Biological Activities Related to Plant Protection and Environmental Effects of Coumarin Derivatives: QSAR and Molecular Docking Studies
Abstract
1. Introduction
2. Results
2.1. Synthesis of Coumarin Derivatives
2.2. Biological Activity Evaluation
2.2.1. Antifungal Activity
2.2.2. Antibacterial Activity
2.2.3. Nematicidal Activity
2.3. Estimation of Toxicity
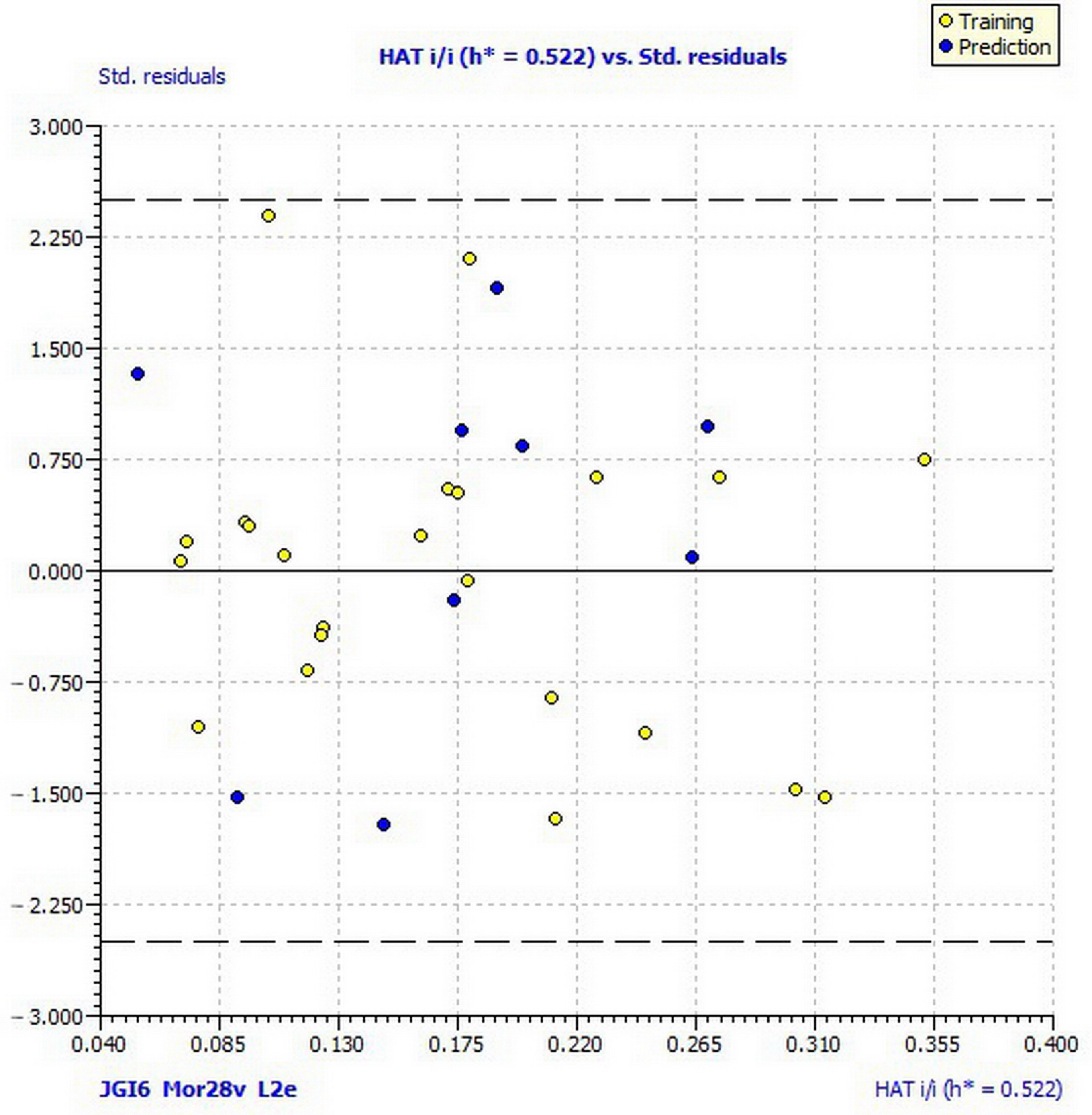
2.4. QSAR and Pharmacophore Mapping for Antifungal Activity
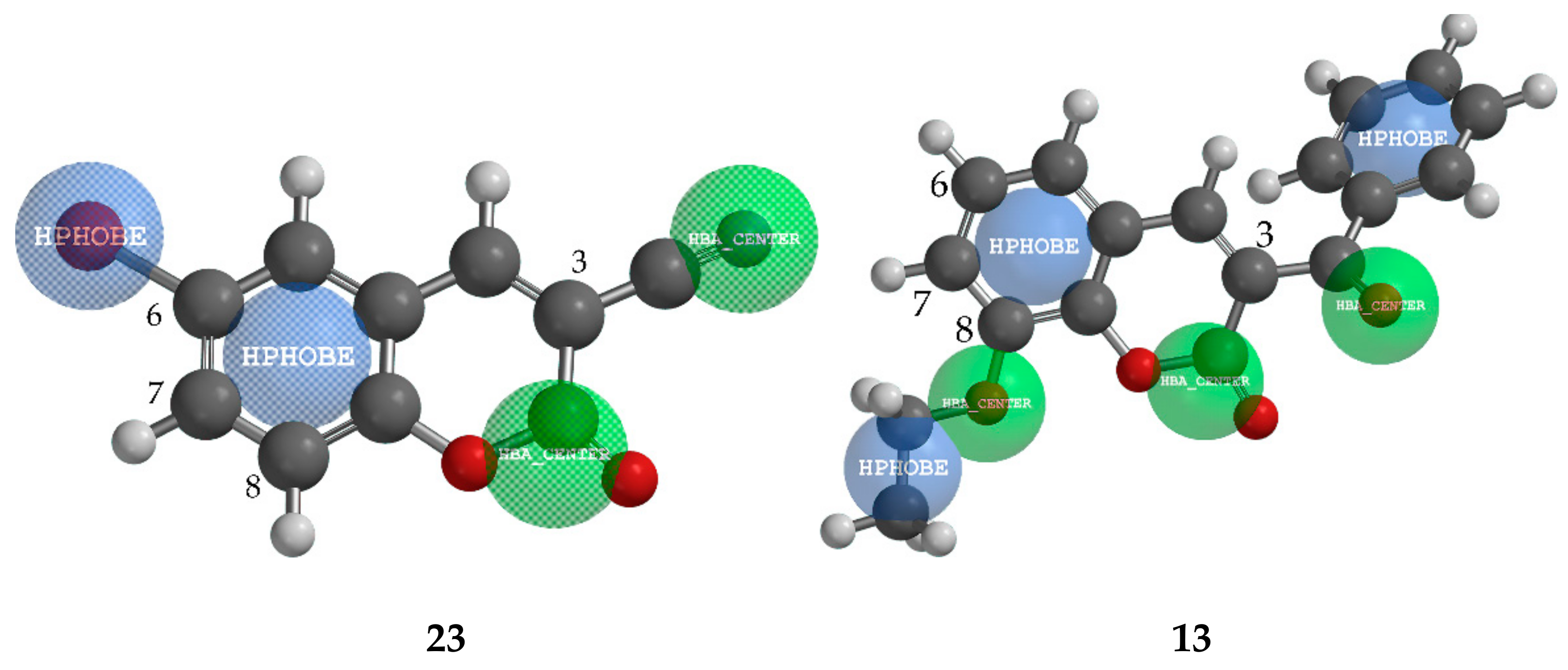
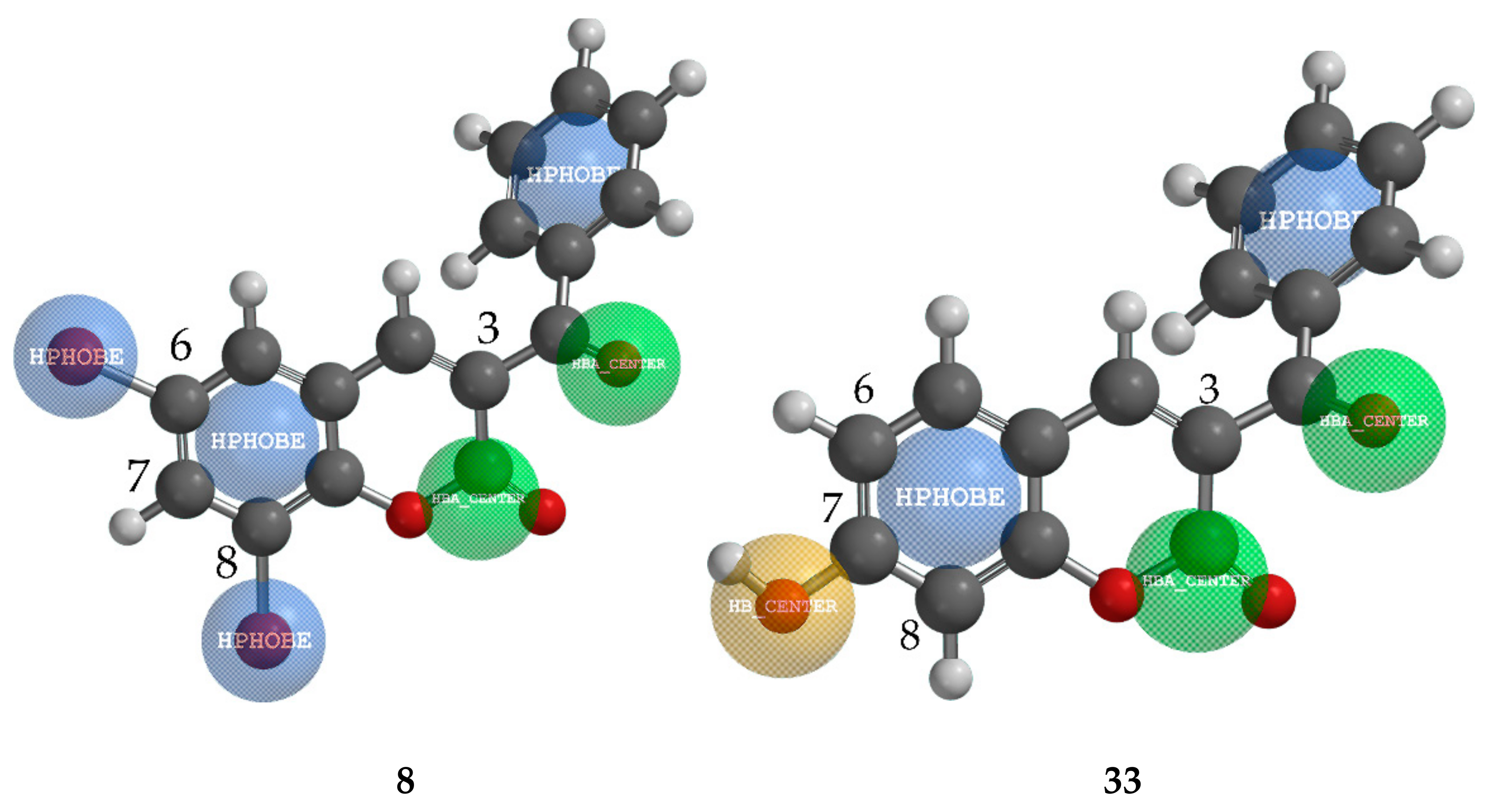
2.4.1. QSAR and Pharmacophore Mapping for Activity against M. phaseolina
2.4.2. QSAR and Pharmacophore Mapping for Activity against S. sclerotiorum
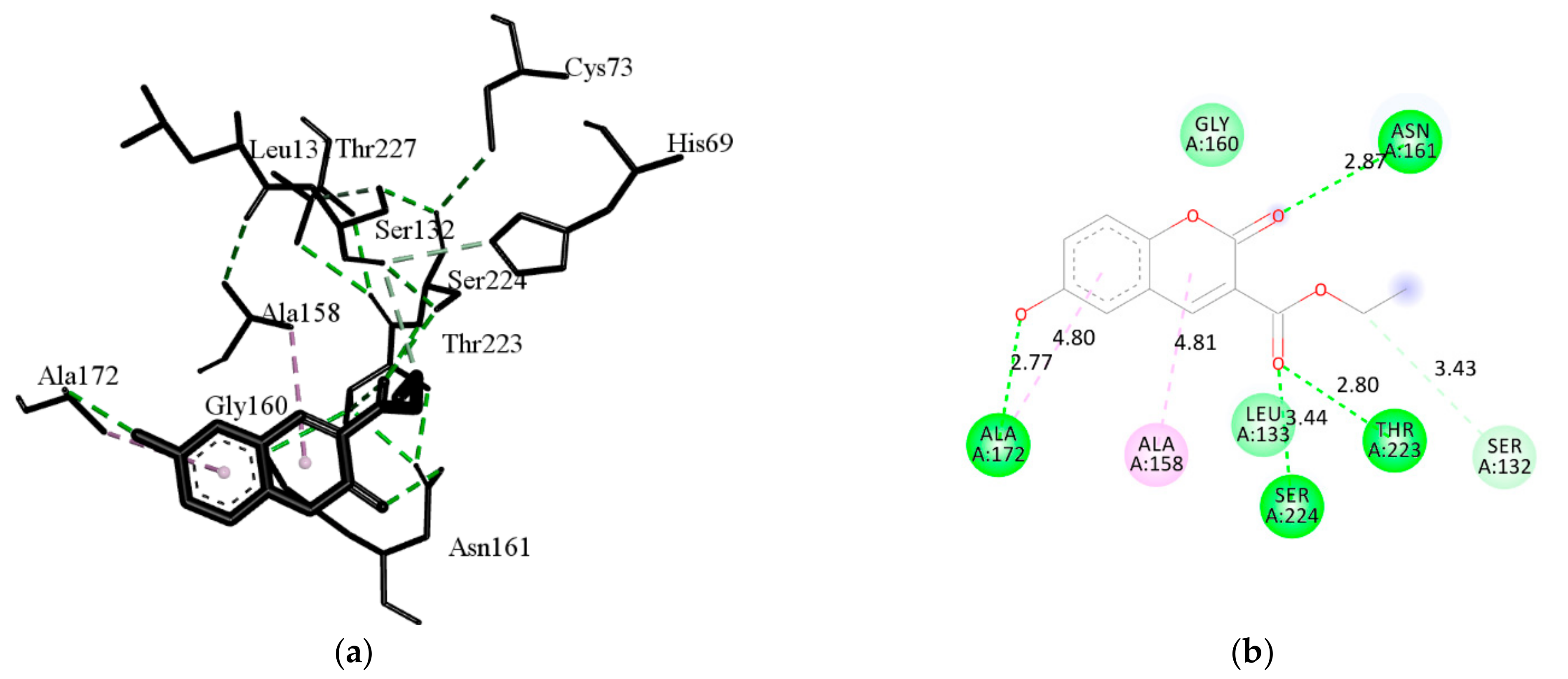
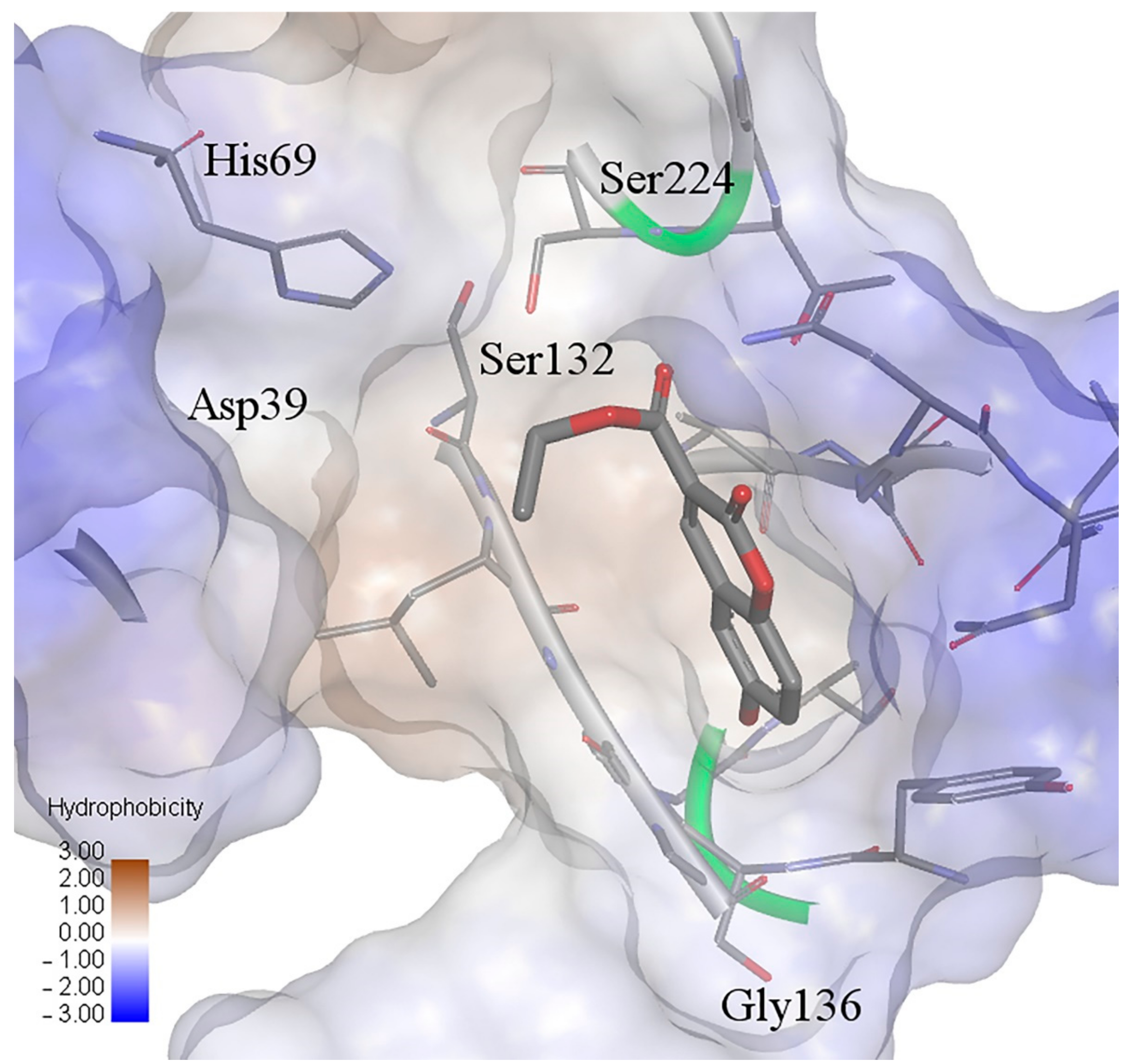
2.5. Molecular Docking Study
3. Discussion
4. Materials and Methods
4.1. Biological Asay
4.1.1. Antifungal Assay
4.1.2. Antibacterial Activity
4.1.3. Nematicidal Activity
4.2. Computational Methods
4.2.1. Calculation of Toxicity
4.2.2. QSAR Methods
4.2.3. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pogăcean, M.O.; Gavrilescu, M. Plant protection products and their sustainable and environmentally friendly use. Environ. Eng. Manag. J. 2009, 8, 608–627. [Google Scholar]
- Young, C.S.; Smith, J.A.; Watling, M.; Clarkson, J.P.; Whipps, J.M. Environmental conditions influencing apothecial production and lettuce infection by Sclerotinia sclerotiorum in field conditions. In Sclerotinia 2001, Proceedings of the Xl International Sclerotinia Workshop, York, UK, 8–12 July 2001; Young, C.S., Hughes, K.J.D., Eds.; Central Science Laboratory: York, UK, 2001; pp. 181–182. [Google Scholar]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. Fusarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef]
- Khan, S. Addition to the host range of Sclerotinia sclerotiorum in West Bengal. Sch. Acad. J. Biosci. Mycopath. 2007, 5, 111–118. [Google Scholar]
- Mondal, B.; Khatua, D.C.; Hansda, S.; Sharma, R. Addition to the host range of Sclerotinia sclerotiorum in West Bengal. Sch. Acad. J. Biosci. 2015, 3, 361–364. [Google Scholar]
- Srinivas, C.; Devi, D.N.; Murthy, K.N.; Mohan, C.D.; Lakshmeesha, T.; Singh, B.P.; Kalagatur, N.K.; Niranjana, S.; Hashem, A.; Alqarawi, A.A.; et al. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: Biology to diversity—A review. Saudi. J. Biol. Sci. 2019, 26, 1315–1324. [Google Scholar] [CrossRef]
- Smith, K.; Evans, D.A.; El-Hiti, G.A. Role of modern chemistry in sustainable arable crop protection. Phil. Trans. R. Soc. B 2008, 363, 623–637. [Google Scholar] [CrossRef]
- Song, P.P.; Zhao, J.; Liu, Z.-L.; Duan, Y.-B.; Hou, Y.-P.; Zhao, C.-Q.; Wu, M.; Wei, M.; Wang, N.-H.; Lv, Y.; et al. Evaluation of antifungal activities and structure–activity relationships of coumarin derivatives. Pest Manag. Sci. 2017, 73, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Peng, W.; Wanf, D.; Hao, S.-H.; Li, W.-W.; Ding, F. Design, synthesis, antifungal activity, and 3D-QSAR of coumarin derivatives. J. Pestic. Sci. 2018, 43, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, P.A.; Santos Junior, M.C.; Sousa, C.S.; Góes-Neto, A.; Luz, E.D.N.; Damaceno, V.O.; Niella, A.R.R.; Filho, J.M.B.; Assis, S.A. Study of sodium 3-hydroxycoumarin as inhibitors in vitro, in vivo and in silico of Moniliophthora perniciosa fungus. Eur. J. Plant Pathol. 2019, 153, 15–27. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, S.; Ding, W. Resveratrol and coumarin: Novel agricultural antibacterial agent against Ralstonia solanacearum in vitro and in vivo. Molecules 2016, 21, 1501. [Google Scholar] [CrossRef]
- Rehman, S.; Ikram, M.; Baker, R.J.; Zubair, M.; Azad, E.; Min, S.; Riaz, K.; Mok, K.H.; Rehman, S.-U. Synthesis, characterization, in vitro antimicrobial, and U2OS tumoricidal activities of different coumarin derivatives. Chem. Cent J. 2013, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Dekić, B.D.; Radulović, N.S.; Dekić, V.S.; Vukićević, R.D.; Palić, R.M. Synthesis and antimicrobial activity of new 4-heteroarylamino coumarin derivatives containing nitrogen and sulfur as heteroatoms. Molecules 2010, 15, 2246–2256. [Google Scholar] [CrossRef]
- Vincent, J. A Manual for the Practical Study of Root Nodule Bacteria; Blackwell Scientific Publications: Oxford, UK, 1970. [Google Scholar]
- Cui, H.; Jin, H.; Liu, Q.; Yan, Z.; Ding, L.; Qin, B. Nematicidal metabolites from roots of Stellera chamaejasme against Bursaphelenchus xylophilus and Bursaphelenchus mucronatus. Pest Manag. Sci. 2014, 70, 827–835. [Google Scholar] [CrossRef]
- Pan, L.; Li, X.-Z.; Sun, S.-A.; Guo, H.-R.; Qin, B. Design and synthesis of novel coumarin analogs and their nematicidal activity against five phytonematodes. Chin. Chem. Lett. 2016, 27, 375–379. [Google Scholar] [CrossRef]
- Santhi, V.S.; Salame, L.; Muklada, H.; Azaizeh, H.; Haj-Zaroubi, M.; Awwad, S.; Landau, S.Y.; Glazer, I. Toxicity of phenolic compounds to entomopathogenic nematodes: A case study with Heterorhabditis bacteriophora exposed to lentisk (Pistacia lentiscus) extracts and their chemical components. J. Invertebr. Pathol. 2019, 160, 43–53. [Google Scholar] [CrossRef]
- Jiao, Y.; Preston, S.; Song, H.; Jabbar, A.; Liu, Y.; Baell, J.; Hofmann, A.; Hutchinson, D.; Wang, T.; Koehler, A.V.; et al. Assessing the anthelmintic activity of pyrazole-5-carboxamide derivatives against Haemonchus contortus. Parasit. Vectors 2017, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Adams, B.J.; Ciche, T.A.; Clifton, S.; Gaugler, R.; Kim, K.S.; Spieth, J.; Sternberg, P.W.; Wilson, R.K.; Grewal, P.S. A lover and a fighter: The genome sequence of an entomopathogenic nematode Heterorhabditis bacteriophora. PLoS ONE 2013, 8, 69618. [Google Scholar] [CrossRef]
- Özdemir, E.; İnak, E.; Evlice, E.; Laznik, Z. Compatibility of entomopathogenic nematodes with pesticides registered in vegetable crops under laboratory conditions. J. Plant Dis. Prot. 2020, 127, 529–535. [Google Scholar] [CrossRef]
- Kar, S.; Roy, K.; Leszczynski, J. On Applications of QSARs in Food and Agricultural Sciences: History and Critical Review of Recent Developments. In Advances in QSAR Modeling, Challenges and Advances in Computational Chemistry and Physics; Roy, K., Ed.; Springer: Cham, Switzerland, 2017; Volume 24, pp. 203–300. [Google Scholar] [CrossRef]
- Du, H.; Wang, J.; Hu, Z.; Yao, X.; Zhang, X. Prediction of fungicidal activities of rice blast disease based on least-squares support vector machines and project pursuit regression. J. Agric. Food Chem. 2008, 56, 10785–10792. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xu, S.; Li, X.; Shen, X.; Zhang, Q.; Li, J.; Chen, C. N-Nitrourea derivatives as novel potential fungicides against Rhizoctonia solani: Synthesis, antifungal activities, and 3D-QSAR. Chem. Biol. Drug Des. 2012, 80, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Tyndall, J.D.A.; Sabherwal, M.; Sagatova, A.A.; Keniya, M.V.; Negroni, J.; Wilson, R.K.; Woods, M.A.; Tietjen, K.; Monk, B.C. Structural and functional elucidation of yeast lanosterol 14α-demethylase in complex with agrochemical antifungals. PLoS ONE 2016, 11, e0167485. [Google Scholar] [CrossRef] [PubMed]
- Ayine-Tora, D.M.; Kingsford-Adaboh, R.; Asomaning, W.A.; Harrison, J.J.E.K.; Mills-Robertson, F.C.; Bukari, Y.; Sakyi, P.O.; Kaminta, S.; Reynisson, J. Coumarin antifungal lead compounds from Millettia thonningii and heir predicted mechanism of action. Molecules 2016, 21, 1369. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.L.; Colombo, A.L.; de Almeida Junior, J.N. Fungal cell wall: Emerging antifungals and drug resistance. Front. Microbiol. 2019, 10, 2573. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.M.; Gally, M.; Szapiro, G.; Itzcovich, T.; Carabajal, M.; Levin, L. In Vitro growth and cell wall degrading enzyme production by Argentinean isolates of Macrophomina phaseolina, the causative agent of charcoal rot in corn. Rev. Argent. Microbiol. 2016, 21, 267–273. [Google Scholar] [CrossRef]
- Lockhart, D.E.A.; Schuettelkopf, A.; Blair, D.E.; van Aalten, D.M.F. Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors. FEBS Lett. 2014, 588, 3282–3290. [Google Scholar] [CrossRef][Green Version]
- Riou, C.; Freyssinet, G.; Fevre, M. Production of cell wall degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum. Appl. Environ. Microbiol. 1991, 57, 1478–1484. [Google Scholar] [CrossRef]
- ECHA-11-R-004.2-EN, The Use of Alternatives to Testing on Animals for the REACH Regulation 2011; European Chemicals Agency: Helsinki, Finland, 2011.
- Lonačarić, M.; Sušjenka, M.; Molnar, M. An extensive study of coumarin synthesis via Knoevenagel condensation in choline chloride based deep eutectic solvents. Curr. Org. Synth. 2020, 17, 98–108. [Google Scholar] [CrossRef]
- U.S. EPA. User’s Guide for T.E.S.T. (version 5.1) (Toxicity Estimation Software Tool): A Program to Estimate Toxicity from Molecular Structure; U.S. Environmental Protection Agency: Cincinnati, OH, USA, 2020.
- Schultz, T.W.; Sparfkin, C.L.; Aptula, A.O. Reactivity-based toxicity modelling of five-membered heterocyclic compounds: Application to Tetrahymena pyriformis. SAR QSAR Environ. Res. 2010, 21, 681–691. [Google Scholar] [CrossRef]
- Hansen, K.; Mika, S.; Schroeter, T.; Sutter, A.; Ter Laak, A.; Steger-Hartmann, T.; Heinrich, N.; Müller, K.-R. Benchmark Data set for in silico prediction of ames mutagenicity. J. Chem. Inf. Model. 2009, 49, 2077–2081. [Google Scholar] [CrossRef]
- Arnot, J.A.; Gobas, F.A.P.C. A review of bioconcentration factor (BCF) and bioaccumulation factor (BAF) assessments for organic chemicals in aquatic organisms. Environ. Rev. 2006, 14, 257–297. [Google Scholar] [CrossRef]
- Bulgheroni, A.; Kinsner-Ovaskainen, A.; Hoffmann, S.; Hartung, T.; Prieto, P. Estimation of acute oral toxicity using the No Observed Adverse Effect Level (NOAEL) from the 28 day repeated dose toxicity studies in rats. Regul. Toxicol. Pharmacol. 2009, 53, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Masand, H.V.; El-Sayed, N.N.E.; Rastija, V.; Rathore, M.M.; Karnaš, M. Identification of prodigious and under-privileged structural features for RG7834 analogs as Hepatitis B virus expression inhibitor. Med. Chem. Res. 2019, 28, 2270–2278. [Google Scholar] [CrossRef]
- Zaki, M.E.A.; Al-Hussain, S.A.; Masand, V.H.; Akasapu, S.; Lewaa, I. QSAR and pharmacophore modeling of nitrogen heterocycles as potent human N-myristoyltransferase (Hs-NMT) inhibitors. Molecules 2021, 26, 1834. [Google Scholar] [CrossRef]
- Masand, V.H.; El-Sayed, N.N.E.; Bambole, M.U.; Patil, V.R. Multiple quantitative structure-activity relationships (QSARs) analysis for orally active trypanocidal N-myristoyltransferase inhibitors. J. Mol. Struct. 2019, 1175, 481–487. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V.; Maiocchi, A. The K correlation index: Theory development and its application in chemometrics. Chemom. Intell. Lab. Syst. 1999, 46, 13–29. [Google Scholar] [CrossRef]
- Chirico, N.; Gramatica, P. Real external predictivity of QSAR models. Part 2. New intercomparable thresholds for different validation criteria and the need for sccatter plot inspection. J. Chem. Inf. Model. 2012, 52, 2044–2058. [Google Scholar] [CrossRef] [PubMed]
- Kiralj, R.; Ferreira, M.M.C. Basic validation procedures for regression models in QSAR and QSPR studies: Theory and application. J. Braz. Chem. Soc. 2009, 20, 770–787. [Google Scholar] [CrossRef]
- Masand, V.H.; Mahajan, D.T.; Nazeruddin, G.M.; Hadda, T.B.; Rastija, V.; Alfeefy, A.M. Effect of information leakage and method of splitting (rational and random) on external predictive ability and behavior of different statistical parameters of QSAR model. Med. Chem. Res. 2015, 24, 1241–1264. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Roy, P.P.; Paul, S.; Mitr, I.; Roy, K. On two novel parameters for validation of predictive QSAR models. Molecules 2009, 14, 1660–1701. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatics, 2nd ed.; Wiley-VCH: Weinheim, Germany, 2009. [Google Scholar]
- Devinyak, O.; Havrylyuk, D.; Lesyk, R. 3D-MoRSE descriptors explained. J. Mol. Graph. Model. 2014, 54, 194–203. [Google Scholar] [CrossRef]
- Todeschini, R.; Lasagni, M.; Marengo, E. New molecular descriptors for 2D and 3D structures. Theory. J. Chemom. 1994, 8, 263–273. [Google Scholar] [CrossRef]
- Todeschini, R.; Gramatica, P. New 3D molecular descriptors: The WHIM theory and QSAR applications. Perspect. Drug Discov. 1998, 9–11, 355–380. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V.; Pavan, M. Structure/response correlations and similarity/diversity analysis by GETAWAY descriptors. 1. Theory of the novel 3D molecular descriptors. J. Chem. Inf. Comput. Sci. 2002, 42, 682–692. [Google Scholar] [CrossRef]
- Wu, J.; Tao, Y.; Zhang, M.; Howard, M.H.; Gutteridge, S.; Ding, J. Crystal structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoyl-CoA and inhibitors reveal the functional roles of the N-terminal region. J. Biol. Chem. 2007, 282, 22185–22194. [Google Scholar] [CrossRef]
- Sulzenbacher, G.; Schülein, M.; Davies, G.J. Structure of the endoglucanase I from Fusarium oxysporum: Native, cellobiose, and 3,4-epoxybutyl β-D-cellobioside-inhibited forms, at 2.3 Å resolution. Biochemistry 1997, 36, 5902–5911. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Zanetti, E.; Oliva, C.R.; Covarrubias, A.A.; Casalongué, C.A. Characterization of an extracellular serine protease of Fusarium eumartii and its action on pathogenesis related proteins. Eur. J. Plant Pathol. 2002, 108, 63–72. [Google Scholar] [CrossRef]
- Santen, Y.; Benen, J.A.E.; Schröter, K.-H.; Kalk, K.H.; Armand, S.; Visser, J.; Dijkstra, B.W. 1.68-Å Crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J. Biol. Chem. 1999, 274, 30474–30480. [Google Scholar] [CrossRef]
- Kiani, A.; Jalili-baleh, L.; Abdolahi, Z.; Nadri, H.; Foroumadi, A.; Ebrahimi, S.E.S.; Khoobi, M. Cholinesterase inhibition activity and docking simulation study of coumarin mannich base derivatives. J. Sci. Islamic Repub. Iran 2019, 30, 5–12. [Google Scholar] [CrossRef]
- Yike, I. Fungal proteases and their pathophysiological effects. Mycopathologia 2011, 171, 299–323. [Google Scholar] [CrossRef]
- Bajorath, J.; Saenger, W.; Pal, G.P. Autolysis and inhibition of proteinase K, a subtilisin-related serine proteinase isolated from the fungus Tritirachium album Limber. Biochim. Biophys. Acta 1988, 954, 176–182. [Google Scholar] [CrossRef]
- De Araújo, R.S.A.; Guerra, F.Q.S.; De Lima, E.O.; De Simone, C.A.; Tavares, J.F.; Scotti, L.; Scotti, M.T.; De Aquino, T.M.; De Moura, R.O.; Mendonça, F.J.B.; et al. Synthesis, structure-activity relationships (SAR) and in silico studies of coumarin derivatives with antifungal activity. Int. J. Mol. Sci. 2013, 14, 1293–1309. [Google Scholar] [CrossRef]
- Montagner, C.; de Souza, S.M.; Groposo, C.; Monache, F.D.; Smánia, E.F.A.; Smánia, A. Antifungal activity of coumarins. Z. Naturforsch C 2008, 63, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.M.; Saify, Z.S.; Khan, M.Z.; Choudhary, Z.-U.M.I.; Rahman, A.; Perveen, S.; Chohan, Z.H.; Supuran, C.T. Synthesis of coumarin derivatives with cytotoxic, antibacterial and antifungal activity. J. Enzym. Inhib. Med. Chem. 2004, 19, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Thangavelu, B.; Chun, S.; Sathiyabama, M. Proteases from phytopathogenic fungi and their importance in phytopathogenicity. J. Gen. Plant. Pathol. 2016, 82, 233–239. [Google Scholar] [CrossRef]
- Dobinson, K.F.; Lecomte, N.; Lazarovits, G. Production of an extracellular trypsin- like protease by the fungal plant pathogen Verticillum dahliae. Can. J. Microbiol. 1997, 43, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wei, W.; Fu, Y.; Cheng, J.; Xie, J.; Li, G.; Yi, X.; Kang, Z.; Dickman, M.B.; Jiamg, D. A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 2013, 8, e53901. [Google Scholar] [CrossRef]
- Dattagupta, J.K.; Fujiwara, T.; Grishin, E.V.; Lindner, K.; Manor, P.C.; Pieniazek, N.J.; Saenger, W.; Suck, D. Crystallization of the fungal enzyme proteinase K and amino acid composition. J. Mol. Biol. 1975, 97, 267–271. [Google Scholar] [CrossRef]
- Cera, E.D. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef]
- Mahajan, R.; Kaur, D.J.; Bajaj, K.L. Nematicidal activity of phenolic compounds against Meloidogyne incognita. Nematol. Medit. 1992, 20, 217–219. [Google Scholar]
- Wang, X.B.; Li, G.H.; Li, L.; Zheng, L.J.; Huang, R.; Zhang, K.Q. Nematicidal coumarins from Heracleum candicans Wall. Nat. Prod. Res. 2008, 22, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Ntalli, N.G.; Caboni, P. Botanical nematicides: A review. J. Agric. Food Chem. 2012, 60, 9929–9940. [Google Scholar] [CrossRef]
- Liu, F.; Yang, Z.; Zheng, X.; Luo, S.; Zhang, K.; Li, G. Nematicidal coumarin from Ficus carica L. J. Asia Pac. Entomol. 2011, 14, 79–81. [Google Scholar] [CrossRef]
- Lifschitz, A.; Lanusse, C.; Alvarez, L. Host pharmacokinetics and drug accumulation of anthelmintics within target helminth parasites of ruminants. N. Z. Vet. J. 2017, 65, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Lacy, A.; O’Kennedy, R. Studies on coumarines and coumarin related compounds to determine their therapeutic role in the treatment of cancer. Curr. Pharm. Des. 2004, 10, 3797–3811. [Google Scholar] [CrossRef]
- Fentem, J.H.; Hammond, A.H.; Garle, M.J.; Fry, M.J. Toxicity of coumarin and various methyl derivatives in cultures of rat hepatocytes and V79 cells. Toxicol. In Vitro 1992, 6, 21–25. [Google Scholar] [CrossRef]
- Loprinzi, C.L.; Sloan, J.; Kugler, J. Coumarin-induced hepatotoxicity. J. Clin. Oncol. 1997, 15, 3167–3168. [Google Scholar] [CrossRef] [PubMed]
- Weigta, S.; Hueblera, N.; Streckerb, R.; Braunbeckb, T.; Broscharda, T.H. Developmental effects of coumarin and the anticoagulant coumarin derivative warfarin on zebrafish (Danio rerio) embryos. Reprod. Toxicol. 2012, 33, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Popp, D.; Plugge, C.M.; Kleinsteuber, S.; Harms, H.; Sträuber, H. Inhibitory effect of coumarin on syntrophic fatty acid-oxidizing and methanogenic cultures and biogas reactor microbiomes. Appl. Environ. Microbiol. 2017, 83, e00438-17. [Google Scholar] [CrossRef] [PubMed]
- Maistro, E.L.; de Souza Marques, E.; Fedato, R.P.; Tolentino, F.; da Silva, C.D.A.C.; Tsuboy, M.S.F.; Aparecida, F.; Varanda, E.A. In vitro assessment of mutagenic and genotoxic effects of coumarin derivatives 6,7-dihydroxycoumarin and 4-methylesculetin. J. Toxicol. Environ. Health Part A 2014, 78, 109–118. [Google Scholar] [CrossRef]
- Lešnik, S.; Furlan, V.; Bren, U. Rosemary (Rosmarinus officinalis L.): Extraction techniques, analytical methods and health-promoting biological effects. Phytochem. Rev. 2021, 1–56. [Google Scholar] [CrossRef]
- Štern, A.; Furlan, V.; Novak, M.; Štampar, M.; Kolenc, Z.; Kores, K.; Filipić, M.; Bren, U.; Žegura, B. Chemoprotective effects of xanthohumol against the carcinogenic mycotoxin aflatoxin B1. Foods 2021, 10, 1331. [Google Scholar] [CrossRef] [PubMed]
- Siber, T.; Bušić, V.; Zobundžija, D.; Roca, S.; Vikić Topić, D.; Vrandečić, K.; Gašo Sokač, D. An improved method for the quaternization of nicotinamide and antifungal activities of its derivatives. Molecules 2019, 24, 1001. [Google Scholar] [CrossRef] [PubMed]
- Bušić, V.; Vrandečić, K.; Siber, T.; Roca, S.; Vikić-Topić, D.; Gašo-Sokač, D. A rapid microwave induced synthesis of isonicotinamide derivatives and their antifungal activity. Croat. Chem. Acta 2019, 92, 125–135. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hanckok, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Hocquet, A.; Langgård, M. An Evaluation of the MM+ Force Field. J. Mol. Model. 1998, 4, 94–112. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Optimization of parameters for semiempirical methods I. Method. J. Comput. Chem. 1989, 10, 209–220. [Google Scholar] [CrossRef]
- Tetko, I.V.; Gasteiger, J.; Todeschini, R.; Mauri, A.; Livingstone, D.; Ertl, P.; Palyulin, V.A.; Radchenko, E.V.; Zefirov, N.S.; Makarenko, A.S.; et al. Virtual computational chemistry laboratory—Design and description. J. Comput. Aided Mol. Des. 2005, 19, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Gramatica, P.; Chirico, N.; Papa, E.; Cassani, S.; Kovarich, S. QSARINS: A new software for the development, analysis, and validation of QSAR MLR models. J. Comput. Chem. 2013, 34, 2121–2132. [Google Scholar] [CrossRef]
- Gramatica, O. Principles of QSAR modeling: Comments and suggestions from personal experience. IJQSPR 2020, 5, 61–97. [Google Scholar] [CrossRef]
- Eriksson, L.; Jaworska, J.; Worth, A.P.; Cronin, M.T.D.; McDowell, R.M.; Gramatica, P. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ. Health Perspect. 2003, 111, 1361–1375. [Google Scholar] [CrossRef] [PubMed]

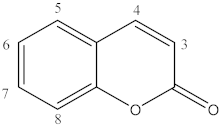 | ||
|---|---|---|
| No. mol | Mol ID | Substituents |
| 1 | A3 | 3-COOC2H5 |
| 2 | A5 | 3-COC6H5 |
| 3 | C5 | 3-COC6H5; 7-OCH2C6H5 |
| 4 | G3 | 3-COOC2H5; 6-Br |
| 5 | J2 | 3-COOCH3; 6-OH |
| 6 | J3 | 3-COOC2H5; 6-OH |
| 7 | K3 | 3-COOC2H5; 6-Cl |
| 8 | L5 | 3-COC6H5; 6-Br; 8-Br |
| 9 | M2 | 3-COOCH3; 7-OCH3 |
| 10 | M3 | 3-COOC2H5; 7-OCH3 |
| 11 | N2 | 3-COOCH3; 6-OCH3 |
| 12 | O3 | 3-COOC2H5; 8-OC2H5 |
| 13 | O5 | 3-COC6H5; 8-OC2H5 |
| 14 | P2 | 3-COOCH3; 6-N+OO− |
| 15 | P3 | 3-COOC2H5; 6-N+OO− |
| 16 | A1 | 3-COCH3 |
| 17 | A4 | 3-CN |
| 18 | D1 | 3-COCH3; 8-OH |
| 19 | D4 | 3-CN; 8-OH |
| 20 | E1 | 3-COCH3; 7-OH |
| 21 | F1 | 3-COCH3; 7-N(C2H5)2 |
| 22 | G1 | 3-COCH3; 6-Br |
| 23 | G4 | 3-CN; 6-Br |
| 24 | J1 | 3-COCH3; 6-OH |
| 25 | J4 | 3-CN; 6-OH |
| 26 | M4 | 3-CN; 7-OCH3 |
| 27 | N4 | 3-CN; 6-OCH3 |
| 28 | O1 | 3-COCH3; 8-OC2H5 |
| 29 | O4 | 3-CN; 8-OC2H5 |
| 30 | A2 | 3-COOCH3 |
| 31 | C4 | 3-CN; 7-OCH2C6H5 |
| 32 | E2 | 3-COOCH3; 7-OH |
| 33 | E5 | 3-COC6H5; 7-OH |
| 34 | G2 | 3-COOCH3; 6-Br |
| 35 | K5 | 3-COC6H5; 6-Cl |
| 36 | M5 | 3-COC6H5; 7-OCH3 |
| 37 | J5 | 3-COC6H5; 6-OH |
| 38 | L3 | 3-COOC2H5; 6-Br; 8-Br |
| Antifungal Activity a | Antibacterial Activity b | Nematicidal Activity c | ||||||
|---|---|---|---|---|---|---|---|---|
| No. mol | Macrophomina phaseolina | Sclerotinia sclerotiorum | Fusarium oxysporum f. sp. lycopersici | F. culmorum | Bacillus mycoides | Bradhrizobium japonicum | Heterorhabditis bacteriophora | Steinernema feltiae |
| 1 | 53.63 | 73.09 | −3.64 | −9.53 | >512 | >512 | 8.00 | 10.00 |
| 2 | 57.67 | 39.62 | 1.21 | −8.67 | >512 | >512 | 29.75 | 18.75 |
| 3 | 53.63 | 64.89 | 4.85 | −7.80 | >512 | >512 | 0.00 | 0.00 |
| 4 | 56.52 | 77.87 | 1.21 | −0.87 | >512 | >512 | 0.00 | 3.75 |
| 5 | 64.01 | 76.50 | −6.07 | −6.07 | >512 | 64 | 0.00 | 0.00 |
| 6 | 66.32 | 82.65 | 14.56 | 26.86 | >512 | >512 | 31.25 | 20.75 |
| 7 | 65.74 | 84.02 | 10.92 | 13.86 | >512 | 512 | 0.00 | 0.00 |
| 8 | 66.32 | 84.70 | 13.35 | 6.93 | >512 | >512 | 0.00 | 0.00 |
| 9 | 74.39 | 84.70 | 13.35 | 10.40 | >512 | 512 | 0.00 | 0.00 |
| 10 | 72.09 | 64.89 | 19.42 | 13.86 | >512 | >512 | 56.75 | 64.00 |
| 11 | 53.06 | 51.91 | 6.07 | −6.93 | >512 | >512 | 0.00 | 4.25 |
| 12 | 69.78 | 59.43 | −7.28 | −27.73 | >512 | >512 | 24.75 | 21.00 |
| 13 | 24.80 | 67.62 | −6.07 | −32.93 | >512 | >512 | 0.00 | 0.00 |
| 14 | 55.94 | 81.97 | −10.92 | −4.33 | >512 | 512 | 0.00 | 0.00 |
| 15 | 59.98 | 76.50 | 0.00 | −6.93 | >512 | 512 | 0.00 | 2.00 |
| 16 | 61.71 | 43.72 | 27.91 | 30.33 | >512 | >512 | 0.00 | 0.00 |
| 17 | 61.71 | 38.93 | 23.06 | 26.00 | >512 | >512 | 8.75 | 14.75 |
| 18 | 59.98 | 27.32 | 27.91 | 9.53 | >512 | >512 | 0.00 | 0.00 |
| 19 | 69.78 | 54.64 | 32.77 | 13.00 | 512 | >512 | 20.75 | 11.25 |
| 20 | 65.74 | 4.78 | 29.13 | −3.47 | 512 | >512 | 5.50 | 17.00 |
| 21 | 74.97 | 40.98 | 40.05 | 26.00 | >512 | >512 | 13.00 | 22.50 |
| 22 | 70.36 | 43.72 | 29.13 | 33.80 | >512 | >512 | 0.00 | 0.00 |
| 23 | 83.62 | −15.03 | 27.91 | −9.53 | >512 | >512 | 0.00 | 2.75 |
| 24 | 80.16 | 55.33 | 64.32 | 70.19 | >512 | >512 | 44.50 | 43.00 |
| 25 | 74.39 | 54.64 | 66.75 | 62.39 | >512 | >512 | 0.00 | 1.25 |
| 26 | 72.66 | 23.91 | 40.05 | 45.06 | 512 | >512 | 12.00 | 14.25 |
| 27 | 75.55 | 61.48 | 71.60 | 63.26 | 512 | 512 | 0.00 | 0.00 |
| 28 | 76.12 | 79.92 | 65.53 | 65.86 | >512 | >512 | 0.00 | 0.00 |
| 29 | 77.28 | 66.26 | 65.53 | 70.19 | >512 | >512 | 0.00 | 0.00 |
| 30 | 70.93 | −7.51 | 20.63 | −17.33 | >512 | >512 | 0.00 | 0.00 |
| 31 | 69.20 | 0.68 | 32.77 | −1.73 | >512 | >512 | 0.00 | 0.00 |
| 32 | 79.58 | −4.10 | 24.27 | −11.27 | >512 | >512 | 17.50 | 10.50 |
| 33 | 68.63 | 6.83 | 27.91 | −10.40 | >512 | >512 | 0.00 | 10.50 |
| 34 | 66.90 | −15.71 | 27.91 | −10.40 | >512 | >512 | 16.50 | 40.25 |
| 35 | 78.43 | −26.64 | 35.19 | 6.93 | >512 | >512 | 0.00 | 0.00 |
| 36 | 79.01 | 51.91 | 44.90 | 23.40 | >512 | >512 | 0.00 | 9.00 |
| 37 | 64.01 | 13.66 | 31.55 | 0.00 | >512 | >512 | 0.00 | 4.75 |
| 38 | 80.16 | 10.25 | 35.19 | 1.73 | >512 | >512 | 0.00 | 0.00 |
| control | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| No. mol | Oral Rat LD50 (mg/kg bw) a | Tetrahymena pyriformis pIGC50 48-h (mol/L) b | Fathead Minnow pLC50 96-h (mol/L) c | Mutagenicity Value (Result) d | Bioaccumulation (logBAF/L kg−1) e |
|---|---|---|---|---|---|
| 1 | 978.2 | 4.03 | 4.59 | 0.45 (neg) | 0.85 |
| 2 | 386.97 | 4.97 | 5.68 | 0.43 (neg) | 1.52 |
| 3 | 147.37 | 5.86 | 7.12 | 0.18 (neg) | 1.48 |
| 4 | 2223.31 | 4.66 | 5.24 | 0.14 (neg) | 1.14 |
| 5 | 1146.8 | 4.37 | 4.43 | 0.28 (neg) | 0.79 |
| 6 | 1592.38 | 4.15 | 4.78 | 0.36 (neg) | 0.76 |
| 7 | 1251.57 | 4.54 | 5.08 | 0.26 (neg) | 1.31 |
| 8 | 634.37 | 5.87 | 6.82 | 0.35 (neg) | 2.09 |
| 9 | 1882.79 | 3.96 | 4.29 | 0.49 (neg) | 0.68 |
| 10 | 2471.53 | 4.37 | 4.66 | 0.45 (neg) | 0.77 |
| 11 | 975.12 | 3.74 | 4.52 | 0.44 (neg) | 1.03 |
| 12 | 2358.27 | 4.14 | 5.07 | 0.45 (neg) | 1.12 |
| 13 | 387.72 | 4.74 | 5.64 | 0.40 (neg) | 1.42 |
| 14 | 1799.19 | 4.18 | 4.93 | 0.63 (pos) | 0.45 |
| 15 | 1169.36 | 4.78 | 5.20 | 0.61 (pos) | 0.48 |
| 16 | 774.04 | 5.01 | 4.27 | 0.03 (neg) | 0.90 |
| 17 | 519.66 | 4.37 | 3.74 | 0.36 (neg) | 1.24 |
| 18 | 1163.28 | 4.96 | 3.90 | 0.02 (neg) | 0.78 |
| 19 | 313.76 | 4.37 | 3.71 | 0.34 (neg) | 1.17 |
| 20 | 1212.38 | 5.10 | 4.17 | 0.03 (neg) | 0.61 |
| 21 | 1882.68 | 4.96 | 4.87 | 0.75 (pos) | 1.03 |
| 22 | 2307.19 | 5.11 | 4.63 | 0.23 (neg) | 1.14 |
| 23 | 1733.09 | 4.14 | 4.20 | 0.30 (neg) | 1.51 |
| 24 | 725.86 | 4.99 | 4.10 | 0.01 (neg) | 0.79 |
| 25 | 418.02 | 4.31 | 3.90 | 0.37 (neg) | 1.21 |
| 26 | 1097.06 | 4.2 | 3.96 | 0.47 (neg) | 0.98 |
| 27 | 718.52 | 3.78 | 3.74 | 0.35 (neg) | 1.29 |
| 28 | 1187.42 | 4.21 | 4.83 | 0.57 (pos) | 1.06 |
| 29 | 320.31 | 4.12 | 3.69 | 0.50 (pos) | 1.22 |
| 30 | 695.39 | 3.87 | 4.43 | 0.46 (neg) | 0.83 |
| 31 | 190.96 | 4.41 | 5.60 | 0.61 (pos) | 1.61 |
| 32 | 1515.42 | 4.15 | 4.63 | 0.33 (neg) | 0.49 |
| 33 | 190.83 | 5.21 | 5.43 | 0.43 (neg) | 1.32 |
| 34 | 2158.86 | 4.85 | 5.31 | 0.08 (neg) | 1.18 |
| 35 | 100.83 | 5.27 | 5.87 | 0.54 (pos) | 1.92 |
| 36 | 451.79 | 4.91 | 5.34 | 0.39 (neg) | 1.29 |
| 37 | 139.64 | 5.00 | 5.68 | 0.57 (pos) | 1.43 |
| 38 | 749.5 | 5.17 | 6.34 | 0.49 (neg) | 0.85 |
| Statistical Parameters | Model 1 |
|---|---|
| Ntr | 23 |
| Nex | 9 |
| R2 | 0.78 |
| R2adj | 0.75 |
| s | 0.03 |
| F | 23.46 |
| Kxx | 0.20 |
| ΔK | 0.12 |
| RMSEtr | 0.02 |
| MAEtr | 0.02 |
| CCCtr | 0.88 |
| Q2LOO | 0.67 |
| RMSEcv | 0.03 |
| MAEcv | 0.02 |
| CCCcv | 0.82 |
| R2Yscr | 0.14 |
| Q2Yscr | −0.28 |
| RMSEAV Yscr | 0.09 |
| RMSEext | 0.03 |
| MAEext | 0.02 |
| R2ext | 0.67 |
| CCCext | 0.81 |
| Q2F1 | 0.62 |
| Q2F2 | 0.61 |
| Q2F3 | 0.68 |
| r2m average | 0.54 |
| r2m difference | 0.02 |
| Applicability domain | h* = 0.522 |
| N compounds outlier | - |
| N compounds out of app.dom. | - |
| JGI6 | Mor28v | L2e | |
|---|---|---|---|
| JGI6 | 1.00 | ||
| Mor28v | −0.16 | 1.00 | |
| L2e | 0.29 | −0.22 | 1.00 |
| Statistical Parameters | Model 2 | Model 2a * |
|---|---|---|
| Ntr | 28 | 28 |
| Nex | 10 ** | |
| R2 | 0.78 | 0.78 |
| R2adj | 0.75 | 0.75 |
| s | 0.07 | 0.07 |
| F | 28.84 | 28.84 |
| Kxx | 0.22 | 0.22 |
| ΔK | 0.17 | 0.17 |
| RMSEtr | 0.07 | 0.07 |
| MAEtr | 0.06 | 0.06 |
| CCCtr | 0.88 | 0.88 |
| Q2LOO | 0.71 | 0.71 |
| RMSEcv | 0.08 | 0.08 |
| MAEcv | 0.07 | 0.07 |
| CCCcv | 0.84 | 0.84 |
| R2Yscr | 0.11 | 0.11 |
| Q2Yscr | −0.24 | −0.24 |
| RMSEAV Yscr | 0.14 | 0.14 |
| RMSEext | 1.51 | |
| MAEext | 1.44 | |
| R2ext | 0.03 | |
| CCCext | 0.01 | |
| Q2F1 | −0.03 | |
| Q2F2 | −9.06 | |
| Q2F3 | −102.96 | |
| r2m average | −0.21 | |
| r2m difference | 0.47 | |
| Applicability domain | h* = 0.429 | |
| N compounds outlier | 1 (9) | 10 (20, 23, 30–35, 37, 38) |
| N compounds out of app.dom. | 1 (8) | 2 (8, 38) |
| SEigm | P2s | R1e+ | |
|---|---|---|---|
| SEigm | 1.00 | ||
| P2s | 0.20 | 1.00 | |
| R1e+ | 0.08 | 0.53 | 1.00 |
| Demethylase | Chitinase | Transferase | Endoglucanase I | Proteinase K | Pectinase | AChE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Comp. (Pose) | Total E | Comp. (Pose) | Total E | Comp. (Pose) | Total E | Comp. (Pose) | Total E | Comp. (Pose) | Total E | Comp. (Pose) | Total E | Comp. (Pose) | Total E |
| 3 (2) | −100.41 | 3 (1) | −122.83 | 3 (2) | −99.82 | 3 (1) | −127.50 | 6 (0) | −114.21 | 3 (1) | −96.65 | 3 (0) | −118.95 |
| 5lw * (1) | −96.19 | 37 (2) | −121.58 | 15 (1) | −93.73 | 31 (1) | −123.92 | 13 (1) | −114.07 | 15 (0) | −89.25 | 13 (0) | −112.30 |
| 33 (0) | −86.71 | 13 (0) | −115.99 | 3lp * (2) | −91.27 | 15 (1) | −113.50 | 5 (1) | −110.68 | 14 (2) | −86.21 | 31 (2) | −107.19 |
| 31 (1) | −86.05 | 33 (2) | −115.37 | 14 (0) | −88.30 | 13 (1) | −107.84 | 36 (2) | −109.91 | 31 (2) | −84.93 | 8 (1) | −106.67 |
| 13 (0) | −85.79 | 38f * (0) | −112.76 | 13 (1) | −85.82 | 14 (1) | −105.32 | 37 (1) | −109.92 | 13 (1) | −84.49 | 33 (0) | −105.41 |
| 15 (0) | −84.84 | 6 (0) | −112.29 | 37 (1) | −84.52 | 36 (0) | −104.68 | 33 (1) | −107.92 | 6 (0) | −84.14 | 15 (1) | −104.48 |
| 37 (1) | −84.18 | 36 (0) | −110.99 | 31 (0) | −84.51 | 12 (2) | −104.19 | 2 (1) | −104.19 | 36 (2) | −81.44 | 36 (1) | −104.32 |
| 36 (1) | −81.35 | 8 (1) | −110.43 | 12 (2) | −83.12 | 21 (2) | −104.11 | 1 (1) | −104.13 | 21 (1) | −81.19 | 37 (1) | −103.03 |
| 8 (0) | −81.14 | 31 (0) | −109.87 | 35 (1) | −82.71 | 33 (2) | −104.07 | 12 (2) | −103.34 | 33 (0) | −80.92 | 12 (1) | −102.67 |
| 14 (0) | −80.10 | 5 (1) | −107.96 | 8 (1) | −82.32 | 6 (0) | −102.75 | 8 (2) | −102.34 | 5 (1) | −80.27 | 2 (1) | −101.18 |
| H Bond | Energy | Van der Waals Interaction | Energy |
|---|---|---|---|
| M-GLY-160 | −3.76 | S-HIS-69 | −1.28 |
| S-ASN-161 | −9.68 | M-SER-132 | −3.58 |
| M-SER-170 | −5.61 | M-LEU-133 | −8.08 |
| M-PRO-171 | −3.50 | M-GLY-134 | −10.23 |
| M-ALA-172 | −3.50 | M-GLY-135 | −4.94 |
| S-THR-223 | −2.50 | M-ALA-158 | −4.13 |
| M-SER-224 | −3.50 | M-ALA-159 | −6.16 |
| S-SER-224 | −4.71 | M-GLY-160 | −7.28 |
| M-ASN-161 | −3.49 | ||
| S-ASN-161 | −4.87 | ||
| S-ASN-162 | −2.68 | ||
| S-TYR-169 | −2.09 | ||
| M-SER-170 | −0.14 | ||
| M-SER-224 | −1.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rastija, V.; Vrandečić, K.; Ćosić, J.; Majić, I.; Šarić, G.K.; Agić, D.; Karnaš, M.; Lončarić, M.; Molnar, M. Biological Activities Related to Plant Protection and Environmental Effects of Coumarin Derivatives: QSAR and Molecular Docking Studies. Int. J. Mol. Sci. 2021, 22, 7283. https://doi.org/10.3390/ijms22147283
Rastija V, Vrandečić K, Ćosić J, Majić I, Šarić GK, Agić D, Karnaš M, Lončarić M, Molnar M. Biological Activities Related to Plant Protection and Environmental Effects of Coumarin Derivatives: QSAR and Molecular Docking Studies. International Journal of Molecular Sciences. 2021; 22(14):7283. https://doi.org/10.3390/ijms22147283
Chicago/Turabian StyleRastija, Vesna, Karolina Vrandečić, Jasenka Ćosić, Ivana Majić, Gabriella Kanižai Šarić, Dejan Agić, Maja Karnaš, Melita Lončarić, and Maja Molnar. 2021. "Biological Activities Related to Plant Protection and Environmental Effects of Coumarin Derivatives: QSAR and Molecular Docking Studies" International Journal of Molecular Sciences 22, no. 14: 7283. https://doi.org/10.3390/ijms22147283
APA StyleRastija, V., Vrandečić, K., Ćosić, J., Majić, I., Šarić, G. K., Agić, D., Karnaš, M., Lončarić, M., & Molnar, M. (2021). Biological Activities Related to Plant Protection and Environmental Effects of Coumarin Derivatives: QSAR and Molecular Docking Studies. International Journal of Molecular Sciences, 22(14), 7283. https://doi.org/10.3390/ijms22147283







