Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients
Abstract
:1. Introduction
2. Results
2.1. Database Mining for ALS Transcriptomic Studies
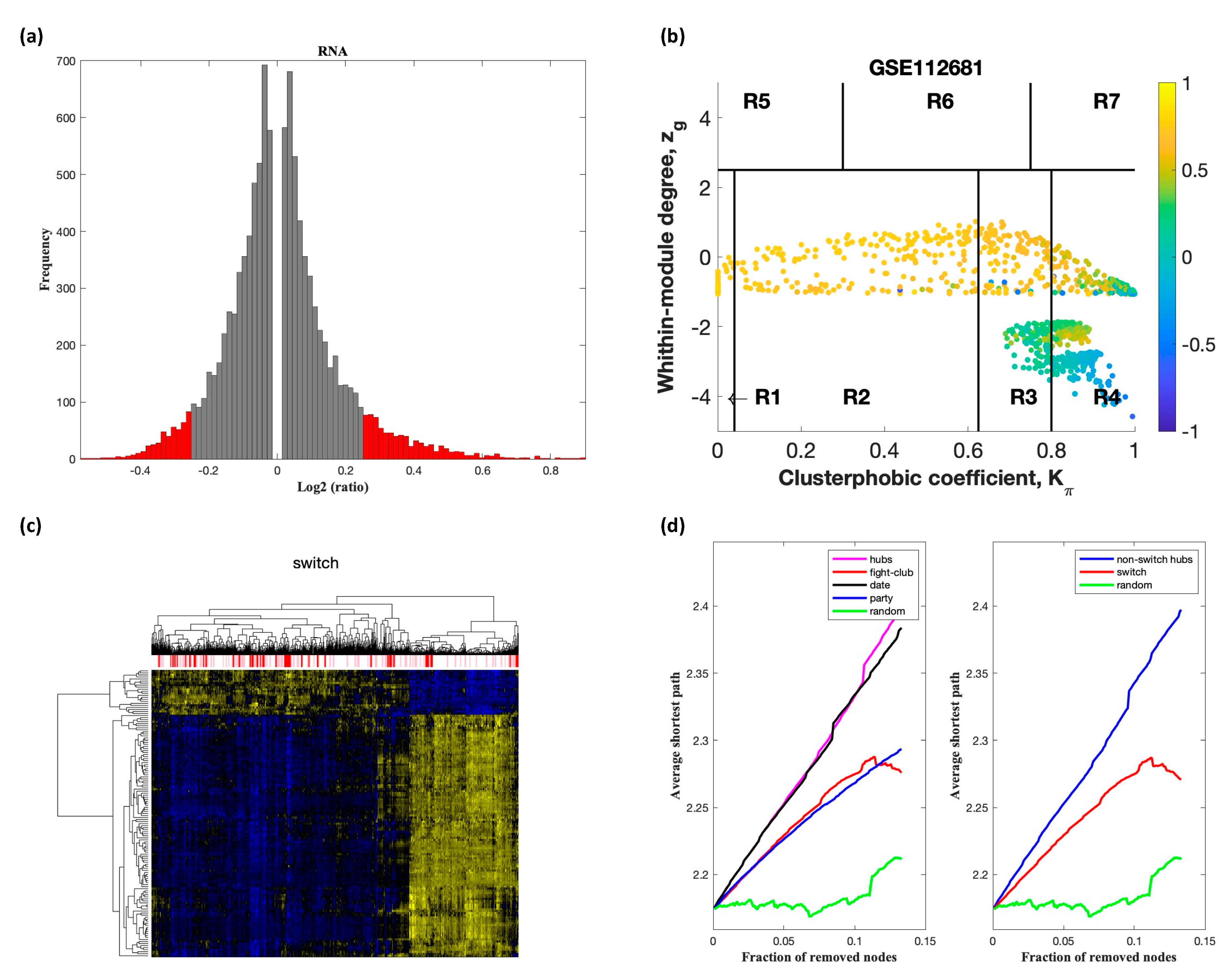
2.2. Identification of Switch Genes in ALS
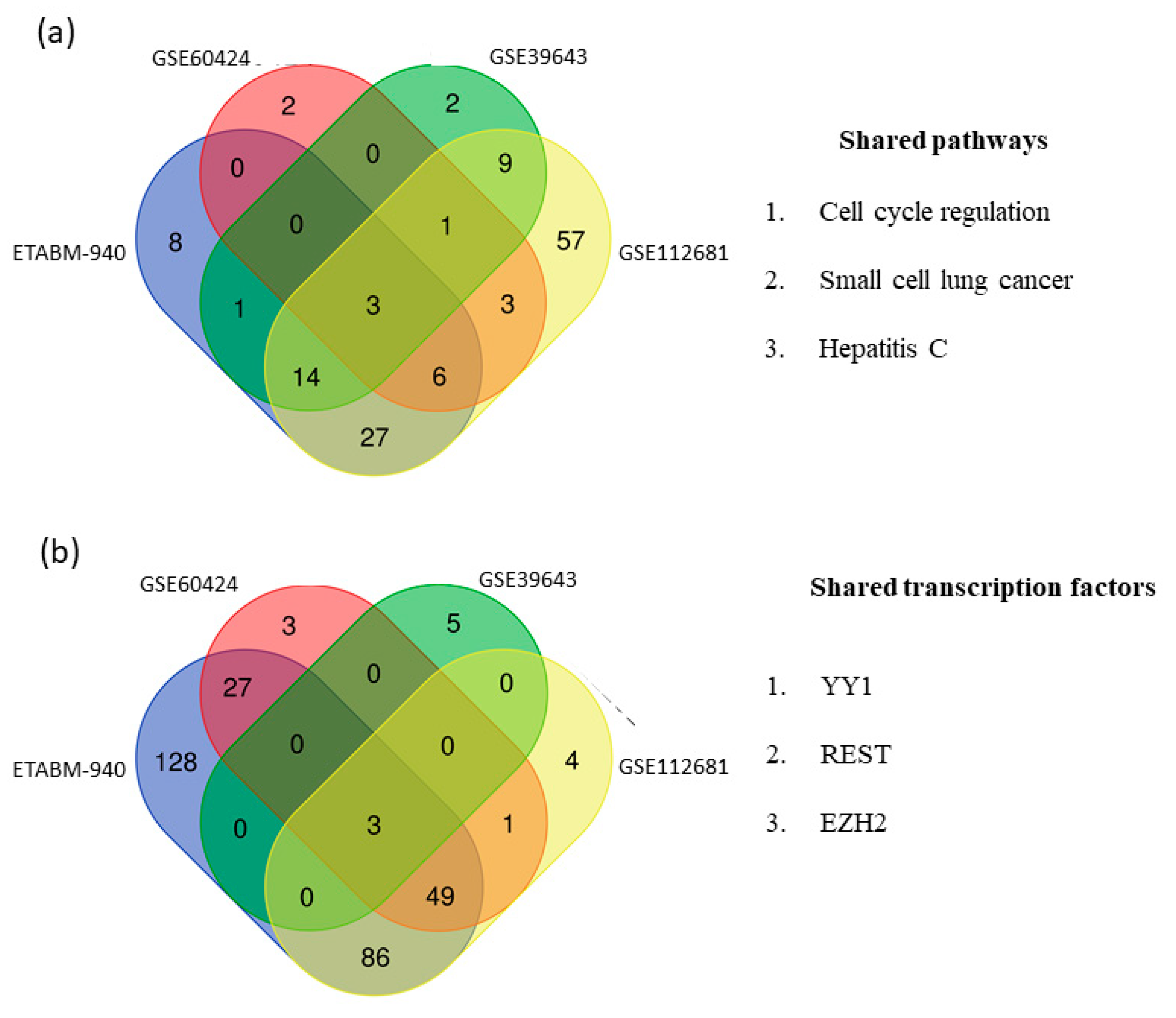
2.3. Network and Pathway Analysis of Switch Genes in ALS
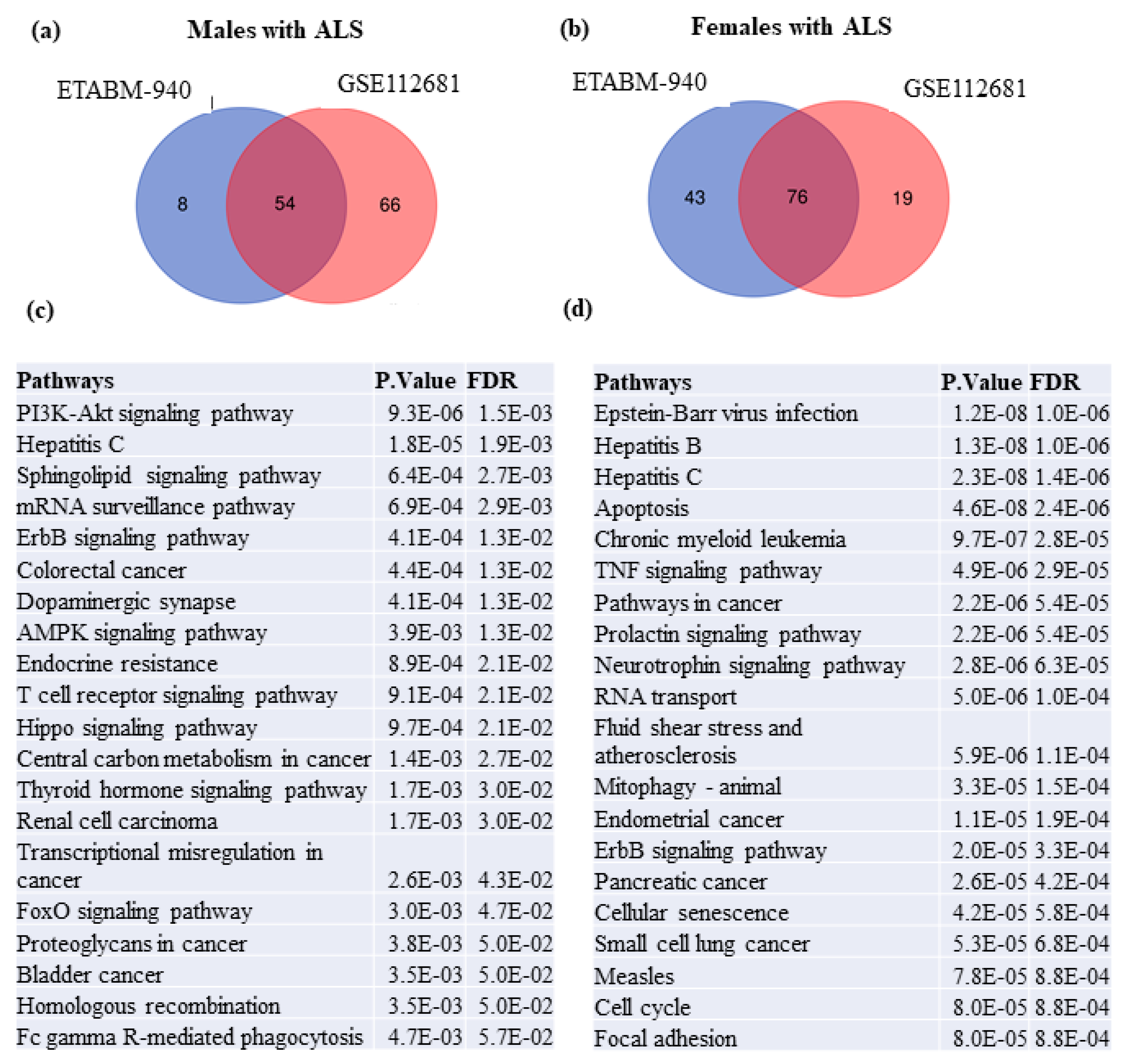
2.4. Sex-Specific Gene Expression in the Blood of ALS Patients
2.5. Identification of Sex-Associated Switch Genes, Biological Pathways, and Transcription Factors
2.6. Protein–Chemical Interaction Analysis
3. Discussion
3.1. Identification of Switch Genes in ALS
3.2. Sex-Specific Gene Expression Patterns in ALS Switch Genes
3.3. Biological and Functional Analyses of Switch Genes
3.4. Sex-Associated Switch Genes, Biological Pathways, and Transcription Factors
3.5. Protein–Chemical Network Analysis Identifies Potential Therapeutics for ALS
3.6. Conclusions
4. Methods
4.1. Database Mining
4.2. Clinical and Demographic Characteristics of Participants Analyzed in the Study
4.3. Identification of Switch Genes by SWIM Analysis
4.4. Identification and Analysis of Switch Genes by Sex
4.5. Pathway and Transcription Factor Analyses
4.6. Genes–Chemical Interaction Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grad, L.I.; Rouleau, G.A.; Ravits, J.; Cashman, N.R. Clinical Spectrum of Amyotrophic Lateral Sclerosis (ALS). Cold Spring Harb. Perspect. Med. 2017, 7, a024117. [Google Scholar] [CrossRef] [Green Version]
- Renton, A.E.; Chio, A.; Traynor, B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014, 17, 17–23. [Google Scholar] [CrossRef]
- Ingre, C.; Roos, P.M.; Piehl, F.; Kamel, F.; Fang, F. Risk factors for amyotrophic lateral sclerosis. Clin. Epidemiol. 2015, 7, 181–193. [Google Scholar]
- Li, R.; Strykowski, R.; Meyer, M.; Mulcrone, P.; Krakora, D.; Suzuki, M. Male-specific differences in proliferation, neurogenesis, and sensitivity to oxidative stress in neural progenitor cells derived from a rat model of ALS. PLoS ONE 2012, 7, e48581. [Google Scholar] [CrossRef] [PubMed]
- Martineau, E.; Di Polo, A.; Vande Velde, C.; Robitaille, R. Sex-Specific Differences in Motor-Unit Remodeling in a Mouse Model of ALS. eNeuro 2020, 7, ENEURO.0388-19.2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCombe, P.A.; Henderson, R.D. Effects of gender in amyotrophic lateral sclerosis. Gend Med. 2010, 7, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Couratier, P.; Corcia, P.; Lautrette, G.; Nicol, M.; Preux, P.M.; Marin, B. Epidemiology of amyotrophic lateral sclerosis: A review of literature. Rev. Neurol. 2016, 172, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Benatar, M.; Wuu, J.; Andersen, P.M.; Lombardi, V.; Malaspina, A. Neurofilament light: A candidate biomarker of presymptomatic amyotrophic lateral sclerosis and phenoconversion. Ann. Neurol. 2018, 84, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Preische, O.; Schultz, S.A.; Apel, A.; Kuhle, J.; Kaeser, S.A.; Barro, C.; Graber, S.; Kuder-Buletta, E.; LaFougere, C.; Laske, C.; et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat. Med. 2019, 25, 277–283. [Google Scholar] [CrossRef]
- Boylan, K.B.; Glass, J.D.; Crook, J.E.; Yang, C.; Thomas, C.S.; Desaro, P.; Johnston, A.; Overstreet, K.; Kelly, C.; Polak, M.; et al. Phosphorylated neurofilament heavy subunit (pNF-H) in peripheral blood and CSF as a potential prognostic biomarker in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2013, 84, 467–472. [Google Scholar] [CrossRef]
- Ehrhart, J.; Smith, A.J.; Kuzmin-Nichols, N.; Zesiewicz, T.A.; Jahan, I.; Shytle, R.D.; Kim, S.H.; Sanberg, C.D.; Vu, T.H.; Gooch, C.L.; et al. Humoral factors in ALS patients during disease progression. J. Neuroinflammation 2015, 12, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago, J.A.; Potashkin, J.A. A network approach to clinical intervention in neurodegenerative diseases. Trends Mol. Med. 2014, 20, 694–703. [Google Scholar] [CrossRef]
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Dissecting the Molecular Mechanisms of Neurodegenerative Diseases through Network Biology. Front. Aging Neurosci. 2017, 9, 166. [Google Scholar] [CrossRef] [Green Version]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Computational identification of key genes that may regulate gene expression reprogramming in Alzheimer’s patients. PLoS ONE 2019, 14, e0222921. [Google Scholar] [CrossRef]
- Potashkin, J.A.; Bottero, V.; Santiago, J.A.; Quinn, J.P. Bioinformatic Analysis Reveals Phosphodiesterase 4D-Interacting Protein as a Key Frontal Cortex Dementia Switch Gene. Int. J. Mol. Sci. 2020, 21, 3787. [Google Scholar] [CrossRef]
- Fiscon, G.; Conte, F.; Licursi, V.; Nasi, S.; Paci, P. Computational identification of specific genes for glioblastoma stem-like cells identity. Sci. Rep. 2018, 8, 7769. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- Plath, K.; Mlynarczyk-Evans, S.; Nusinow, D.A.; Panning, B. Xist RNA and the mechanism of X chromosome inactivation. Annu. Rev. Genet. 2002, 36, 233–278. [Google Scholar] [CrossRef]
- Dunford, A.; Weinstock, D.M.; Savova, V.; Schumacher, S.E.; Cleary, J.P.; Yoda, A.; Sullivan, T.J.; Hess, J.M.; Gimelbrant, A.A.; Beroukhim, R.; et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 2017, 49, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Berletch, J.B.; Deng, X.; Nguyen, D.K.; Disteche, C.M. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013, 9, e1003489. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y.; Golden, L.C.; Itoh, N.; Matsukawa, M.A.; Ren, E.; Tse, V.; Arnold, A.P.; Voskuhl, R.R. The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. J. Clin. Investig. 2019, 129, 3852–3863. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.J. Destabilizing influences in apoptosis: Sowing the seeds of IAP destruction. Cell 2002, 109, 793–796. [Google Scholar] [CrossRef] [Green Version]
- Perrelet, D.; Perrin, F.E.; Liston, P.; Korneluk, R.G.; MacKenzie, A.; Ferrer-Alcon, M.; Kato, A.C. Motoneuron resistance to apoptotic cell death in vivo correlates with the ratio between X-linked inhibitor of apoptosis proteins (XIAPs) and its inhibitor, XIAP-associated factor 1. J. Neurosci. 2004, 24, 3777–3785. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Cao, Y.; Stucchi, R.; Hooikaas, P.J.; Portegies, S.; Will, L.; Martin, M.; Akhmanova, A.; Harterink, M.; Hoogenraad, C.C. MAP7D2 Localizes to the Proximal Axon and Locally Promotes Kinesin-1-Mediated Cargo Transport into the Axon. Cell Rep. 2019, 26, 1988–1999.e6. [Google Scholar] [CrossRef] [Green Version]
- Young-Baird, S.K.; Lourenco, M.B.; Elder, M.K.; Klann, E.; Liebau, S.; Dever, T.E. Suppression of MEHMO Syndrome Mutation in eIF2 by Small Molecule ISRIB. Mol. Cell 2020, 77, 875–886.e7. [Google Scholar] [CrossRef] [PubMed]
- Chanda, K.; Mukhopadhyay, D. LncRNA Xist, X-chromosome Instability and Alzheimer’s Disease. Curr. Alzheimer Res. 2020, 17, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, C.; Geng, C.; Zhao, K. LncRNA XIST knockdown attenuates Abeta25-35-induced toxicity, oxidative stress, and apoptosis in primary cultured rat hippocampal neurons by targeting miR-132. Int. J. Clin. Exp. Pathol. 2018, 11, 3915–3924. [Google Scholar] [PubMed]
- Du, Y.; Gao, G.; Li, J.; Suo, S.; Liu, S.; Sun, Y.; Zhang, M.; Yin, P.; Lu, C.; Zhang, Z.; et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 2020, 44, 630–636. [Google Scholar]
- Greenfield, A.; Carrel, L.; Pennisi, D.; Philippe, C.; Quaderi, N.; Siggers, P.; Steiner, K.; Tam, P.P.; Monaco, A.P.; Willard, H.F.; et al. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 1998, 7, 737–742. [Google Scholar] [CrossRef] [Green Version]
- Davis, E.J.; Broestl, L.; Abdulai-Saiku, S.; Worden, K.; Bonham, L.W.; Minones-Moyano, E.; Moreno, A.J.; Wang, D.; Chang, K.; Williams, G.; et al. A second X chromosome contributes to resilience in a mouse model of Alzheimer’s disease. Sci. Transl. Med. 2020, 12, eaaz5677. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Q.; Shi, Q.; Liu, Y.; Zhao, K.; Shen, Q.; Shi, Y.; Liu, X.; Wang, C.; Li, N.; et al. Demethylase Kdm6a epigenetically promotes IL-6 and IFN-beta production in macrophages. J. Autoimmun. 2017, 80, 85–94. [Google Scholar] [CrossRef]
- Schneider-Gadicke, A.; Beer-Romero, P.; Brown, L.G.; Nussbaum, R.; Page, D.C. ZFX has a gene structure similar to ZFY, the putative human sex determinant, and escapes X inactivation. Cell 1989, 57, 1247–1258. [Google Scholar] [CrossRef]
- Xu, S.; Duan, P.; Li, J.; Senkowski, T.; Guo, F.; Chen, H.; Romero, A.; Cui, Y.; Liu, J.; Jiang, S.W. Zinc Finger and X-Linked Factor (ZFX) Binds to Human SET Transcript 2 Promoter and Transactivates SET Expression. Int. J. Mol. Sci. 2016, 17, 1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arif, M.; Wei, J.; Zhang, Q.; Liu, F.; Basurto-Islas, G.; Grundke-Iqbal, I.; Iqbal, K. Cytoplasmic retention of protein phosphatase 2A inhibitor 2 (I2PP2A) induces Alzheimer-like abnormal hyperphosphorylation of Tau. J. Biol. Chem. 2014, 289, 27677–27691. [Google Scholar] [CrossRef] [Green Version]
- Manjaly, Z.R.; Scott, K.M.; Abhinav, K.; Wijesekera, L.; Ganesalingam, J.; Goldstein, L.H.; Janssen, A.; Dougherty, A.; Willey, E.; Stanton, B.R.; et al. The sex ratio in amyotrophic lateral sclerosis: A population based study. Amyotroph. Lateral Scler. 2010, 11, 439–442. [Google Scholar] [CrossRef]
- Curtis, A.F.; Masellis, M.; Hsiung, G.R.; Moineddin, R.; Zhang, K.; Au, B.; Millett, G.; Mackenzie, I.; Rogaeva, E.; Tierney, M.C. Sex differences in the prevalence of genetic mutations in FTD and ALS: A meta-analysis. Neurology 2017, 89, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Gershoni, M.; Pietrokovski, S. The landscape of sex-differential transcriptome and its consequent selection in human adults. BMC Biol. 2017, 15, 7. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, H.; Han, S.M.; Yang, Y.; Tong, C.; Lin, Y.Q.; Mohan, K.; Haueter, C.; Zoghbi, A.; Harati, Y.; Kwan, J.; et al. The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell 2008, 133, 963–977. [Google Scholar] [CrossRef] [Green Version]
- Gupta, G.; Qin, H.; Song, J. Intrinsically unstructured domain 3 of hepatitis C Virus NS5A forms a “fuzzy complex” with VAPB-MSP domain which carries ALS-causing mutations. PLoS ONE 2012, 7, e39261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhvlediani, T.; Kvirkvelia, N.; Shakarishvili, R.; Tsertsvadze, T. Als-like syndrome in the patient with chronic hepatitis C. Georgian Med. News 2009, 172–173, 70–72. [Google Scholar]
- Tannahill, D.; Melton, D.A. Localized synthesis of the Vg1 protein during early Xenopus development. Development 1989, 106, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Liachko, N.F.; McMillan, P.J.; Guthrie, C.R.; Bird, T.D.; Leverenz, J.B.; Kraemer, B.C. CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann. Neurol. 2013, 74, 39–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas-Prats, E.; Martinez-Gonzalez, L.; Gonzalo-Consuegra, C.; Liachko, N.F.; Perez, C.; Ramirez, D.; Kraemer, B.C.; Martin-Requero, A.; Perez, D.I.; Gil, C.; et al. Targeting nuclear protein TDP-43 by cell division cycle kinase 7 inhibitors: A new therapeutic approach for amyotrophic lateral sclerosis. Eur. J. Med. Chem. 2021, 210, 112968. [Google Scholar] [CrossRef] [PubMed]
- Vaca, G.; Martinez-Gonzalez, L.; Fernandez, A.; Rojas-Prats, E.; Porras, G.; Cuevas, E.P.; Gil, C.; Martinez, A.; Martin-Requero, A. Therapeutic potential of novel Cell Division Cycle Kinase 7 inhibitors on TDP-43-related pathogenesis such as Frontotemporal Lobar Degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). J. Neurochem. 2021, 156, 379–390. [Google Scholar] [CrossRef]
- Schoenherr, C.J.; Anderson, D.J. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 1995, 267, 1360–1363. [Google Scholar] [CrossRef]
- Lu, T.; Aron, L.; Zullo, J.; Pan, Y.; Kim, H.; Chen, Y.; Yang, T.H.; Kim, H.M.; Drake, D.; Liu, X.S.; et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature 2014, 507, 448–454. [Google Scholar] [CrossRef] [Green Version]
- Formisano, L.; Guida, N.; Cocco, S.; Secondo, A.; Sirabella, R.; Ulianich, L.; Paturzo, F.; Di Renzo, G.; Canzoniero, L.M. The repressor element 1-silencing transcription factor is a novel molecular target for the neurotoxic effect of the polychlorinated biphenyl mixture aroclor 1254 in neuroblastoma SH-SY5Y cells. J. Pharmacol. Exp. Ther. 2011, 338, 997–1003. [Google Scholar] [CrossRef]
- Recabarren-Leiva, D.; Alarcon, M. New insights into the gene expression associated to amyotrophic lateral sclerosis. Life Sci. 2018, 193, 110–123. [Google Scholar] [CrossRef]
- Yin, X.; Ren, M.; Jiang, H.; Cui, S.; Wang, S.; Jiang, H.; Qi, Y.; Wang, J.; Wang, X.; Dong, G.; et al. Downregulated AEG-1 together with inhibited PI3K/Akt pathway is associated with reduced viability of motor neurons in an ALS model. Mol. Cell. Neurosci. 2015, 68, 303–313. [Google Scholar] [CrossRef]
- Ranno, E.; D’Antoni, S.; Spatuzza, M.; Berretta, A.; Laureanti, F.; Bonaccorso, C.M.; Pellitteri, R.; Longone, P.; Spalloni, A.; Iyer, A.M.; et al. Endothelin-1 is over-expressed in amyotrophic lateral sclerosis and induces motor neuron cell death. Neurobiol. Dis. 2014, 65, 160–171. [Google Scholar] [CrossRef]
- D’Antoni, S.; Ranno, E.; Spatuzza, M.; Cavallaro, S.; Catania, M.V. Endothelin-1 Induces Degeneration of Cultured Motor Neurons Through a Mechanism Mediated by Nitric Oxide and PI3K/Akt Pathway. Neurotox. Res. 2017, 32, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Ascherio, A.; Munger, K.L.; Lunemann, J.D. The initiation and prevention of multiple sclerosis. Nat. Rev. Neurol. 2012, 8, 602–612. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago, J.A.; Bottero, V.; Potashkin, J.A. Transcriptomic and Network Analysis Highlight the Association of Diabetes at Different Stages of Alzheimer’s Disease. Front. Neurosci. 2019, 13, 1273. [Google Scholar] [CrossRef]
- Cui, A.; Fan, H.; Zhang, Y.; Zhang, Y.; Niu, D.; Liu, S.; Liu, Q.; Ma, W.; Shen, Z.; Shen, L.; et al. Dexamethasone-induced Kruppel-like factor 9 expression promotes hepatic gluconeogenesis and hyperglycemia. J. Clin. Investig. 2019, 129, 2266–2278. [Google Scholar] [CrossRef] [PubMed]
- Gans, I.; Hartig, E.I.; Zhu, S.; Tilden, A.R.; Hutchins, L.N.; Maki, N.J.; Graber, J.H.; Coffman, J.A. Klf9 is a key feedforward regulator of the transcriptomic response to glucocorticoid receptor activity. Sci. Rep. 2020, 10, 11415. [Google Scholar] [CrossRef] [PubMed]
- Apara, A.; Galvao, J.; Wang, Y.; Blackmore, M.; Trillo, A.; Iwao, K.; Brown, D.P., Jr.; Fernandes, K.A.; Huang, A.; Nguyen, T.; et al. KLF9 and JNK3 Interact to Suppress Axon Regeneration in the Adult CNS. J. Neurosci. 2017, 37, 9632–9644. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zeng, W.; Sun, H.; Huang, F.; Yang, C.; Cai, X.; Lu, Y.; Zeng, J.; Yang, K. Bioinformatical Analysis of Gene Expression Omnibus Database Associates TAF7/CCNB1, TAF7/CCNA2, and GTF2E2/CDC20 Pathways with Glioblastoma Development and Prognosis. World Neurosurg. 2020, 138, e492–e514. [Google Scholar] [CrossRef] [PubMed]
- Dziuba, N.; Ferguson, M.R.; O’Brien, W.A.; Sanchez, A.; Prussia, A.J.; McDonald, N.J.; Friedrich, B.M.; Li, G.; Shaw, M.W.; Sheng, J.; et al. Identification of cellular proteins required for replication of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 2012, 28, 1329–1339. [Google Scholar] [CrossRef]
- Kuschal, C.; Botta, E.; Orioli, D.; Digiovanna, J.J.; Seneca, S.; Keymolen, K.; Tamura, D.; Heller, E.; Khan, S.G.; Caligiuri, G.; et al. GTF2E2 Mutations Destabilize the General Transcription Factor Complex TFIIE in Individuals with DNA Repair-Proficient Trichothiodystrophy. Am. J. Hum. Genet. 2016, 98, 627–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Zhou, J.; Liao, X.; Lu, Y.; Deng, C.; Huang, P.; Chen, Q.; Yang, X. Disruption of Smad5 gene induces mitochondria-dependent apoptosis in cardiomyocytes. Exp. Cell Res. 2005, 306, 85–93. [Google Scholar] [CrossRef]
- Nie, M.; Yu, S.; Peng, S.; Fang, Y.; Wang, H.; Yang, X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol. Reprod. 2015, 93, 98. [Google Scholar] [CrossRef]
- Wong, Y.L.; Behringer, R.R.; Kwan, K.M. Smad1/Smad5 signaling in limb ectoderm functions redundantly and is required for interdigital programmed cell death. Dev. Biol. 2012, 363, 247–257. [Google Scholar] [CrossRef]
- Hegarty, S.V.; O’Keeffe, G.W.; Sullivan, A.M. BMP-Smad 1/5/8 signalling in the development of the nervous system. Prog. Neurobiol. 2013, 109, 28–41. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frakes, A.E.; Ferraiuolo, L.; Haidet-Phillips, A.M.; Schmelzer, L.; Braun, L.; Miranda, C.J.; Ladner, K.J.; Bevan, A.K.; Foust, K.D.; Godbout, J.P.; et al. Microglia induce motor neuron death via the classical NF-kappaB pathway in amyotrophic lateral sclerosis. Neuron 2014, 81, 1009–1023. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Lee, C.; Kodama, L.; Gan, L. Sex Differences in Neurodegeneration: The Role of the Immune System in Humans. Biol. Psychiatry 2021. [Google Scholar] [CrossRef]
- McCauley, M.E.; O’Rourke, J.G.; Yanez, A.; Markman, J.L.; Ho, R.; Wang, X.; Chen, S.; Lall, D.; Jin, M.; Muhammad, A.; et al. C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature 2020, 585, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Kakita, A.; Takahashi, H.; Ihara, Y.; Nobukuni, K.; Fujimura, H.; Sakoda, S.; Sasaki, S.; Iwata, M.; Morikawa, S.; et al. Activation of signal transducer and activator of transcription-3 in the spinal cord of sporadic amyotrophic lateral sclerosis patients. Neurodegener. Dis. 2009, 6, 118–126. [Google Scholar] [CrossRef]
- Gal-Oz, S.T.; Maier, B.; Yoshida, H.; Seddu, K.; Elbaz, N.; Czysz, C.; Zuk, O.; Stranger, B.E.; Ner-Gaon, H.; Shay, T. ImmGen report: Sexual dimorphism in the immune system transcriptome. Nat. Commun. 2019, 10, 4295. [Google Scholar] [CrossRef]
- Long, Z.; Zheng, M.; Zhao, L.; Xie, P.; Song, C.; Chu, Y.; Song, W.; He, G. Valproic acid attenuates neuronal loss in the brain of APP/PS1 double transgenic Alzheimer’s disease mice model. Curr. Alzheimer Res. 2013, 10, 261–269. [Google Scholar] [CrossRef]
- Long, Z.; Zeng, Q.; Wang, K.; Sharma, A.; He, G. Gender difference in valproic acid-induced neuroprotective effects on APP/PS1 double transgenic mice modeling Alzheimer’s disease. Acta Biochim. Biophys. Sin. 2016, 48, 930–938. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.S.; Bate, C. An in vitro model for synaptic loss in neurodegenerative diseases suggests a neuroprotective role for valproic acid via inhibition of cPLA2 dependent signalling. Neuropharmacology 2016, 101, 566–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.Z.; Liu, Y.J.; Zhang, W.; Luo, S.F.; Zhou, X.; He, G.Q. Combined treatment with valproic acid and estrogen has neuroprotective effects in ovariectomized mice with Alzheimer’s disease. Neural. Regen. Res. 2021, 16, 2078–2085. [Google Scholar] [PubMed]
- Agrawal, S.; Dixit, A.; Singh, A.; Tripathi, P.; Singh, D.; Patel, D.K.; Singh, M.P. Cyclosporine A and MnTMPyP Alleviate alpha-Synuclein Expression and Aggregation in Cypermethrin-Induced Parkinsonism. Mol. Neurobiol. 2015, 52, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Kulbe, J.R.; Hill, R.L.; Singh, I.N.; Wang, J.A.; Hall, E.D. Synaptic Mitochondria Sustain More Damage than Non-Synaptic Mitochondria after Traumatic Brain Injury and Are Protected by Cyclosporine A. J. Neurotrauma 2017, 34, 1291–1301. [Google Scholar] [CrossRef] [Green Version]
- Butovsky, O.; Siddiqui, S.; Gabriely, G.; Lanser, A.J.; Dake, B.; Murugaiyan, G.; Doykan, C.E.; Wu, P.M.; Gali, R.R.; Iyer, L.K.; et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J. Clin. Investig. 2012, 122, 3063–3087. [Google Scholar] [CrossRef]
- Lincecum, J.M.; Vieira, F.G.; Wang, M.Z.; Thompson, K.; De Zutter, G.S.; Kidd, J.; Moreno, A.; Sanchez, R.; Carrion, I.J.; Levine, B.A.; et al. From transcriptome analysis to therapeutic anti-CD40L treatment in the SOD1 model of amyotrophic lateral sclerosis. Nat. Genet. 2010, 42, 392–399. [Google Scholar] [CrossRef] [PubMed]
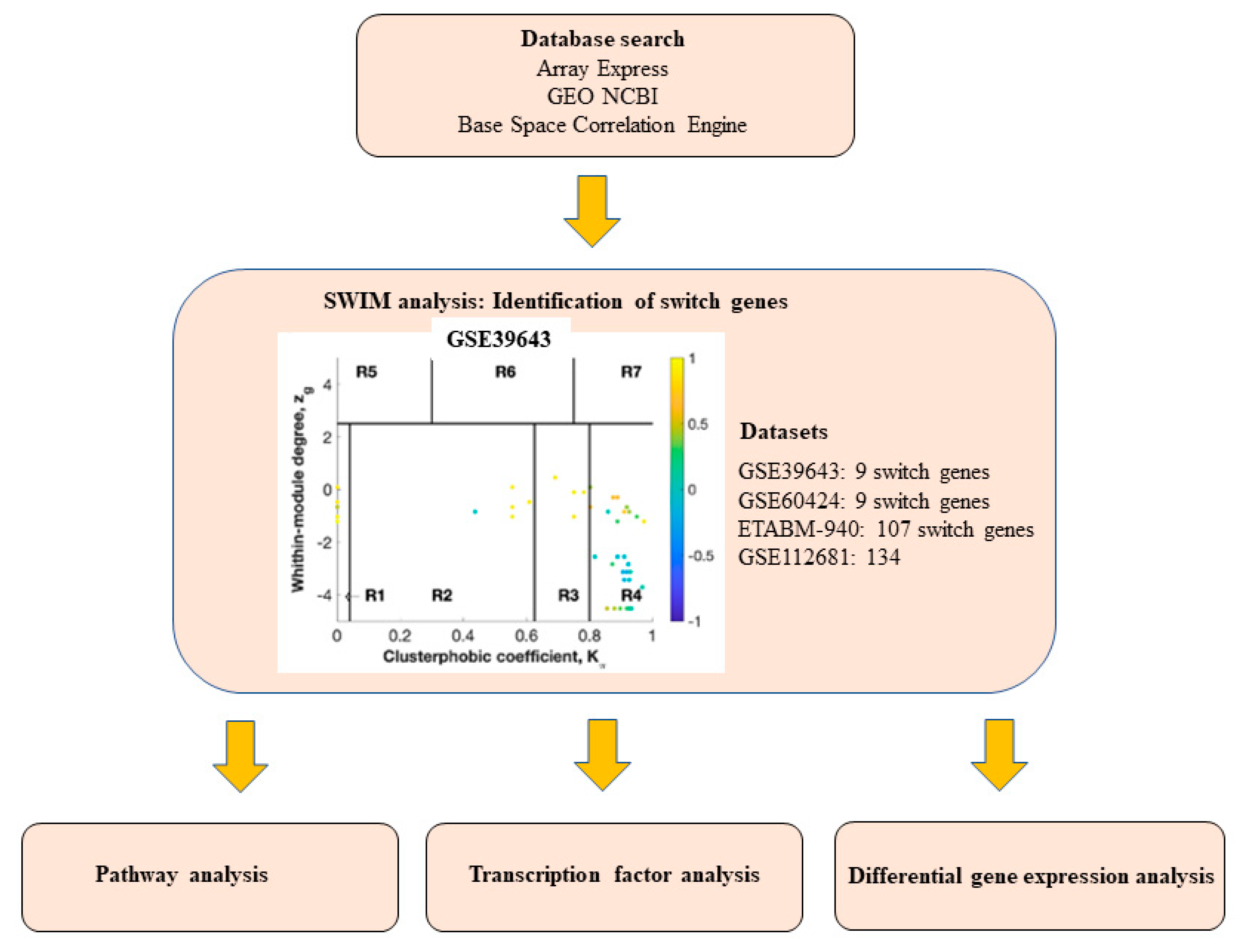
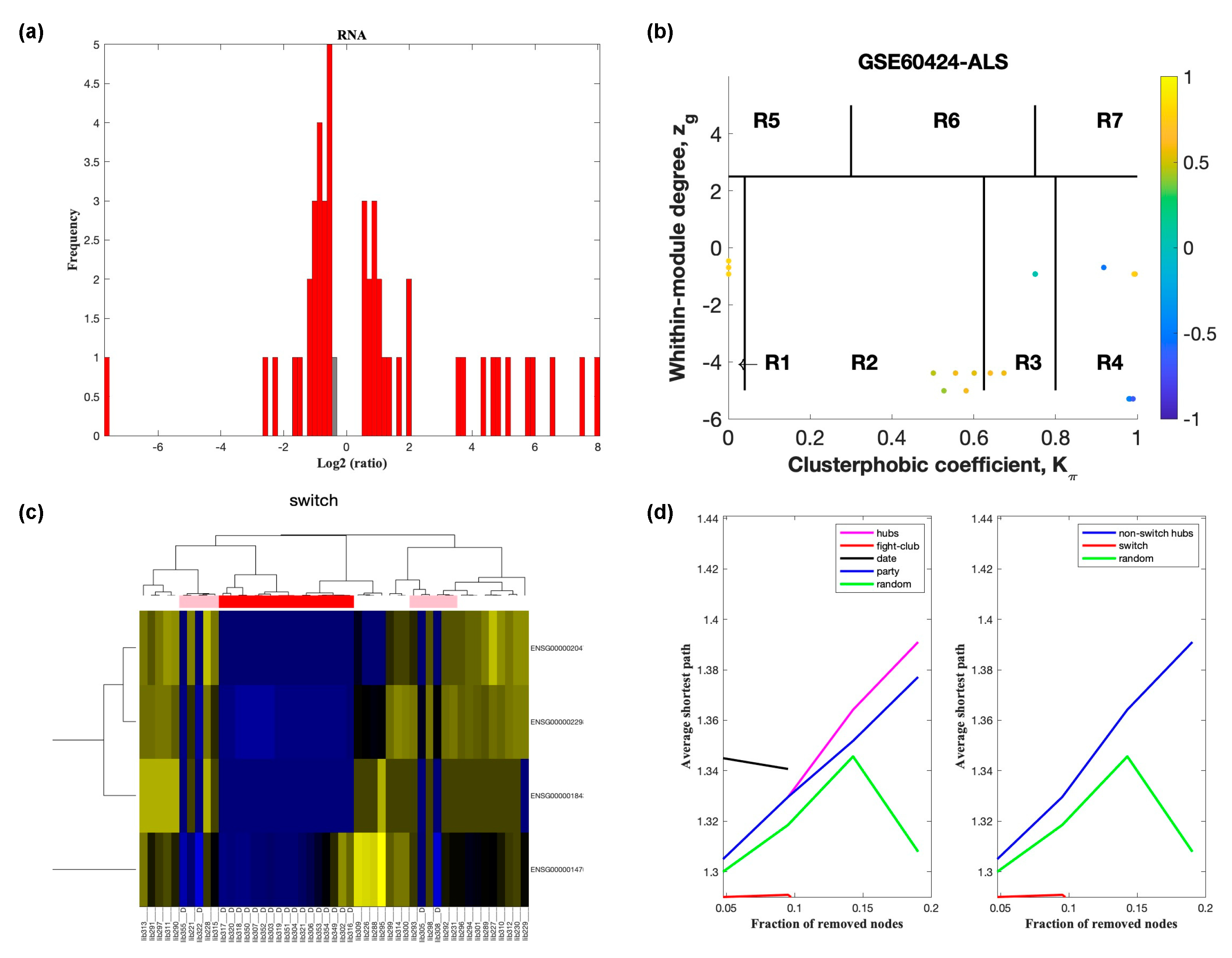
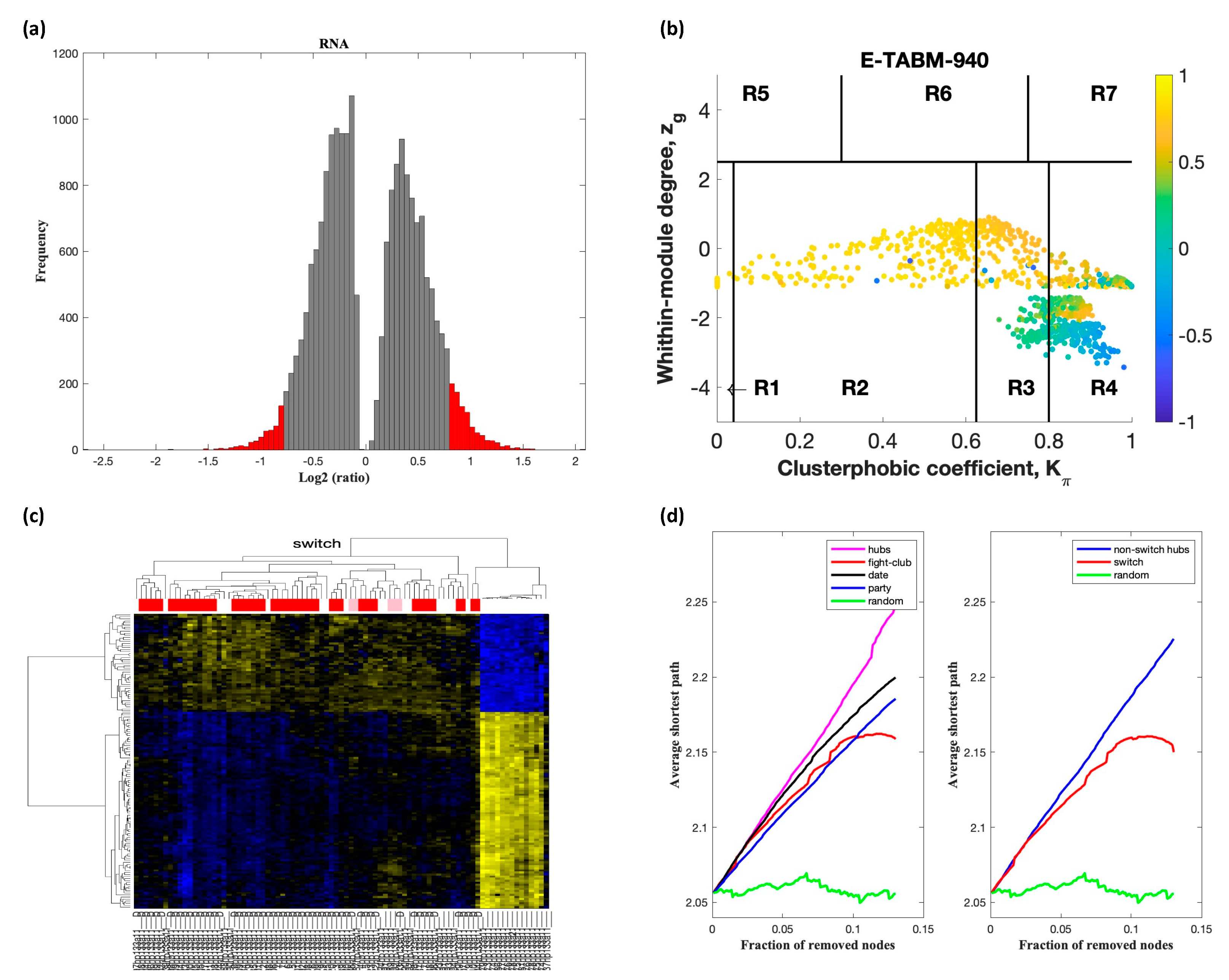
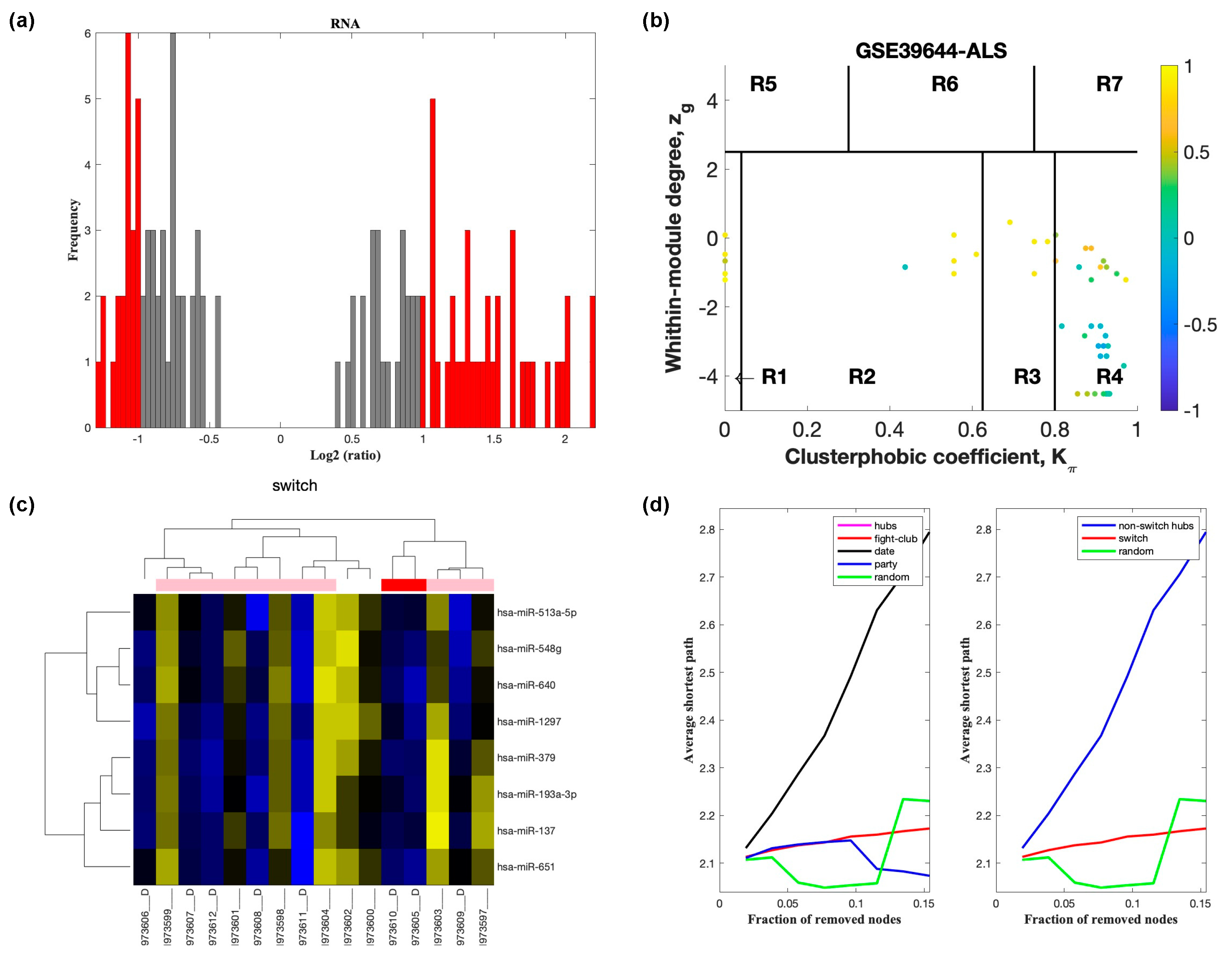
| Dataset | Tissue/Cells | Description of Cases and Controls | Platform |
|---|---|---|---|
| GSE39643 | Blood monocytes | 8 sporadic ALS, 8 healthy controls | NanoString nCounter Human miRNA assay |
| GSE60424 | Whole blood | 6 sporadic ALS | Illumina HiScanSQ |
| E-TABM-940 | Whole blood | 56 sporadic ALS, 28 healthy controls | Affymetrix GeneChip Human Genome U133 2.0 |
| GSE28253 | Blood lymphocytes | 11 sporadic ALS, 11 healthy controls | Agilent-014850 Whole Human Genome Microarray |
| GSE112681 GSE112676 GSE112680 | Whole blood | 233 sporadic ALS, 508 healthy controls | Illumina HumanHT-12 v4.0 |
| Switch gene | Males vs. Healthy | Females vs. Healthy | Males vs. Females |
|---|---|---|---|
| XIST | Not significant | Up: FC = 3.58 (p = 9.7 × 10−7) | Down: FC = −489 (p = 1.8 × 10−31) |
| KDM6A | Not significant | Up: FC = 2.08 (p = 3.9 × 10−5) | Down: FC = −1.57 (p = 3.5 × 10−10) |
| ZFX | Not significant | Up: FC = 2.4 (p = 7.5 × 10−9) | Down: FC = −1.68 (p = 7.4 × 10−9) |
| XAF1 | Not significant | Up: FC = 1.75 (p = 7.5 × 10−7) | Not significant |
| MAP7D2 | Not significant | Up: FC = 1.59 (p = 1.7 × 10−3) | Down: FC= –3.80 (p = 2.0 × 10−17) |
| EIF2S3 | Not significant | Not significant | Down: FC= −1.37 (p = 1.0 × 10−4) |
| IFI44L | Not significant | Up: FC = 2.24 (p = 3.0 × 10−4) | Not significant |
| ERCC6L | Not significant | Not significant | Down: FC = −1.24 (p = 6.0 × 10−4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santiago, J.A.; Quinn, J.P.; Potashkin, J.A. Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients. Int. J. Mol. Sci. 2021, 22, 7150. https://doi.org/10.3390/ijms22137150
Santiago JA, Quinn JP, Potashkin JA. Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients. International Journal of Molecular Sciences. 2021; 22(13):7150. https://doi.org/10.3390/ijms22137150
Chicago/Turabian StyleSantiago, Jose A., James P. Quinn, and Judith A. Potashkin. 2021. "Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients" International Journal of Molecular Sciences 22, no. 13: 7150. https://doi.org/10.3390/ijms22137150
APA StyleSantiago, J. A., Quinn, J. P., & Potashkin, J. A. (2021). Network Analysis Identifies Sex-Specific Gene Expression Changes in Blood of Amyotrophic Lateral Sclerosis Patients. International Journal of Molecular Sciences, 22(13), 7150. https://doi.org/10.3390/ijms22137150






