport of toxic dinoflagellate cysts via ships’ ballast water. Mar. Pollut. Bull. 1991, 22, 27–30. [Google Scholar] [CrossRef]
- Smayda, T.J. Reflections on the ballast water dispersal—Harmful algal bloom paradigm. Harmful Algae 2007, 6, 601–622. [Google Scholar] [CrossRef]
- Tang, Y.Z.; Gobler, C.J. The toxic dinoflagellate Cochlodinium polykrikoides (Dinophyceae) produces resting cysts. Harmful Algae 2012, 20, 71–80. [Google Scholar] [CrossRef]
- Tang, Y.Z.; Gobler, C.J. Sexual resting cyst production by the dinoflagellate Akashiwo sanguinea: A potential mechanism contributing to the ubiquitous distribution of a harmful alga. J. Phycol. 2015, 51, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Bibby, B.; Dodge, J. The encystment of a freshwater dinoflagellate: A light and electron-microscopical study. Br. Phycol. J. 1972, 7, 85–100. [Google Scholar] [CrossRef]
- Binder, B.J.; Anderson, D.M. Biochemical composition and tetabolic activity of Scrippsiella trochoidea (Dinophyceae) resting cysts 1. J. Phycol. 1990, 26, 289–298. [Google Scholar] [CrossRef]
- Lirdwitayaprasit, T.; Okaichi, T.; Montani, S.; Ochi, T.; Anderson, D.M. Changes in cell chemical composition during the life cycle of Scrippsiella trochoidea (Dinophyceae) 1. J. Phycol. 1990, 26, 299–306. [Google Scholar] [CrossRef]
- Rintala, J.-M.; Spilling, K.; Blomster, J. Temporary cyst enables long-term dark survival of Scrippsiella hangoei (Dinophyceae). Mar. Biol. 2007, 152, 57–62. [Google Scholar] [CrossRef]
- Kang, Y.; Tang, Y.Z.; Taylor, G.T.; Gobler, C.J. Discovery of a resting stage in the harmful, brown-tide-causing pelagophyte, Aureoumbra lagunensis: A mechanism potentially facilitating recurrent blooms and geographic expansion. J. Phycol. 2017, 53, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Hu, Z.; Shang, L.; Peng, Q.; Tang, Y.Z. Transcriptomic analyses of Scrippsiella trochoidea reveals processes regulating encystment and dormancy in the life cycle of a dinoflagellate, with a particular attention to the role of abscisic acid. Front. Microbiol. 2017, 8, 2450. [Google Scholar] [CrossRef]
- Roy, S.; Letourneau, L.; Morse, D. Cold-induced cysts of the photosynthetic dinoflagellate Lingulodinium polyedrum have an arrested circadian bioluminescence rhythm and lower levels of protein phosphorylation. Plant. Physiol. 2014, 164, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Lundholm, N.; Ribeiro, S.; Andersen, T.J.; Koch, T.; Godhe, A.; Ekelund, F.; Ellegaard, M. Buried alive–germination of up to a century-old marine protist resting stages. Phycologia 2011, 50, 629–640. [Google Scholar] [CrossRef]
- Capaldi, R.A.; Aggeler, R. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 2002, 27, 154–160. [Google Scholar] [CrossRef]
- Pullman, M.; Penefski, H.; Datta, A.; Racker, E. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphate. J. Biol. Chem. 1960, 235, 3322–3329. [Google Scholar] [CrossRef]
- Walker, J.E.; Fearnley, I.; Gay, N.J.; Gibson, B.; Northrop, F.; Powell, S.; Runswick, M.J.; Saraste, M.; Tybulewicz, V. Primary structure and subunit stoichiometry of F1-ATPase from bovine mitochondria. J. Mol. Biol. 1985, 184, 677–701. [Google Scholar] [CrossRef]
- Boyer, P.D. The ATP synthase—A splendid molecular machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Richter, M.L.; Hein, R.; Huchzermeyer, B. Important subunit interactions in the chloroplast ATP synthase. Biochim. Biophys. Acta Bioenerg. 2000, 1458, 326–342. [Google Scholar] [CrossRef][Green Version]
- Yoshida, M.; Muneyuki, E.; Hisabori, T. ATP synthase—A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001, 2, 669–677. [Google Scholar] [CrossRef]
- Esch, F.S.; Allison, W.S. Identification of a tyrosine residue at a nucleotide binding site in the beta subunit of the mitochondrial ATPase with p-fluorosulfonyl [14C]-benzoyl-5’-adenosine. J. Biol. Chem. 1978, 253, 6100–6106. [Google Scholar] [CrossRef]
- Stock, D.; Leslie, A.G.; Walker, J.E. Molecular architecture of the rotary motor in ATP synthase. Science 1999, 286, 1700–1705. [Google Scholar] [CrossRef]
- Jie, C.; Dechang, X.; Binsheng, L.; Qian, Y.; Jinghan, S. Cloning, sequence analysis and evolution of gene atpB sugar beet ATP synthase beta subunit. Zhi Wu Yan Jiu 2006, 26, 583–588. [Google Scholar]
- Kane, L.A.; Youngman, M.J.; Jensen, R.E.; Van Eyk, J.E. Novelty and Significance. Circ. Res. 2010, 106, 504–513. [Google Scholar] [CrossRef]
- Lai, C.; Lai, Z.; Fang, Z.; Lin, Y.; Jiang, S. Cloning of mitochondrial F1-ATPase beta subunit gene from embryogenic callus and its expression analysis by qRT-PCR during somatic embryogenesis in longan. Sci. Agric. Sin. 2010, 43, 3392–3401. [Google Scholar]
- Deng, Y.; Hu, Z.; Shang, L.; Chai, Z.; Tang, Y.Z. Transcriptional responses of the heat shock protein 20 (Hsp20) and 40 (Hsp40) genes to temperature stress and alteration of life cycle stages in the harmful alga Scrippsiella trochoidea (Dinophyceae). Biology 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed]
- Akopyants, N.S.; Fradkov, A.; Diatchenko, L.; Hill, J.E.; Siebert, P.D.; Lukyanov, S.A.; Sverdlov, E.D.; Berg, D.E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 1998, 95, 13108–13113. [Google Scholar] [CrossRef]
- Lisitsyn, N.; Rosenberg, M.; Launer, G.; Wagner, L.; Potapov, V.; Kolesnik, T.; Sverdlov, E. A method for isolation of sequences missing in one of two related genomes. Mol. Genet. Mikrobiol. I Virusol. 1993, 3, 26–29. [Google Scholar]
- Lisitsyn, N.; Wigler, M. Cloning the differences between two complex genomes. Science 1993, 259, 946–951. [Google Scholar] [CrossRef]
- Straus, D.; Ausubel, F.M. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 1990, 87, 1889–1893. [Google Scholar] [CrossRef] [PubMed]
- Lamar, E.E.; Palmer, E. Y-encoded, species-specific DNA in mice: Evidence that the Y chromosome exists in two polymorphic forms in inbred strains. Cell 1984, 37, 171–177. [Google Scholar] [CrossRef]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Li, L.; Zhang, Y.; Zhang, H.; Ji, Z. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef]
- Kanazawa, H.; Kayano, T.; Mabuchi, K.; Futai, M. Nucleotide sequence of the genes coding for α, β and γ subunits of the proton-translocating ATPase of Escherichia coli. Biochem. Biophys. Res. Commun. 1981, 103, 604–612. [Google Scholar] [CrossRef]
- Hudson, G.S.; Mason, J.G.; Holton, T.A.; Koller, B.; Cox, G.B.; Whitfeld, P.R.; Bottomley, W. A gene cluster in the spinach and pea chloroplast genomes encoding one CF1 and three CF0 subunits of the H+-ATP synthase complex and the ribosomal protein S2. J. Mol. Biol. 1987, 196, 283–298. [Google Scholar] [CrossRef]
- Masaike, T.; Suzuki, T.; Tsunoda, S.P.; Konno, H.; Yoshida, M. Probing conformations of the β subunit of F0F1-ATP synthase in catalysis. Biochem. Biophys. Res. Commun. 2006, 342, 800–807. [Google Scholar] [CrossRef]
- Dressel, D.M.; Heinle, D.R.; Grote, M.C. Vital staining to sort dead and live copepods 1, 2, 3. Chesap. Sci. 1972, 13, 156–159. [Google Scholar] [CrossRef]
- Crippen, R.W.; Perrier, J. The use of neutral red and evans blue for live-dead determinations of marine plankton (with comments on the use of rotenone for inhibition of grazing). Stain Technol. 1974, 49, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.C. Patterns of cell viability in the diatom, Skeletonema costatum, in batch culture and in natural populations. Estuaries 1984, 7, 98–101. [Google Scholar] [CrossRef]
- Weber, H.; Borisjuk, L.; Wobus, U. Molecular physiology of legume seed development. Annu. Rev. Plant. Biol. 2005, 56, 253–279. [Google Scholar] [CrossRef]
- Shin, H.; Hong, S.-J.; Yoo, C.; Han, M.-A.; Lee, H.; Choi, H.-K.; Cho, S.; Lee, C.-G.; Cho, B.-K. Genome-wide transcriptome analysis revealed organelle specific responses to temperature variations in algae. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Law, R.D.; Crafts-Brandner, S.J. Inhibition and acclimation of photosynthesis to heat stress is closely correlated with activation of ribulose-1, 5-bisphosphate carboxylase/oxygenase. Plant. Physiol. 1999, 120, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef]
- Bolton, M.D. Primary metabolism and plant defense—Fuel for the fire. Mol. Plant Microbe Interact. 2009, 22, 487–497. [Google Scholar] [CrossRef]
- Ribeiro, S.; Berge, T.; Lundholm, N.; Andersen, T.J.; Abrantes, F.; Ellegaard, M. Phytoplankton growth after a century of dormancy illuminates past resilience to catastrophic darkness. Nat. Commun. 2011, 2, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals; Smith, W.L., Chanley, M.H., Eds.; Plenum: New York, NY, USA, 1975; 338p. [Google Scholar]
- Yang, A.; Hu, Z.; Tang, Y. Solid sand particle addition can enhance the production of resting cysts in dinoflagellates. J. Oceanol. Limnol. 2018, 36, 273–280. [Google Scholar] [CrossRef]
- Su, J.; Yang, X.; Zheng, T.; Hong, H. An efficient method to obtain axenic cultures of Alexandrium tamarense—A PSP-producing dinoflagellate. J. Microbiol. Methods 2007, 69, 425–430. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34 (Suppl. S2), W293–W297. [Google Scholar] [CrossRef]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar] [CrossRef]
- Deng, Y.; Hu, Z.; Zhan, Z.; Ma, Z.; Tang, Y. Differential expressions of an Hsp70 gene in the dinoflagellate Akashiwo sanguinea in response to temperature stress and transition of life cycle and its implications. Harmful Algae 2015, 50, 57–64. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- de Almeida, M.R.; Ruedell, C.M.; Ricachenevsky, F.K.; Sperotto, R.A.; Pasquali, G.; Fett-Neto, A.G. Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol. Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, N.V.; Logacheva, M.D.; Penin, A.A. Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE 2011, 6, e19434. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Zakrajsek, B.A.; Mills, A.G.; Gorn, V.; Singer, M.J.; Reed, M.W. Quantitative reverse transcription–polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal. Biochem. 2000, 285, 194–204. [Google Scholar] [CrossRef]
- Reynolds, A.E.; Mackiernan, G.B.; Van Valkenburg, S.D. Vital and mortal staining of algae in the presence of chlorine-produced oxidants. Estuaries 1978, 1, 192–196. [Google Scholar] [CrossRef]
- Onji, M.; Sawabe, T.; Ezura, Y. An evaluation of viable staining dyes suitable for marine phytoplankton. Bull. Fac. Fish. Hokkaido Univ. 2000, 51, 153–157. [Google Scholar]
- Zetsche, E.-M.; Meysman, F.J. Dead or alive? Viability assessment of micro-and mesoplankton. J. Plankton Res. 2012, 34, 493–509. [Google Scholar] [CrossRef]
- Greenstein, K.E.; Wert, E.C. Using rapid quantification of adenosine triphosphate (ATP) as an indicator for early detection and treatment of cyanobacterial blooms. Water Res. 2019, 154, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Curto, A.L.; Stehouwer, P.; Gianoli, C.; Schneider, G.; Raymond, M.; Bonamin, V. Ballast water compliance monitoring: A new application for ATP. J. Sea Res. 2018, 133, 124–133. [Google Scholar] [CrossRef]


| Database | Number of Comments | Comment Percentage (%) |
|---|---|---|
| NR | 204 | 72.86 |
| KOG | 40 | 14.29 |
| KO | 112 | 40 |
| KEGG | 58 | 20.71 |
| GO | 112 | 40 |
| SWISS-PROT | 108 | 38.57 |
| All Databases | 21 | 7.50 |
| At least one Database | 208 | 74.29 |
| Unannotated | 72 | 25.71 |
| Total | 280 | 100 |
| Classification of GO | GO-Id | Gene Function | Sequence Number | Percentage of GO-Annotated Genes | Percentage of Total Sequence |
|---|---|---|---|---|---|
| Cellular component | GO:0005623 | cell | 41 | 36.61 | 14.64 |
| GO:0044464 | cell part | 41 | 36.61 | 14.64 | |
| GO:0032991 | macromolecular complex | 4 | 3.57 | 1.43 | |
| GO:0043226 | organelle | 3 | 2.68 | 1.07 | |
| GO:0031975 | envelope | 2 | 1.79 | 0.71 | |
| Molecular function | GO:0003824 | catalytic activity | 61 | 54.46 | 21.79 |
| GO:0005488 | binding | 40 | 35.71 | 14.29 | |
| GO:0060089 | molecular transducer activity | 19 | 16.96 | 6.79 | |
| GO:0005215 | transporter activity | 12 | 10.71 | 4.29 | |
| GO:0005198 | structural molecule activity | 6 | 5.36 | 2.14 | |
| GO:0030528 | transcription regulator activity | 2 | 1.79 | 0.71 | |
| GO:0009055 | electron carrier activity | 2 | 1.79 | 0.71 | |
| GO:0003824 | catalytic activity | 61 | 54.46 | 21.79 | |
| Biological process | GO:0008152 | metabolic process | 55 | 49.11 | 19.64 |
| Primer Name | Primer Sequences (5′→3′) | Remarks |
|---|---|---|
| β-F1-ATPase-3′-F | GATGAGGAGGGCCAGGACGTGC | 3′ RACE |
| β-F1-ATPase -3′-R | AGATGAACGAGCCCCCAGGTGC | 3′ RACE |
| β-F1-ATPase -5′-F | ACCTGGGGGCTCGTTCATCTGGC | 5′ RACE |
| β-F1-ATPase -5′-R | CTTGCGTGAAGCGGAGGATGTTG | 5′ RACE |
| qβ-F1-ATPase-F | CCAACAGTGCCGATACCT | Stβ-F1-ATPase qPCR |
| qβ-F1-ATPase-R | GCCAGATGAACGAGCC | Stβ-F1-ATPase qPCR |
| CYC-F | CTACGAATGGTGGGAGACG | CYC qPCR |
| CYC-R | TCGCAAGTTAGCGGGACT | CYC qPCR |
| PEPCK-F | GAATGCCACCGTTGAGTTG | PEPCK qPCR |
| PEPCK-R | CTCCGCGAGTGAATGTGC | PEPCK qPCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Yang, A.; Hu, Z.; Lin, S.; Deng, Y.; Tang, Y.Z. Probing the Energetic Metabolism of Resting Cysts under Different Conditions from Molecular and Physiological Perspectives in the Harmful Algal Blooms-Forming Dinoflagellate Scrippsiella trochoidea. Int. J. Mol. Sci. 2021, 22, 7325. https://doi.org/10.3390/ijms22147325
Li F, Yang A, Hu Z, Lin S, Deng Y, Tang YZ. Probing the Energetic Metabolism of Resting Cysts under Different Conditions from Molecular and Physiological Perspectives in the Harmful Algal Blooms-Forming Dinoflagellate Scrippsiella trochoidea. International Journal of Molecular Sciences. 2021; 22(14):7325. https://doi.org/10.3390/ijms22147325
Chicago/Turabian StyleLi, Fengting, Aoao Yang, Zhangxi Hu, Siheng Lin, Yunyan Deng, and Ying Zhong Tang. 2021. "Probing the Energetic Metabolism of Resting Cysts under Different Conditions from Molecular and Physiological Perspectives in the Harmful Algal Blooms-Forming Dinoflagellate Scrippsiella trochoidea" International Journal of Molecular Sciences 22, no. 14: 7325. https://doi.org/10.3390/ijms22147325
APA StyleLi, F., Yang, A., Hu, Z., Lin, S., Deng, Y., & Tang, Y. Z. (2021). Probing the Energetic Metabolism of Resting Cysts under Different Conditions from Molecular and Physiological Perspectives in the Harmful Algal Blooms-Forming Dinoflagellate Scrippsiella trochoidea. International Journal of Molecular Sciences, 22(14), 7325. https://doi.org/10.3390/ijms22147325







