Overlapping Functions of the Paralogous Proteins AtPAP2 and AtPAP9 in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. The C-Terminal Hydrophobic Motif of AtPAP9 Targets GFP to Chloroplasts
2.2. Overexpression Lines of AtPAP9 Did Not Exhibit Growth-Promoting Phenotypes
2.3. AtPAP9 Modulates the Import Rate of [35S]-Labeled AtSSU1B into Chloroplasts
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Isolation of Chloroplasts from Arabidopsis Leaves
4.3. Generation of Overexpression Lines of AtPAP9
4.4. Western Blotting Analysis
4.5. Subcellular Localization Analysis by GFP
4.6. Yeast Two-Hybrid Assay and Bimolecular Fluorescence Complementation Analysis
4.7. Chloroplast Import Assays
4.8. Chlorophyll Fluorescence Measurements
4.9. 3D Computer Modeling
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, D.P.; Zhu, H.F.; Liu, K.F.; Liu, X.; Leggewie, G.; Udvardi, M.; Wang, D.W. Purple acid phosphatases of Arabidopsis thaliana-Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 2002, 277, 27772–27781. [Google Scholar] [CrossRef]
- Schenk, G.; Guddat, L.T.; Ge, Y.; Carrington, L.E.; Hume, D.A.; Hamilton, S.; de Jersey, J. Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 2000, 250, 117–125. [Google Scholar] [CrossRef][Green Version]
- Lung, S.C.; Leung, A.; Kuang, R.; Wang, Y.; Leung, P.; Lim, B.L. Phytase activity in tobacco (Nicotiana tabacum) root exudates is exhibited by a purple acid phosphatase. Phytochemistry 2008, 69, 365–373. [Google Scholar] [CrossRef]
- Kuang, R.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Kaida, R.; Satoh, Y.; Bulone, V.; Yamada, Y.; Kaku, T.; Hayashi, T.; Kaneko, T.S. Activation of beta-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol. 2009, 150, 1822–1830. [Google Scholar] [CrossRef]
- Liang, C.Y.; Tian, J.; Lam, H.M.; Lim, B.L.; Yan, X.L.; Liao, H. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol. 2010, 152, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Wong, F.-L.; Phang, T.-H.; Cheung, M.-Y.; Francisca Li, W.-Y.; Shao, G.; Yan, X.; Lam, H.-M. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 2003, 318, 103–111. [Google Scholar] [CrossRef]
- Sun, F.; Suen, P.K.; Zhang, Y.; Liang, C.; Carrie, C.; Whelan, J.; Ward, J.L.; Hawkins, N.D.; Jiang, L.; Lim, B.L. A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytol. 2012, 194, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Carrie, C.; Law, S.; Murcha, M.W.; Zhang, R.; Law, Y.S.; Suen, P.K.; Whelan, J.; Lim, B.L. AtPAP2 is a tail-anchored protein in the outer membrane of chloroplasts and mitochondria. Plant Signal. Behav. 2012, 7, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Law, Y.S.; Zhang, R.; Guan, X.; Cheng, S.; Sun, F.; Duncan, O.; Murcha, M.; Whelan, J.; Lim, B.L. Phosphorylation and dephosphorylation of the presequence of pMORF3 during import into mitochondria from Arabidopsis thaliana. Plant Physiol. 2015, 169, 1–12. [Google Scholar] [CrossRef]
- Zhang, R.; Guan, X.; Law, Y.S.; Sun, F.; Chen, S.; Wong, K.B.; Lim, B.L. AtPAP2 modulates the import of the small subunit of Rubisco into chloroplasts. Plant Signal. Behav. 2016, 11, e1239687. [Google Scholar] [CrossRef] [PubMed]
- Ghifari, A.S.; Gill-Hille, M.; Murcha, M.W. Plant mitochondrial protein import: The ins and outs. Biochem. J. 2018, 475, 2191–2208. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.X.; Theg, S.M. The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 2013, 1833, 314–331. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, G.; Gugel, I.L.; Meurer, J.; Soll, J.; Schwenkert, S. The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol. 2011, 157, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Law, Y.S.; Ngan, L.; Yan, J.R.; Kwok, L.Y.; Sun, Y.Z.; Cheng, S.F.; Schwenkert, S.; Lim, B.L. Multiple kinases can phosphorylate the N-terminal sequences of mitochondrial proteins in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 982. [Google Scholar] [CrossRef] [PubMed]
- Waegemann, K.; Soll, J. Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem. 1996, 271, 6545–6554. [Google Scholar] [CrossRef]
- Nickel, C.; Soll, J.; Schwenkert, S. Phosphomimicking within the transit peptide of pHCF136 leads to reduced photosystem II accumulation in vivo. FEBS Lett. 2015, 589, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, Y.; Cheng, S.; Osorio, S.; Sun, Y.; Fernie, A.R.; Cheung, C.Y.M.; Lim, B.L. Impacts of high ATP supply from chloroplasts and mitochondria on the leaf metabolism of Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 922. [Google Scholar] [CrossRef]
- Voon, C.P.; Law, Y.S.; Guan, X.; Lim, S.L.; Xu, Z.; Chu, W.T.; Zhang, R.; Sun, F.; Labs, M.; Leister, D.; et al. Modulating the activities of chloroplasts and mitochondria promotes ATP production and plant growth. Quant. Plant Biol. 2021, 2, e7. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, R.; Yang, M.; Law, Y.-S.; Sun, F.; Hon, N.L.; Ngai, S.M.; Lim, B.L. A balance between the activities of chloroplasts and mitochondria is crucial for optimal plant growth. Antioxidants 2021, 10, 935. [Google Scholar] [CrossRef]
- Zamani, K.; Lohrasebi, T.; Sabet, M.S.; Malboobi, M.A.; Mousavi, A. Expression pattern and subcellular localization of Arabidopsis purple acid phosphatase AtPAP9. Gene Expr. Patterns 2014, 14, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Kleffmann, T.; Russenberger, D.; von Zychlinski, A.; Christopher, W.; Sjolander, K.; Gruissem, W.; Baginsky, S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 2004, 14, 354–362. [Google Scholar] [CrossRef]
- Dhanoa, P.K.; Richardson, L.G.; Smith, M.D.; Gidda, S.K.; Henderson, M.P.; Andrews, D.W.; Mullen, R.T. Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 2010, 5, e10098. [Google Scholar] [CrossRef]
- Liang, C.; Liu, X.; Yiu, S.M.; Lim, B.L. De novo assembly and characterization of Camelina sativa transcriptome by paired-end sequencing. BMC Genom. 2013, 14, 146. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; van den Bergh, E.; Zeng, P.; Zhong, X.; Xu, J.; Liu, X.; Hofberger, J.; de Bruijn, S.; Bhide, A.S.; Kuelahoglu, C.; et al. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 2013, 25, 2813–2830. [Google Scholar] [CrossRef] [PubMed]
- Antonyuk, S.V.; Olczak, M.; Olczak, T.; Ciuraszkiewicz, J.; Strange, R.W. The structure of a purple acid phosphatase involved in plant growth and pathogen defence exhibits a novel immunoglobulin-like fold. IUCrJ 2014, 1, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, L.; Yung, K.F.; Leung, D.Y.; Sun, F.; Lim, B.L. Over-expression of AtPAP2 in Camelina sativa leads to faster plant growth and higher seed yield. Biotechnol. Biofuels 2012, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, F.; Fettke, J.; Schottler, M.A.; Ramsden, L.; Fernie, A.R.; Lim, B.L. Heterologous expression of AtPAP2 in transgenic potato influences carbon metabolism and tuber development. FEBS Lett. 2014, 588, 3726–3731. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Schnell, D.J.; Fitzpatrick, L.; Keegstra, K. In vitro analysis of chloroplast protein import. Curr. Protoc. Cell Biol. 2003, 11.1–11.16. [Google Scholar] [CrossRef]
- Lister, R.; Carrie, C.; Duncan, O.; Ho, L.H.; Howell, K.A.; Murcha, M.W.; Whelan, J. Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 2007, 19, 3739–3759. [Google Scholar] [CrossRef]
- Chen, S.; Songkumarn, P.; Liu, J.; Wang, G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009, 150, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Carrie, C.; Kuhn, K.; Murcha, M.W.; Duncan, O.; Small, I.D.; O’Toole, N.; Whelan, J. Approaches to defining dual-targeted proteins in Arabidopsis. Plant J. 2009, 57, 1128–1139. [Google Scholar] [CrossRef]
- Schweiger, R.; Schwenkert, S. Protein-protein interactions visualized by bimolecular fluorescence complementation in tobacco protoplasts and leaves. J. Vis. Exp. 2014, 85, e51327. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, L.M.; Keegstra, K. A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J. 2001, 27, 59–65. [Google Scholar] [CrossRef]

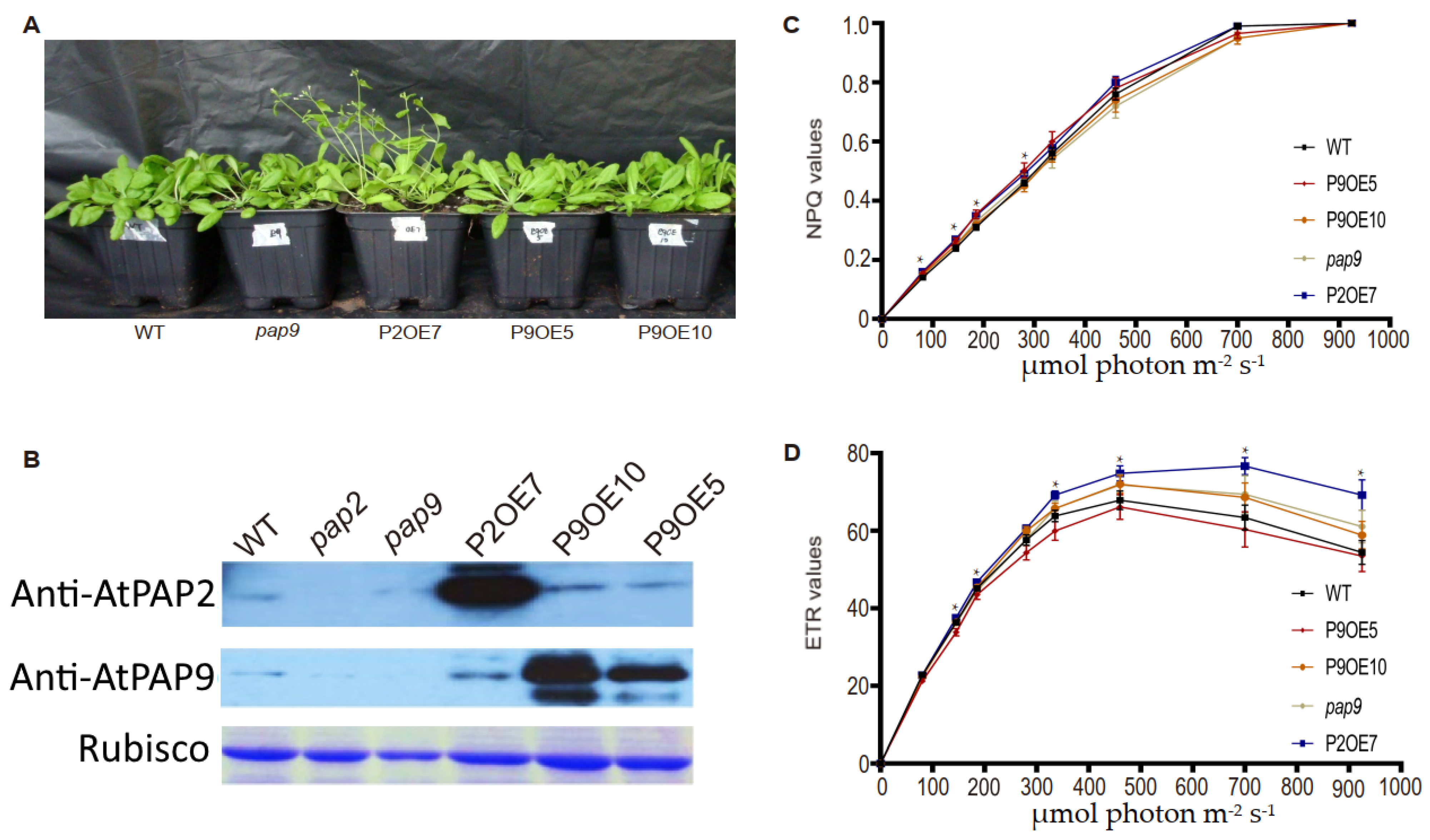
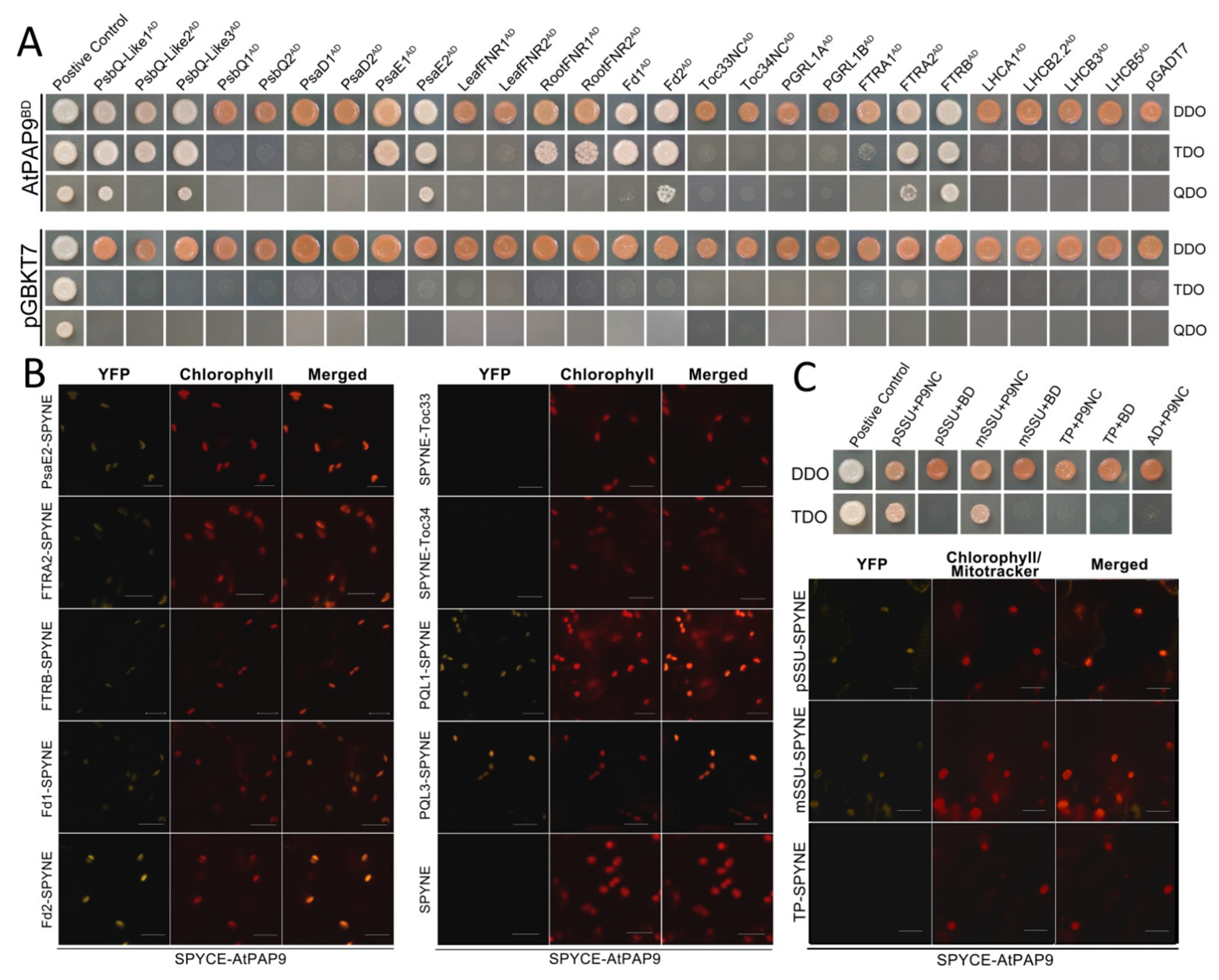

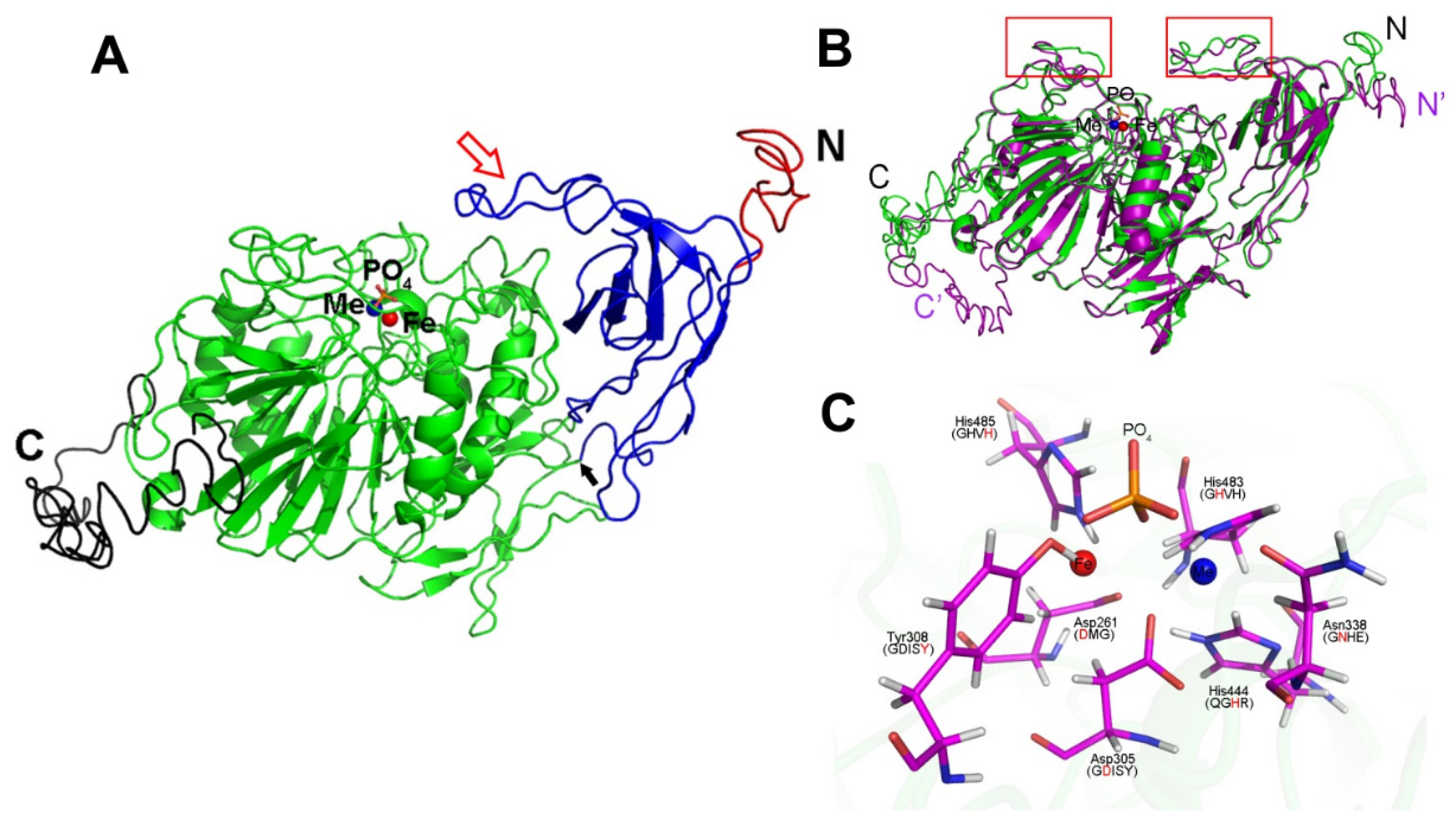
| Lines | Siliques per Plant | Seed Yield (g per Plant) |
|---|---|---|
| WT | 367 ± 50 a | 0.203 ± 0.047 a |
| PAP9 | 353 ± 34 a | 0.198 ± 0.062 a |
| P2OE7 | 577 ± 81 b | 0.313 ± 0.093 b |
| P9OE5 | 350 ± 36 a | 0.209 ± 0.077 a |
| P9OE10 | 358 ± 47 a | 0.230 ± 0.061 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Guan, X.; Yang, M.; Law, Y.-S.; Voon, C.P.; Yan, J.; Sun, F.; Lim, B.L. Overlapping Functions of the Paralogous Proteins AtPAP2 and AtPAP9 in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 7243. https://doi.org/10.3390/ijms22147243
Zhang R, Guan X, Yang M, Law Y-S, Voon CP, Yan J, Sun F, Lim BL. Overlapping Functions of the Paralogous Proteins AtPAP2 and AtPAP9 in Arabidopsis thaliana. International Journal of Molecular Sciences. 2021; 22(14):7243. https://doi.org/10.3390/ijms22147243
Chicago/Turabian StyleZhang, Renshan, Xiaoqian Guan, Meijing Yang, Yee-Song Law, Chia Pao Voon, Junran Yan, Feng Sun, and Boon Leong Lim. 2021. "Overlapping Functions of the Paralogous Proteins AtPAP2 and AtPAP9 in Arabidopsis thaliana" International Journal of Molecular Sciences 22, no. 14: 7243. https://doi.org/10.3390/ijms22147243
APA StyleZhang, R., Guan, X., Yang, M., Law, Y.-S., Voon, C. P., Yan, J., Sun, F., & Lim, B. L. (2021). Overlapping Functions of the Paralogous Proteins AtPAP2 and AtPAP9 in Arabidopsis thaliana. International Journal of Molecular Sciences, 22(14), 7243. https://doi.org/10.3390/ijms22147243







