Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses
Abstract
:1. Introduction
2. Results
2.1. Assessment of Primer Specificity and PCR Amplification Efficiency
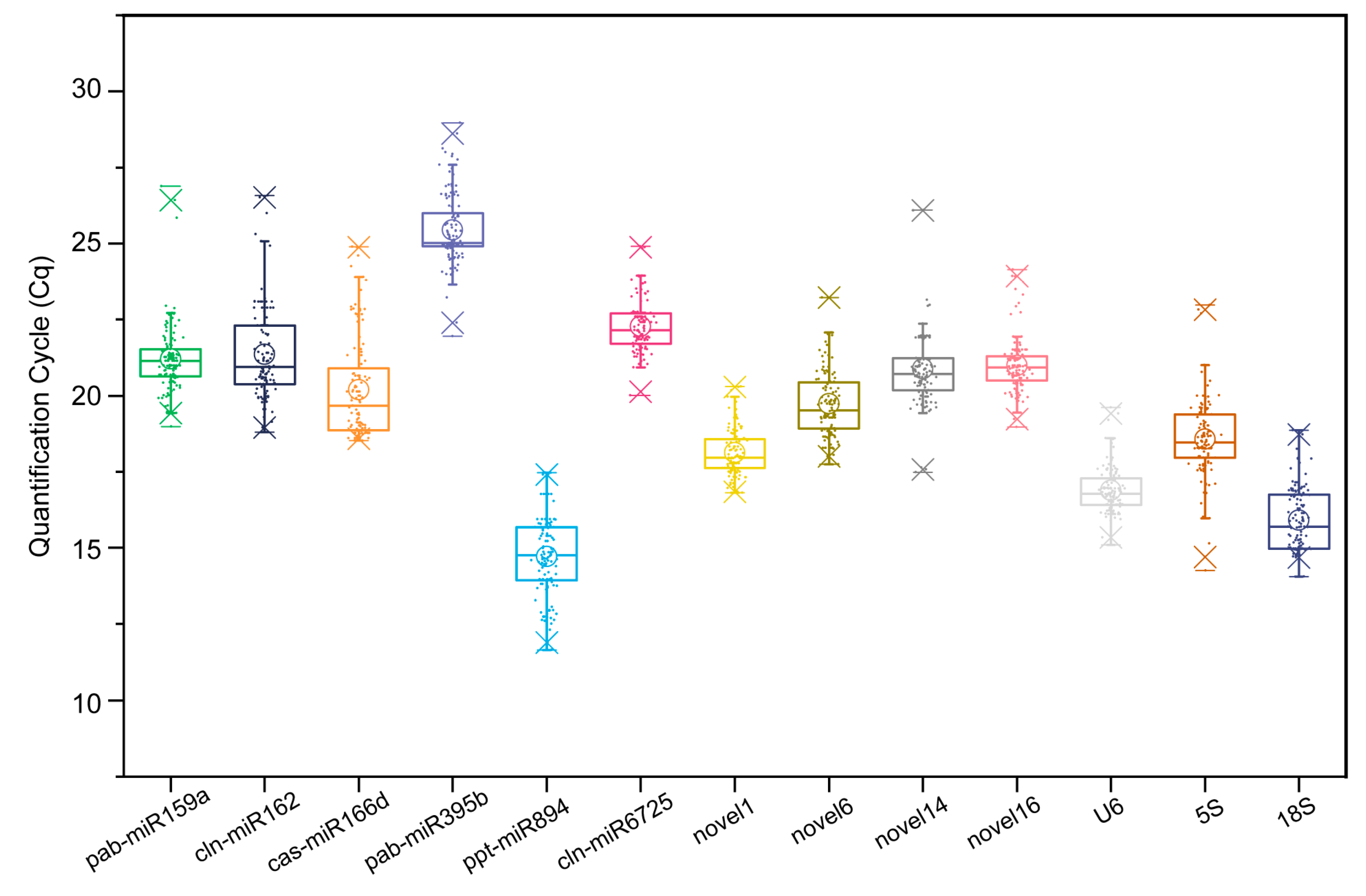
2.2. Expression Levels of Candidate RGs
2.3. Expression Stability of Candidate RGs under Multiple Stresses and in Different Tissues
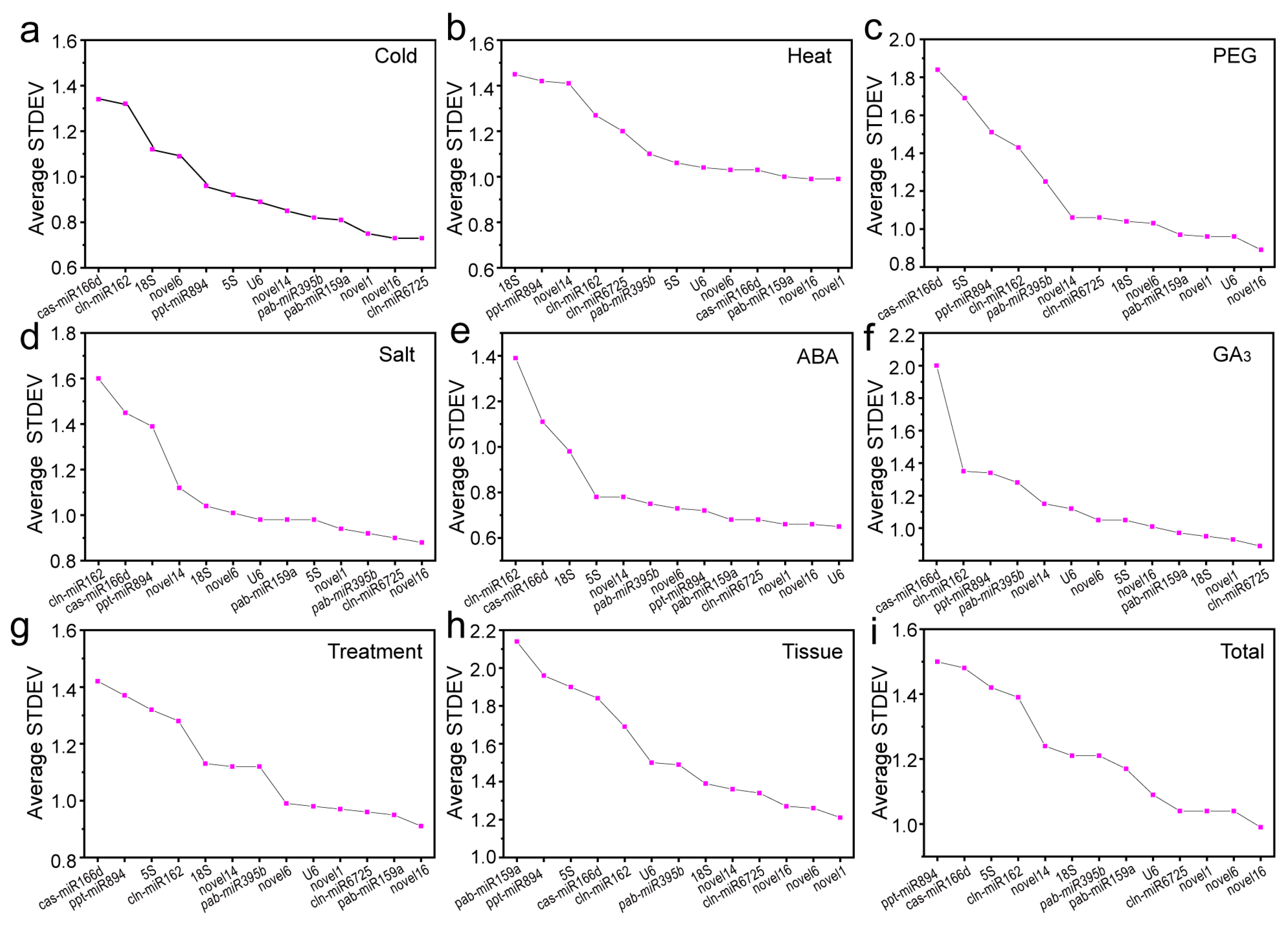
2.3.1. Delta Ct Method
2.3.2. GeNorm Analysis
2.3.3. Norm Finder Analysis
2.3.4. BestKeeper Algorithm
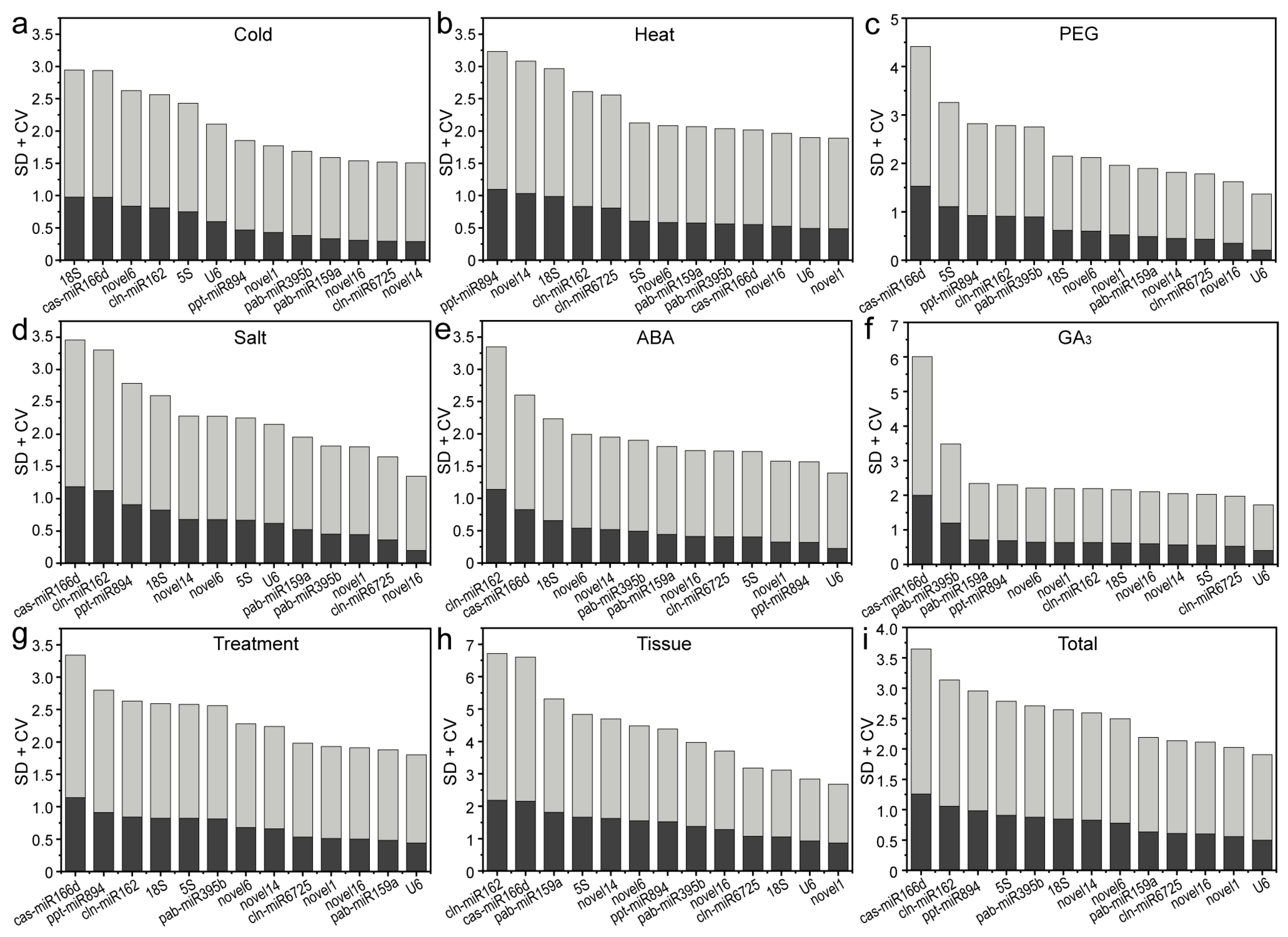
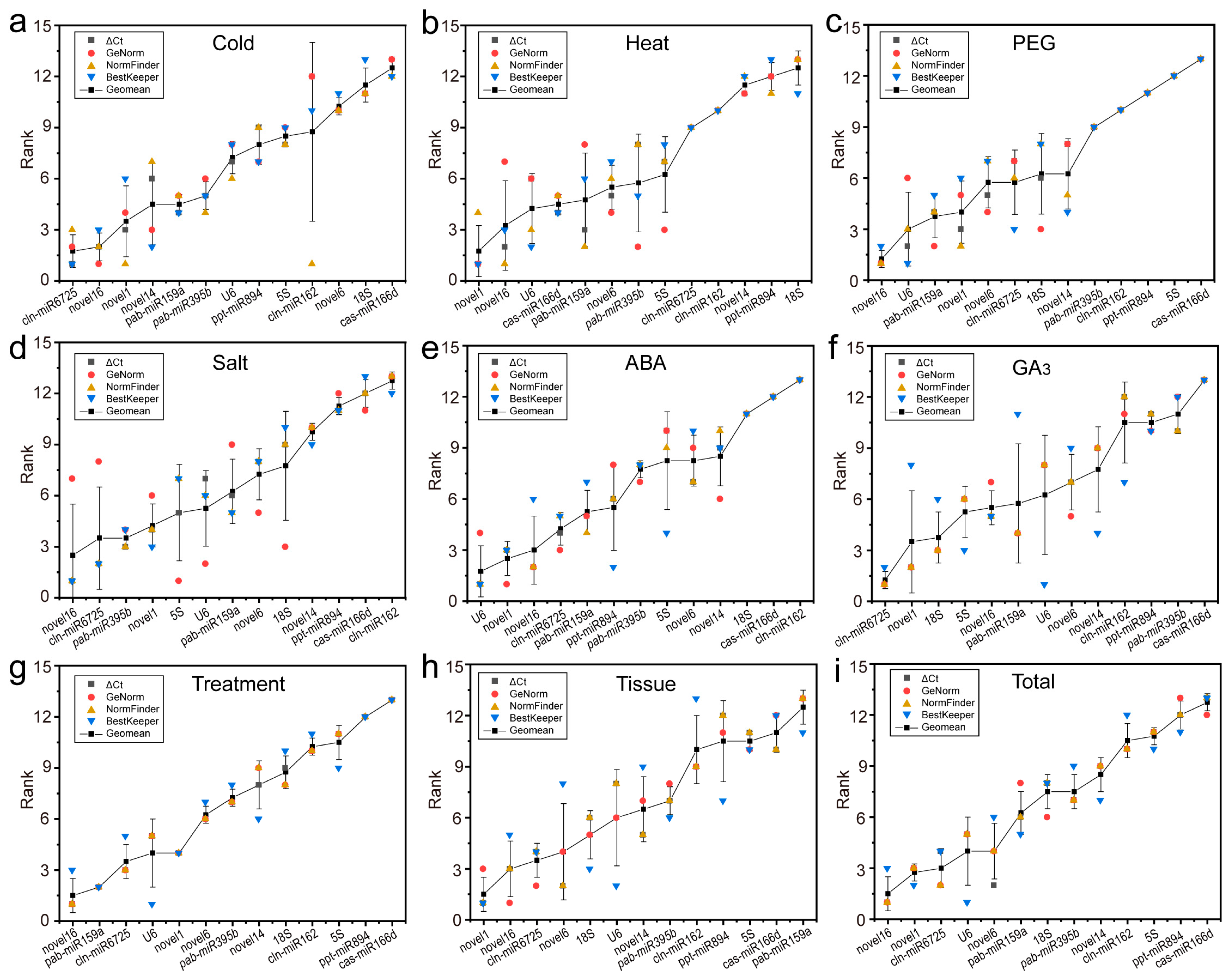
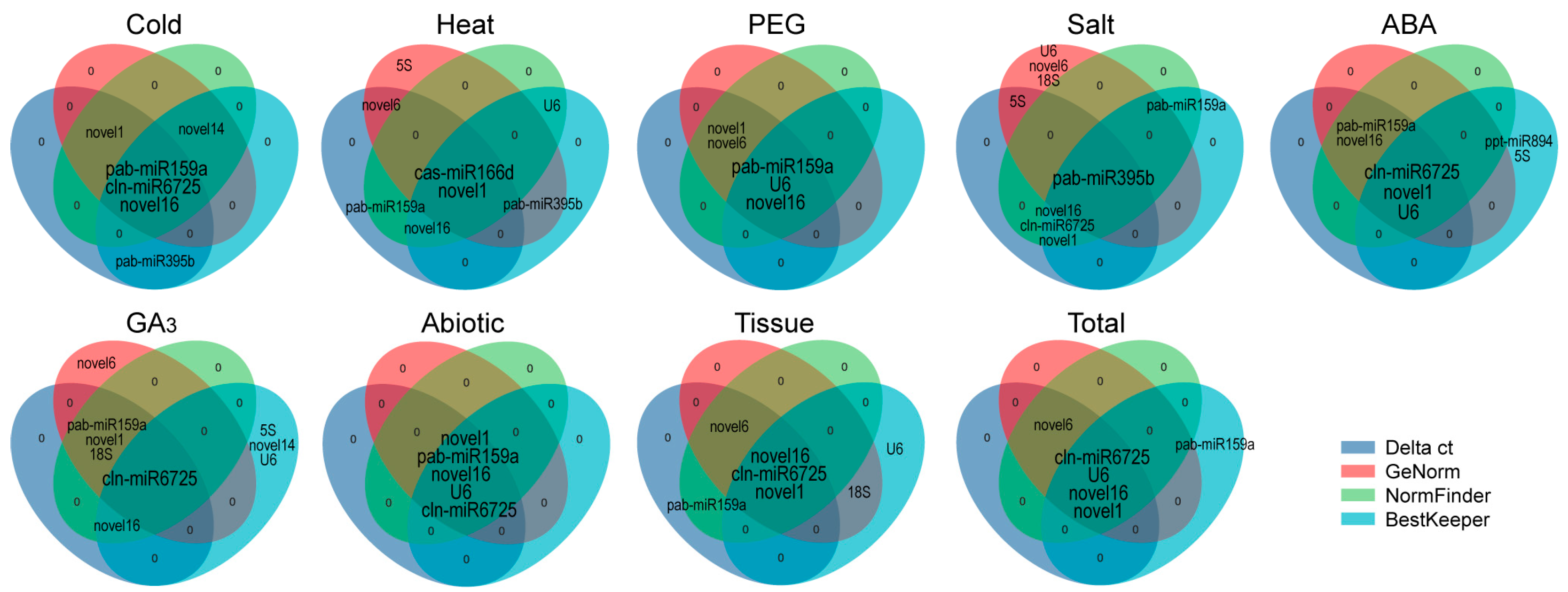
2.4. Comprehensive Stability Analysis of RGs
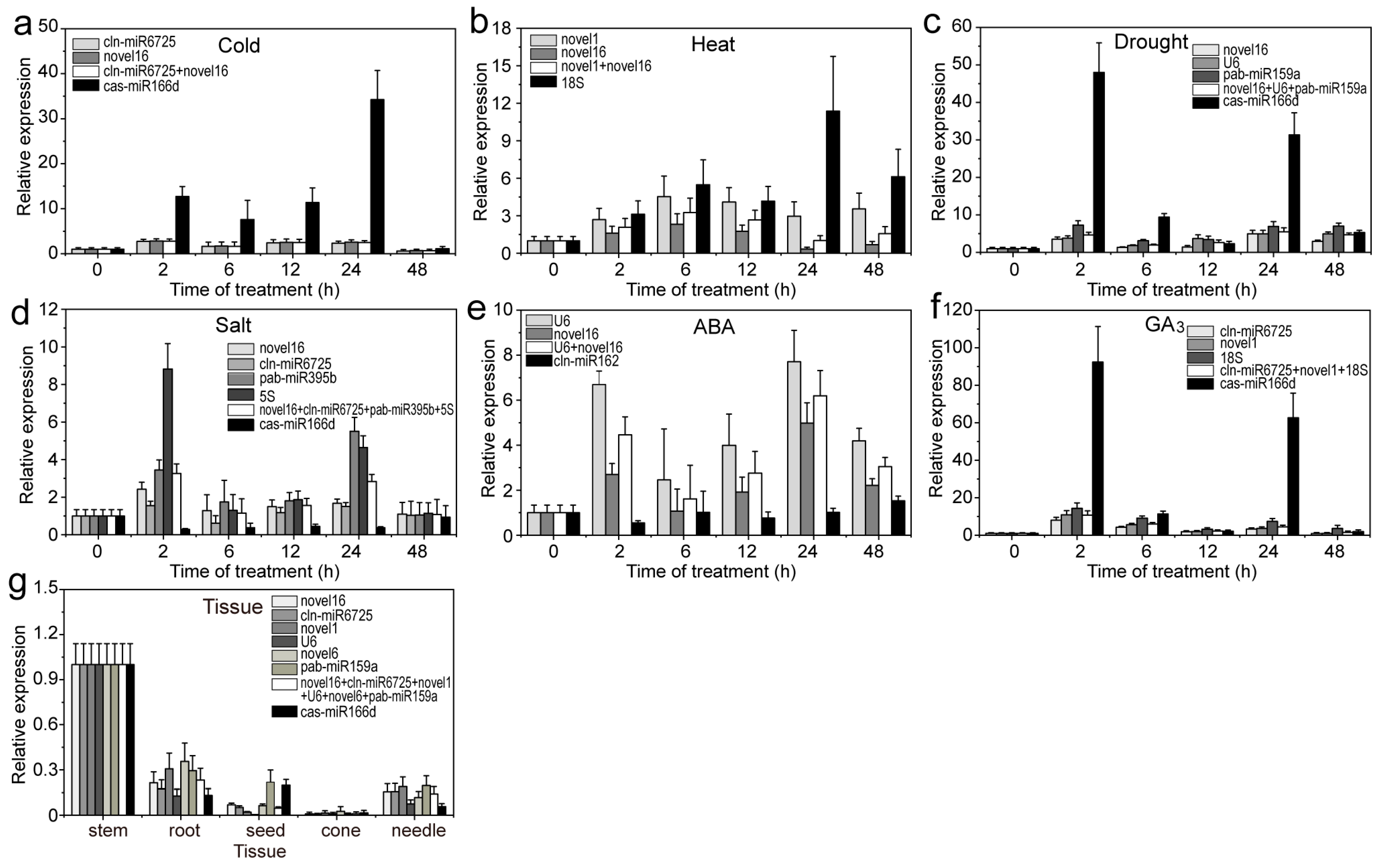
2.5. Validation of the Stability of RGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Selection of Candidate RGs and Primer Design
4.3. RNA Extraction and cDNA Preparation
4.4. RT-PCR and qRT-PCR Analysis
4.5. Gene Expression Stability Analysis
4.6. Validation of RGs by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Takenaka, C.; Yamada, T.; Iwata, S. Ethylene emission from seedlings of hinoki (Chamaecyparis obtusa) and sugi (Cryptomeria japonica) under acidic stress. J. For. Res. 1996, 1, 183–185. [Google Scholar] [CrossRef]
- Ujino-Ihara, T. Transcriptome analysis of heat stressed seedlings with or without pre-heat treatment in Cryptomeria japonica. Mol. Genet. Genom. 2020, 295, 1163–1172. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Q.; Guo, Z.; Yang, J.; Mo, J.; Cui, J.; Hu, H.; Xu, J. Individual Cryptomeria fortunei Hooibrenk clones show varying degrees of chilling stress resistance. Forests 2020, 11, 189. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Hua, H.; Xu, J.; Mo, J.; Zhao, H.; Yang, J. Cloning and functional analysis of lignin biosynthesis genes Cf4CL and CfCCoAOMT in Cryptomeria fortunei. Genes 2019, 10, 619. [Google Scholar] [CrossRef] [Green Version]
- Tsubomura, M.; Kurita, M.; Watanabe, A. Determination of male strobilus developmental stages by cytological and gene expression analyses in Japanese cedar (Cryptomeria japonica). Tree Physiol. 2016, 36, 653–666. [Google Scholar] [CrossRef] [Green Version]
- Ujino-Ihara, T.; Ueno, S.; Uchiyama, K.; Futamura, N. Comprehensive analysis of small RNAs expressed in developing male strobili of Cryptomeria japonica. PLoS ONE 2018, 13, e193665. [Google Scholar] [CrossRef]
- Patade, V.Y.; Suprasanna, P. Short-term salt and PEG stresses regulate expression of microRNA, miR159 in sugarcane leaves. J. Crop. Sci. Biotechnol. 2010, 13, 177–182. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.K. MicroRNAs and siRNAs in abiotic stress responses. Plant Cell 2015, 18, 2051–2065. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Obernosterer, G.; Leuschner, P.J.F.; Alenius, M.; Martinez, J. Post-transcriptional regulation of microRNA expression. RNA 2006, 12, 1161–1167. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Lai, B.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Identification of microRNAs and their target genes related to the accumulation of anthocyanins in litchi chinensis by high-throughput sequencing and degradome analysis. Front. Plant Sci. 2016, 7, 2059. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, Q.; Jiang, Y.; Li, Y.; Zhang, H.; Li, R. Reference genes identification for normalization of qPCR under multiple stresses in Hordeum brevisubulatum. Plant Methods 2018, 14, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, L.; Wang, Z.; Liu, L.; Guo, J.; Gu, C. Selection and verification of candidate reference genes for gene expression by quantitative RT-PCR in Hibiscus hamabo Sieb. et Zucc. Trees 2019, 33, 1591–1601. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006, 27, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Klein, D. Quantification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef]
- Su, Y.; He, W.; Wang, J.; Li, J.; Liu, S.; Wang, X. Selection of endogenous reference genes for gene expression analysis in the mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J. Econ. Entomol. 2013, 106, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Borowski, J.M.; Galli, V.; Messias, R.S.; Perin, E.C.; Buss, J.H.; Dos, A.E.S.S.; Rombaldi, C.V. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 2014, 239, 1187. [Google Scholar] [CrossRef]
- Tang, F.; Chu, L.; Shu, W.; He, X.; Wang, L.; Lu, M. Selection and validation of reference genes for quantitative expression analysis of miRNAs and mRNAs in poplar. Plant Methods 2019, 15, 35. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Chen, Y.; Guo, J.; Ling, H.; Gao, S.; Su, Y.; Que, Y.; Xu, L. Selection of reference genes for normalization of microRNA expression by RT-qPCR in sugarcane buds under cold stress. Front. Plant Sci. 2016, 7, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davoren, P.A.; Mcneill, R.E.; Lowery, A.J.; Kerin, M.J.; Miller, N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol. Biol. 2008, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Hao, J.; Meng, Y.; Luo, L.; Li, J.; Zhang, Y. Identifying optimal reference genes for the normalization of microRNA expression in cucumber under viral stress. PLoS ONE 2018, 13, e194436. [Google Scholar] [CrossRef] [Green Version]
- Feng, H.; Huang, X.; Zhang, Q.; Wei, G.; Wang, X.; Kang, Z. Selection of suitable inner reference genes for relative quantification expression of microRNA in wheat. Plant Physiol. Bioch. 2012, 51, 116–122. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lai, Z.X. Evaluation of suitable reference genes for normalization of microRNA expression by real-time reverse transcription PCR analysis during longan somatic embryogenesis. Plant Physiol. Bioch. 2013, 66, 20–25. [Google Scholar] [CrossRef]
- Luo, X.; Shi, T.; Sun, H.; Song, J.; Ni, Z.; Gao, Z. Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by RT-qPCR in leaves, flowers and young stems of peach. Sci. Hortic. 2014, 165, 281–287. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstraeten, B.; Smet, L.D.; Kyndt, T.; Meyer, T.D. Selection of miRNA reference genes for plant defence studies in rice (Oryza sativa). Planta 2019, 250, 2101–2110. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Liu, S.; Zhang, J.; Wu, Y.; Wu, W.; Zhang, Y.; Liu, B.; Tang, R.; He, L.; Li, R.; et al. Optimization of reference genes for qRT-PCR analysis of microRNA expression under abiotic stress conditions in sweet potato. Plant Physiol. Bioch. 2020, 154, 379–396. [Google Scholar] [CrossRef]
- Song, H.; Zhang, X.; Shi, C.; Wang, S.; Wu, A.; Wei, C. Selection and verification of candidate reference genes for mature microRNA expression by quantitative RT-PCR in the tea plant (Camellia sinensis). Genes 2016, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Kou, S.J.; Wu, X.M.; Liu, Z.; Liu, Y.L.; Xu, Q.; Guo, W.W. Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep. 2012, 31, 2151–2163. [Google Scholar] [CrossRef] [PubMed]
- Valiollahi, E.; Farsi, M.; Kakhki, A.M. Sly-miR166 and sly-miR319 are components of the cold stress response in Solanum lycopersicum. Plant Biotechnol. Rep. 2014, 8, 349–356. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018, 18, 52. [Google Scholar] [CrossRef]
- Cordeiro, D.; Rito, M.; Borges, F.; Canhoto, J.; Correia, S. Selection and validation of reference genes for qPCR analysis of miRNAs and their targets during somatic embryogenesis in tamarillo (Solanum betaceum Cav.). Plant Cell Tiss. Org. 2020, 143, 109–120. [Google Scholar] [CrossRef]
- Li, W.; Cui, X.; Meng, Z.; Huang, X.; Xie, Q.; Wu, H.; Jin, H.; Zhang, D.; Liang, W. Transcriptional regulation of Arabidopsis MIR168a and ARGONAUTE1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.L.; Wen, Z.; Yang, K.; Wen, X.P. Conserved miR396b-GRF regulation is involved in abiotic stress responses in pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2019, 20, 2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Dun, H.; Lian, C.; Zhang, X.; Yin, W.; Xia, X. The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol. Bioch. 2017, 115, 418–438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Wang, M.; Zhang, H.Y.; Liu, J.H. The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene-polyamine homeostasis. Plant Cell Physiol. 2016, 57, 1865–1878. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Gao, Z.; Li, H.; Li, Q.; Zhang, C.; Xu, W.; Song, S.; Ma, C.; Wang, S. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Sci. Rep. 2018, 8, 4444. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Hao, Z.; Zhong, W.; Li, H. Identification of suitable reference genes for RT-qPCR assays in Liriodendron chinense (Hemsl.) Sarg. Forests 2019, 10, 441. [Google Scholar] [CrossRef] [Green Version]
- Zhu, P.; Ma, Y.; Zhu, L.; Chen, Y.; Ji, K. Selection of suitable reference genes in Pinus massoniana Lamb. under different abiotic stresses for qPCR normalization. Forests 2019, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2019, 65, 2119–2135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
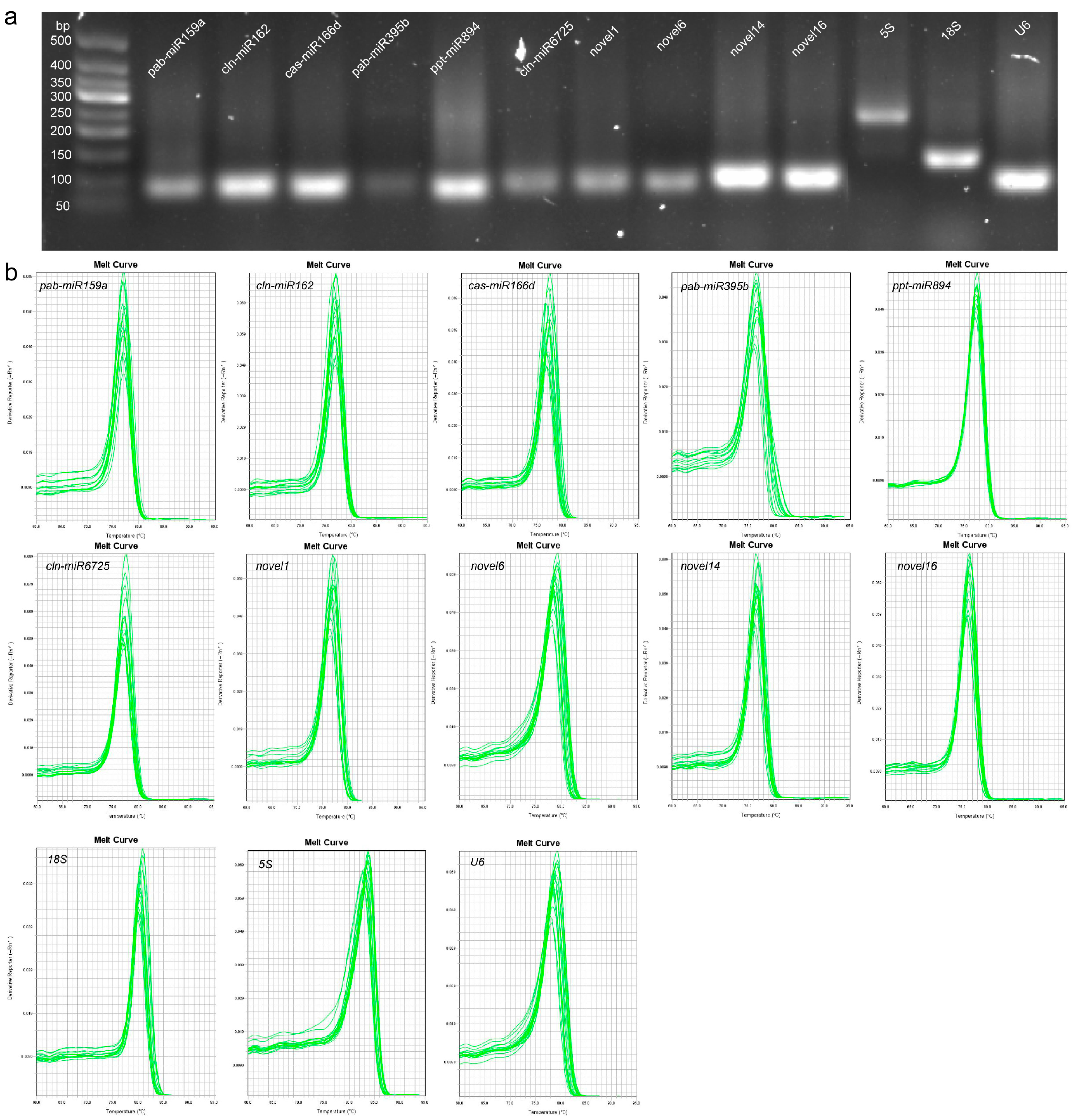



| Gene Symbol | Gene Sequence | Primer Sequence (5′-3′) | Amplicon Length (bp) | E (%) | R2 |
|---|---|---|---|---|---|
| pab-miR159a | UUGGUUUGAAGGGAGCUCUAC | CGTTGGTTTGAAGGGAGCTCTAC | 80–150 | 103.87 | 0.991 |
| cln-miR162 | UUGAUAAACCUCUGCAUCCAG | CGTTGATAAACCTCTGCATCCAG | 113.42 | 0.978 | |
| cas-miR166d | UCGGACCAGGCUUCAUUCCCCU | TCGGACCAGGCTTCATTCC | 108.38 | 0.997 | |
| pab-miR395b | CAGAAGUGUUUGGGGGGACUC | CAGAAGTGTTTGGGGGGACTC | 112.25 | 0.985 | |
| ppt-miR894 | CGUUUCACGUCGGGUUCACC | GTTTCACGTCGGGTTCACC | 103.55 | 0.979 | |
| cln-miR6725 | UGGCAUCUGUCGAGGUCAUCUA | TGGCATCTGTCGAGGTCATCTA | 110.67 | 0.982 | |
| novel1 | UCUUUCCGGAUCCUCCCAUGCC | CTTTCCGGATCCTCCCATGC | 104.29 | 0.999 | |
| novel6 | UUUUACCGAUCCCUCCAAAGCC | TTTACCGATCCCTCCAAAGCC | 92.93 | 0.993 | |
| novel14 | UUUGAGUGAAUCCAGAGUCUCU | CGCTTTGAGTGAATCCAGAGTCTC | 90.04 | 0.995 | |
| novel16 | UUUUUCCAAUACCUCCUAUACC | GCGTTTTTCCAATACCTCCTATACC | 92.93 | 0.993 | |
| U6 | – | Forward: ACAGAGAAGATTAGCATGGCC | 59 | 114.97 | 0.995 |
| Reverse: GACCAATTCTCGATTTGTGCG | |||||
| 5S | – | Forward: CACCAATCCATCCCGAACTT | 193 | 97.98 | 0.998 |
| Reverse: CCGTCTCCACCAGATAACAAATA | |||||
| 18S | – | Forward: TCTGGTCCTGTTCCGTTGG | 124 | 100.36 | 0.995 |
| Reverse: GCTTTCGCAGTGGTTCGTC | |||||
| Target genes | |||||
| aly-miR164c-5p | UGGAGAAGCAGGGCACGUGCG | TGGAGAAGCAGGGCACG | 80–150 | 110.09 | 0.994 |
| aly-miR168a-5p | UCGCUUGGUGCAGGUCGGGAA | TCGCTTGGTGCAGGTCG | 115.20 | 0.987 | |
| smo-miR396 | UUCCACGGCUUUCUUGAACC | GTTCCACGGCTTTCTTGAACC | 104.24 | 0.997 | |
| Treatment | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cold | Gene | novel16 | novel1 | miR6725 | miR159a | novel14 | U6 | miR395b | 5S | miR894 | novel6 | 18S | miR162 | miR166d |
| Stab. | 0.187 | 0.210 | 0.213 | 0.318 | 0.360 | 0.386 | 0.404 | 0.448 | 0.491 | 0.605 | 0.655 | 0.805 | 0.859 | |
| Heat | Gene | novel16 | miR159a | U6 | novel1 | miR166d | novel6 | 5S | miR395b | miR6725 | miR162 | miR894 | novel14 | 18S |
| Stab. | 0.349 | 0.364 | 0.388 | 0.428 | 0.452 | 0.509 | 0.540 | 0.606 | 0.620 | 0.699 | 0.881 | 0.881 | 0.941 | |
| Drought | Gene | novel16 | U6 | novel1 | miR159a | novel6 | miR6725 | novel14 | 18S | miR395b | miR162 | miR894 | 5S | miR166d |
| Stab. | 0.155 | 0.254 | 0.255 | 0.338 | 0.425 | 0.430 | 0.432 | 0.503 | 0.646 | 0.806 | 0.912 | 1.041 | 1.181 | |
| Salt | Gene | novel16 | miR6725 | miR395b | novel1 | miR159a | U6 | 5S | novel6 | 18S | novel14 | miR894 | miR166d | miR162 |
| Stab. | 0.219 | 0.267 | 0.347 | 0.359 | 0.386 | 0.434 | 0.439 | 0.476 | 0.534 | 0.587 | 0.832 | 0.913 | 1.027 | |
| ABA | Gene | U6 | novel16 | novel1 | miR159a | miR6725 | miR894 | novel6 | miR395b | 5S | novel14 | 18S | miR166d | miR162 |
| Stab. | 0.193 | 0.211 | 0.220 | 0.239 | 0.243 | 0.283 | 0.306 | 0.328 | 0.365 | 0.370 | 0.587 | 0.684 | 0.917 | |
| GA3 | Gene | miR6725 | novel1 | 18S | miR159a | novel16 | 5S | novel6 | U6 | novel14 | miR395b | miR894 | miR162 | miR166d |
| Stab. | 0.176 | 0.271 | 0.310 | 0.312 | 0.366 | 0.452 | 0.461 | 0.560 | 0.570 | 0.684 | 0.782 | 0.790 | 1.314 | |
| Treatment | Gene | novel16 | miR159a | miR6725 | novel1 | U6 | novel6 | miR395b | 18S | novel14 | miR162 | 5S | miR894 | miR166d |
| Stab. | 0.280 | 0.344 | 0.356 | 0.372 | 0.373 | 0.410 | 0.563 | 0.586 | 0.588 | 0.733 | 0.737 | 0.816 | 0.864 | |
| Tissue | Gene | novel1 | novel6 | novel16 | miR6725 | novel14 | 18S | miR395b | U6 | miR162 | miR166d | 5S | miR894 | miR159a |
| Stab. | 0.279 | 0.336 | 0.404 | 0.511 | 0.523 | 0.607 | 0.677 | 0.756 | 0.889 | 1.060 | 1.123 | 1.178 | 1.344 | |
| Total | Gene | novel16 | miR6725 | novel1 | novel6 | U6 | miR159a | miR395b | 18S | novel14 | miR162 | 5S | miR894 | miR166d |
| Stab. | 0.304 | 0.373 | 0.381 | 0.390 | 0.452 | 0.556 | 0.601 | 0.612 | 0.634 | 0.789 | 0.799 | 0.876 | 0.877 | |
| Software | Rank | Cold | Heat | Drought | Salt | ABA | GA3 | Treatment | Tissue | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| delta Ct | 1 | cln-miR6725 | novel1 | novel16 | novel16 | U6 | cln-miR6725 | novel16 | novel1 | novel16 |
| 2 | novel16 | novel16 | U6 | cln-miR6725 | novel16 | novel1 | pab-miR159a | novel6 | novel6 | |
| 3 | novel1 | pab-miR159a | novel1 | pab-miR395b | novel1 | 18S | cln-miR6725 | novel16 | novel1 | |
| 4 | pab-miR159a | cas-miR166d | pab-miR159a | novel1 | cln-miR6725 | pab-miR159a | novel1 | cln-miR6725 | cln-miR6725 | |
| 5 | pab-miR395b | novel6 | novel6 | 5S | pab-miR159a | novel16 | U6 | novel14 | U6 | |
| geNorm | 1 | novel16 | novel1 | novel6 | 5S | novel16 | novel1 | novel16 | novel16 | novel16 |
| 2 | cln-miR6725 | pab-miR395b | pab-miR159a | U6 | novel1 | cln-miR6725 | pab-miR159a | cln-miR6725 | cln-miR6725 | |
| 3 | novel1 | 5S | U6 | 18S | cln-miR6725 | 18S | cln-miR6725 | novel1 | novel1 | |
| 4 | pab-miR159a | novel6 | novel16 | pab-miR395b | U6 | pab-miR159a | novel1 | novel6 | novel6 | |
| 5 | novel14 | cas-miR166d | novel1 | novel6 | pab-miR159a | novel6 | U6 | 18S | U6 | |
| NormFinder | 1 | novel16 | novel16 | novel16 | novel16 | U6 | cln-miR6725 | novel16 | novel1 | novel16 |
| 2 | novel1 | pab-miR159a | U6 | cln-miR6725 | novel16 | novel1 | pab-miR159a | novel6 | cln-miR6725 | |
| 3 | cln-miR6725 | U6 | novel1 | pab-miR395b | novel1 | 18S | cln-miR6725 | novel16 | novel1 | |
| 4 | pab-miR159a | novel1 | pab-miR159a | novel1 | pab-miR159a | pab-miR159a | novel1 | cln-miR6725 | novel6 | |
| 5 | novel14 | cas-miR166d | novel6 | pab-miR159a | cln-miR6725 | novel16 | U6 | novel14 | U6 | |
| BestKeeper | 1 | novel14 | novel1 | U6 | novel16 | U6 | U6 | U6 | novel1 | U6 |
| 2 | cln-miR6725 | U6 | novel16 | cln-miR6725 | ppt-miR894 | cln-miR6725 | pab-miR159a | U6 | novel1 | |
| 3 | novel16 | novel16 | cln-miR6725 | novel1 | novel1 | 5S | novel16 | 18S | novel16 | |
| 4 | pab-miR159a | cas-miR166d | novel14 | pab-miR395b | 5S | novel14 | novel1 | cln-miR6725 | cln-miR6725 | |
| 5 | pab-miR395b | pab-miR395b | pab-miR159a | pab-miR159a | cln-miR6725 | novel16 | cln-miR6725 | novel16 | pab-miR159a | |
| RefFinder | 1 | cln-miR6725 | novel1 | novel16 | novel16 | U6 | cln-miR6725 | novel16 | novel1 | novel16 |
| 2 | novel16 | novel16 | U6 | cln-miR6725 | novel16 | novel1 | pab-miR159a | novel16 | cln-miR6725 | |
| 3 | novel1 | U6 | pab-miR159a | pab-miR395b | novel1 | 18S | U6 | cln-miR6725 | novel1 | |
| 4 | novel14 | pab-miR159a | novel1 | 5S | cln-miR6725 | U6 | cln-miR6725 | novel6 | U6 | |
| 5 | pab-miR159a | pab-miR395b | novel6 | U6 | ppt-miR894 | 5S | novel1 | 18S | novel6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Xue, J.; Zhu, L.; Hu, H.; Yang, J.; Cui, J.; Xu, J. Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses. Int. J. Mol. Sci. 2021, 22, 7246. https://doi.org/10.3390/ijms22147246
Zhang Y, Xue J, Zhu L, Hu H, Yang J, Cui J, Xu J. Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses. International Journal of Molecular Sciences. 2021; 22(14):7246. https://doi.org/10.3390/ijms22147246
Chicago/Turabian StyleZhang, Yingting, Jinyu Xue, Lijuan Zhu, Hailiang Hu, Junjie Yang, Jiebing Cui, and Jin Xu. 2021. "Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses" International Journal of Molecular Sciences 22, no. 14: 7246. https://doi.org/10.3390/ijms22147246
APA StyleZhang, Y., Xue, J., Zhu, L., Hu, H., Yang, J., Cui, J., & Xu, J. (2021). Selection and Optimization of Reference Genes for MicroRNA Expression Normalization by qRT-PCR in Chinese Cedar (Cryptomeria fortunei) under Multiple Stresses. International Journal of Molecular Sciences, 22(14), 7246. https://doi.org/10.3390/ijms22147246






