Taking a Step Back: Insights into the Mechanisms Regulating Gut Epithelial Dedifferentiation
Abstract
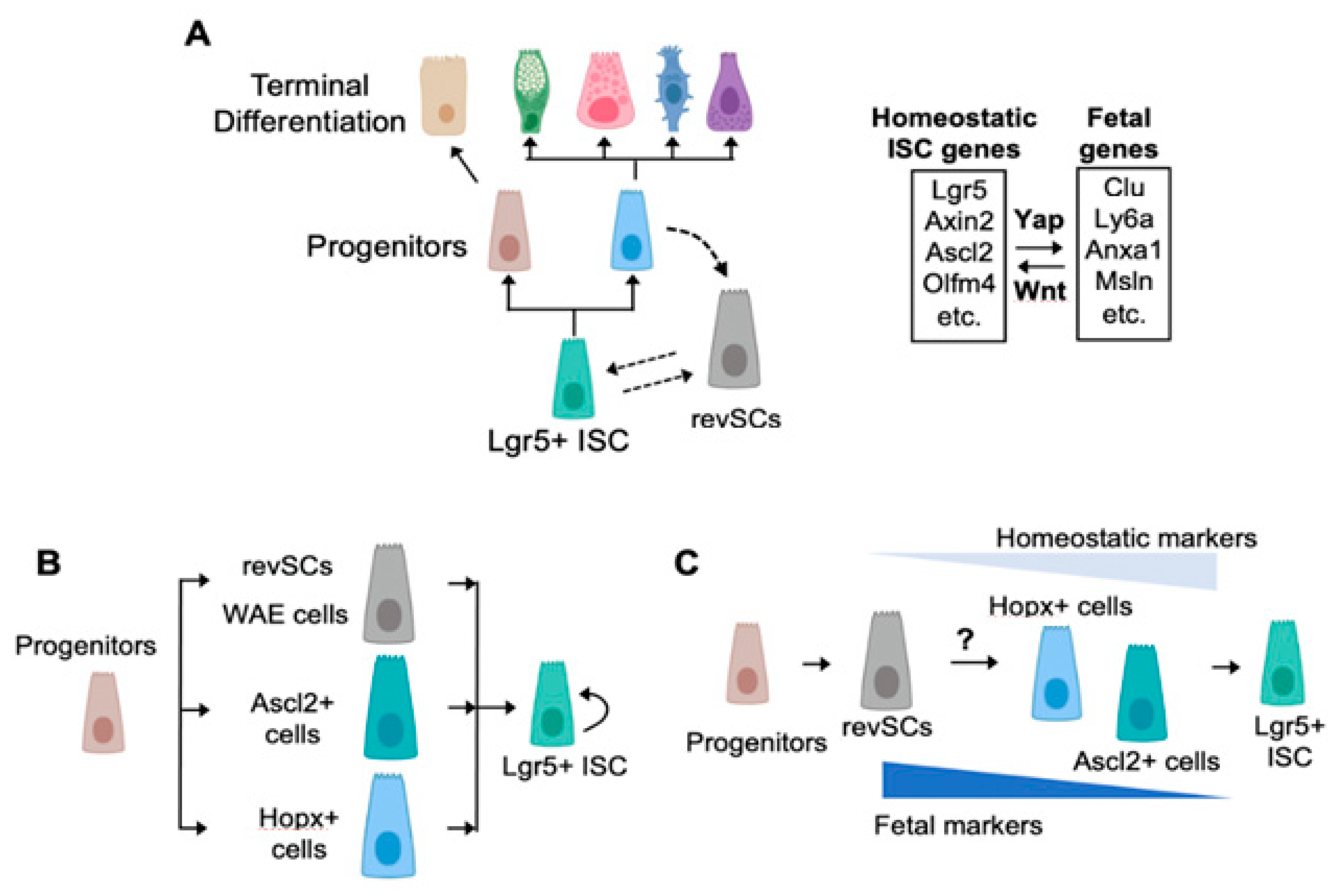
:1. A Brief Survey of the Cellular Components and Drivers of the Homeostatic Crypt
2. Fetal-Like Stem Cells Drive Regeneration
3. How Many Roads Lead to Rome?
4. Regeneration in a Dish
5. Impact of the Microbiota and Infection on Stem-Cell Behavior and Regeneration
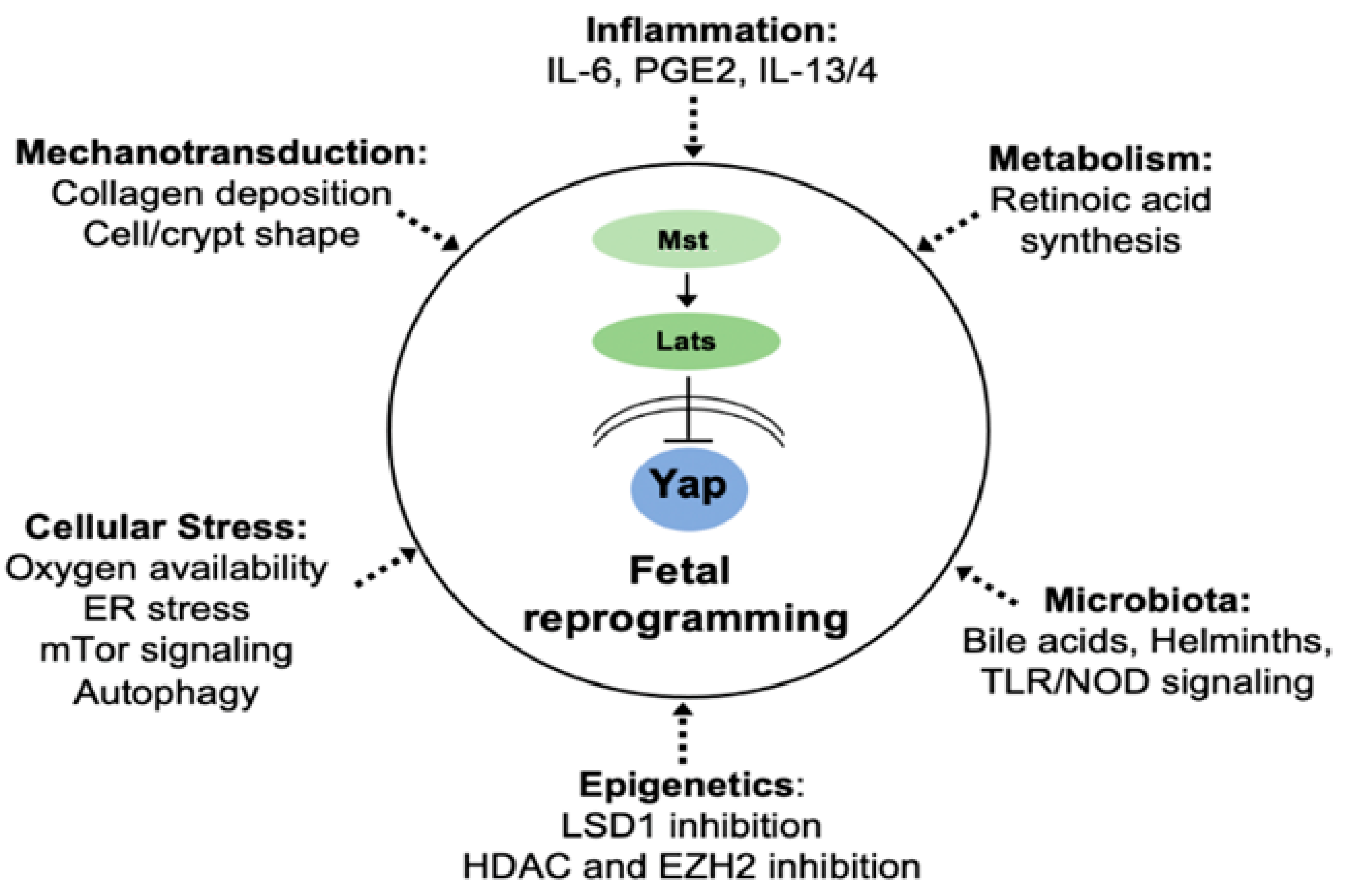
6. Fibro-Inflammatory Signals Drive Fetal Reprogramming
7. Cell-Autonomous Regulation of Epithelial Plasticity
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCracken, V.J.; Lorenz, R.G. The gastrointestinal ecosystem: A precarious alliance among epithelium, immunity and microbiota. Cell. Microbiol. 2001, 3, 1–11. [Google Scholar] [CrossRef]
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019, 567, 49–55. [Google Scholar] [CrossRef]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nature 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Schneider, C.; O’Leary, C.E.; Locksley, R.M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 2019, 19, 584–593. [Google Scholar] [CrossRef]
- Mabbott, N.A.; Donaldson, D.S.; Ohno, H.; Williams, I.R.; Mahajan, A. Microfold (M) cells: Important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013, 6, 666–677. [Google Scholar] [CrossRef] [Green Version]
- Spit, M.; Koo, B.-K.; Maurice, M.M. Tales from the crypt: Intestinal niche signals in tissue renewal, plasticity and cancer. Open Biol. 2018, 8, 180120. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.-J.; Li, X.-G.; Wang, X.-Q. Notch Signaling in Mammalian Intestinal Stem Cells: Determining Cell Fate and Maintaining Homeostasis. Curr. Stem Cell Res. Ther. 2019, 14, 583–590. [Google Scholar] [CrossRef]
- Demitrack, E.S.; Samuelson, L.C. Notch regulation of gastrointestinal stem cells. J. Physiol. 2016, 594, 4791–4803. [Google Scholar] [CrossRef] [Green Version]
- Kriz, V.; Korinek, V. Wnt, RSPO and Hippo Signalling in the Intestine and Intestinal Stem Cells. Genes 2018, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Sphyris, N.; Hodder, M.C.; Sansom, O.J. Subversion of Niche-Signalling Pathways in Colorectal Cancer: What Makes and Breaks the Intestinal Stem Cell. Cancers 2021, 13, 1000. [Google Scholar] [CrossRef]
- Muñoz, J.; Stange, D.E.; Schepers, A.G.; van de Wetering, M.; Koo, B.-K.; Itzkovitz, S.; Volckmann, R.; Kung, K.S.; Koster, J.; Radulescu, S.; et al. The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent “+4” cell markers. EMBO J. 2012, 31, 3079–3091. [Google Scholar] [CrossRef] [Green Version]
- Korinek, V.; Barker, N.; Moerer, P.; van Donselaar, E.; Huls, G.; Peters, P.J.; Clevers, H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998, 19, 379–383. [Google Scholar] [CrossRef]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003, 17, 1709–1713. [Google Scholar] [CrossRef] [Green Version]
- Fevr, T.; Robine, S.; Louvard, D.; Huelsken, J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol. Cell. Biol. 2007, 27, 7551–7559. [Google Scholar] [CrossRef] [Green Version]
- Kuhnert, F.; Davis, C.R.; Wang, H.-T.; Chu, P.; Lee, M.; Yuan, J.; Nusse, R.; Kuo, C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 2004, 101, 266–271. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.-A.; Kakitani, M.; Zhao, J.; Oshima, T.; Tang, T.; Binnerts, M.; Liu, Y.; Boyle, B.; Park, E.; Emtage, P.; et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science 2005, 309, 1256–1259. [Google Scholar] [CrossRef]
- Sansom, O.J.; Reed, K.R.; Hayes, A.J.; Ireland, H.; Brinkmann, H.; Newton, I.P.; Batlle, E.; Simon-Assmann, P.; Clevers, H.; Nathke, I.S.; et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004, 18, 1385–1390. [Google Scholar] [CrossRef] [Green Version]
- Andreu, P.; Colnot, S.; Godard, C.; Gad, S.; Chafey, P.; Niwa-Kawakita, M.; Laurent-Puig, P.; Kahn, A.; Robine, S.; Perret, C.; et al. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development 2005, 132, 1443–1451. [Google Scholar] [CrossRef] [Green Version]
- Van Es, J.H.; Sato, T.; van de Wetering, M.; Lyubimova, A.; Yee Nee, A.N.; Gregorieff, A.; Sasaki, N.; Zeinstra, L.; van den Born, M.; Korving, J.; et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012, 14, 1099–1104. [Google Scholar] [CrossRef] [Green Version]
- Gregorieff, A.; Stange, D.E.; Kujala, P.; Begthel, H.; van den Born, M.; Korving, J.; Peters, P.J.; Clevers, H. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology 2009, 137, 1333–1345. [Google Scholar] [CrossRef]
- Mori-Akiyama, Y.; van den Born, M.; van Es, J.H.; Hamilton, S.R.; Adams, H.P.; Zhang, J.; Clevers, H.; de Crombrugghe, B. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology 2007, 133, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Henderson, J.T.; Beghtel, H.; van den Born, M.M.W.; Sancho, E.; Huls, G.; Meeldijk, J.; Robertson, J.; van de Wetering, M.; Pawson, T.; et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 2002, 111, 251–263. [Google Scholar] [CrossRef] [Green Version]
- VanDussen, K.L.; Carulli, A.J.; Keeley, T.M.; Patel, S.R.; Puthoff, B.J.; Magness, S.T.; Tran, I.T.; Maillard, I.; Siebel, C.; Kolterud, Å.; et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 2012, 139, 488–497. [Google Scholar] [CrossRef] [Green Version]
- Riccio, O.; van Gijn, M.E.; Bezdek, A.C.; Pellegrinet, L.; van Es, J.H.; Zimber-Strobl, U.; Strobl, L.J.; Honjo, T.; Clevers, H.; Radtke, F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008, 9, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Fre, S.; Huyghe, M.; Mourikis, P.; Robine, S.; Louvard, D.; Artavanis-Tsakonas, S. Notch signals control the fate of immature progenitor cells in the intestine. Nature 2005, 435, 964–968. [Google Scholar] [CrossRef]
- Stanger, B.Z.; Datar, R.; Murtaugh, L.C.; Melton, D.A. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA 2005, 102, 12443–12448. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, V.W.Y.; Stange, D.E.; Page, M.E.; Buczacki, S.; Wabik, A.; Itami, S.; van de Wetering, M.; Poulsom, R.; Wright, N.A.; Trotter, M.W.B.; et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012, 14, 401–408. [Google Scholar] [CrossRef] [Green Version]
- Monticelli, L.A.; Osborne, L.C.; Noti, M.; Tran, S.V.; Zaiss, D.M.W.; Artis, D. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 10762–10767. [Google Scholar] [CrossRef] [Green Version]
- Roberts, R.B.; Min, L.; Washington, M.K.; Olsen, S.J.; Settle, S.H.; Coffey, R.J.; Threadgill, D.W. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 1521–1526. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Yu, M.; Lee, E.; Kim, H.; Yang, Y.; Kim, K.; Pannicia, C.; Kurie, J.M.; Threadgill, D.W. Tumor-specific apoptosis caused by deletion of the ERBB3 pseudo-kinase in mouse intestinal epithelium. J. Clin. Invest. 2009, 119, 2702–2713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.; Pearsall, R.S.; Das, S.; Dey, S.K.; Godfrey, V.L.; Threadgill, D.W. Epiregulin is not essential for development of intestinal tumors but is required for protection from intestinal damage. Mol. Cell. Biol. 2004, 24, 8907–8916. [Google Scholar] [CrossRef] [Green Version]
- Kosinski, C.; Li, V.S.W.; Chan, A.S.Y.; Zhang, J.; Ho, C.; Tsui, W.Y.; Chan, T.L.; Mifflin, R.C.; Powell, D.W.; Yuen, S.T.; et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl. Acad. Sci. USA 2007, 104, 15418–15423. [Google Scholar] [CrossRef] [Green Version]
- Hardwick, J.C.H.; Van Den Brink, G.R.; Bleuming, S.A.; Ballester, I.; Van Den Brande, J.M.H.; Keller, J.J.; Offerhaus, G.J.A.; Van Deventer, S.J.H.; Peppelenbosch, M.P. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology 2004, 126, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Haramis, A.-P.G.; Begthel, H.; van den Born, M.; van Es, J.; Jonkheer, S.; Offerhaus, G.J.A.; Clevers, H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [Green Version]
- Howe, J.R.; Bair, J.L.; Sayed, M.G.; Anderson, M.E.; Mitros, F.A.; Petersen, G.M.; Velculescu, V.E.; Traverso, G.; Vogelstein, B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat. Genet. 2001, 28, 184–187. [Google Scholar] [CrossRef]
- He, X.C.; Zhang, J.; Tong, W.-G.; Tawfik, O.; Ross, J.; Scoville, D.H.; Tian, Q.; Zeng, X.; He, X.; Wiedemann, L.M.; et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004, 36, 1117–1121. [Google Scholar] [CrossRef] [Green Version]
- Farin, H.F.; Jordens, I.; Mosa, M.H.; Basak, O.; Korving, J.; Tauriello, D.V.F.; de Punder, K.; Angers, S.; Peters, P.J.; Maurice, M.M.; et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 2016, 530, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012, 143, 1518–1529. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Escudero, S.; Shivdasani, R.A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. USA 2012, 109, 3932–3937. [Google Scholar] [CrossRef] [Green Version]
- Durand, A.; Donahue, B.; Peignon, G.; Letourneur, F.; Cagnard, N.; Slomianny, C.; Perret, C.; Shroyer, N.F.; Romagnolo, B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl. Acad. Sci. USA 2012, 109, 8965–8970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, N.; Manieri, E.; Storm, E.E.; Saadatpour, A.; Luoma, A.M.; Kapoor, V.N.; Madha, S.; Gaynor, L.T.; Cox, C.; Keerthivasan, S.; et al. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 2020, 26, 391–402. [Google Scholar] [CrossRef]
- Degirmenci, B.; Valenta, T.; Dimitrieva, S.; Hausmann, G.; Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 2018, 558, 449–453. [Google Scholar] [CrossRef]
- Shoshkes-Carmel, M.; Wang, Y.J.; Wangensteen, K.J.; Tóth, B.; Kondo, A.; Massasa, E.E.; Itzkovitz, S.; Kaestner, K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 2018, 557, 242–246. [Google Scholar] [CrossRef]
- Stzepourginski, I.; Nigro, G.; Jacob, J.-M.; Dulauroy, S.; Sansonetti, P.J.; Eberl, G.; Peduto, L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA 2017, 114, E506–E513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greicius, G.; Kabiri, Z.; Sigmundsson, K.; Liang, C.; Bunte, R.; Singh, M.K.; Virshup, D.M. PDGFRα + pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. USA 2018, 115, E3173–E3181. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-E.; Fei, L.; Yin, W.-C.; Coquenlorge, S.; Rao-Bhatia, A.; Zhang, X.; Shi, S.S.W.; Lee, J.H.; Hahn, N.A.; Rizvi, W.; et al. Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat. Commun. 2020, 11, 334. [Google Scholar] [CrossRef]
- Kinchen, J.; Chen, H.H.; Parikh, K.; Antanaviciute, A.; Jagielowicz, M.; Fawkner-Corbett, D.; Ashley, N.; Cubitt, L.; Mellado-Gomez, E.; Attar, M.; et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 2018, 175, 372–386. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, N.; Kraiczy, J.; Shivdasani, R.A. Cellular and molecular architecture of the intestinal stem cell niche. Nat. Cell Biol. 2020, 22, 1033–1041. [Google Scholar] [CrossRef]
- Yui, S.; Azzolin, L.; Maimets, M.; Pedersen, M.T.; Fordham, R.P.; Hansen, S.L.; Larsen, H.L.; Guiu, J.; Alves, M.R.P.; Rundsten, C.F.; et al. YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 2018, 22, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregorieff, A.; Liu, Y.; Inanlou, M.R.; Khomchuk, Y.; Wrana, J.L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 2015, 526, 715–718. [Google Scholar] [CrossRef]
- Manning, S.A.; Kroeger, B.; Harvey, K.F. The regulation of Yorkie, YAP and TAZ: New insights into the Hippo pathway. Development 2020, 147. [Google Scholar] [CrossRef]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.-L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustata, R.C.; Vasile, G.; Fernandez-Vallone, V.; Strollo, S.; Lefort, A.; Libert, F.; Monteyne, D.; Pérez-Morga, D.; Vassart, G.; Garcia, M.I. Identification of Lgr5-Independent Spheroid-Generating Progenitors of the Mouse Fetal Intestinal Epithelium. Cell Rep. 2013, 5, 421–432. [Google Scholar] [CrossRef]
- Ayyaz, A.; Kumar, S.; Sangiorgi, B.; Ghoshal, B.; Gosio, J.; Ouladan, S.; Fink, M.; Barutcu, S.; Trcka, D.; Shen, J.; et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019, 569, 121–125. [Google Scholar] [CrossRef]
- Murata, K.; Jadhav, U.; Madha, S.; Van Es, J.; Dean, J.; Cavazza, A.; Wucherpfennig, K.; Michor, F.; Clevers, H.; Shivdasani, R.A. Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells Article Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells. Cell Stem Cell 2020, 26, 377–390. [Google Scholar] [CrossRef] [Green Version]
- De Sousa E Melo, F.; de Sauvage, F.J. Cellular Plasticity in Intestinal Homeostasis and Disease. Cell Stem Cell 2019, 24, 54–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Sase, M.; Ishikawa, S.; Kajita, M.; Asano, J.; Sato, T.; Mori, Y.; Ohteki, T. Characterization of radioresistant epithelial stem cell heterogeneity in the damaged mouse intestine. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Seno, H.; Miyoshi, H.; Brown, S.L.; Geske, M.J.; Colonna, M.; Stappenbeck, T.S. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 256–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chiang, I.-L.; Ohara, T.E.; Fujii, S.; Cheng, J.; Muegge, B.D.; Ver Heul, A.; Han, N.D.; Lu, Q.; Xiong, S.; et al. Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells. Cell 2019, 179, 1144–1159. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Biehs, B.; Warming, S.; Leong, K.G.; Rangell, L.; Klein, O.D.; de Sauvage, F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 2011, 478, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Schuijers, J.; Junker, J.P.; Mokry, M.; Hatzis, P.; Koo, B.-K.; Sasselli, V.; van der Flier, L.G.; Cuppen, E.; van Oudenaarden, A.; Clevers, H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell 2015, 16, 158–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Flier, L.G.; van Gijn, M.E.; Hatzis, P.; Kujala, P.; Haegebarth, A.; Stange, D.E.; Begthel, H.; van den Born, M.; Guryev, V.; Oving, I.; et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 2009, 136, 903–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef]
- Tallapragada, N.P.; Cambra, H.M.; Wald, T.; Keough Jalbert, S.; Abraham, D.M.; Klein, O.D.; Klein, A.M. Inflation-collapse dynamics drive patterning and morphogenesis in intestinal organoids. Cell Stem Cell 2021. [Google Scholar] [CrossRef]
- Fordham, R.P.; Yui, S.; Hannan, N.R.F.; Soendergaard, C.; Madgwick, A.; Schweiger, P.J.; Nielsen, O.H.; Vallier, L.; Pedersen, R.A.; Nakamura, T.; et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell 2013, 13, 734–744. [Google Scholar] [CrossRef] [Green Version]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Naito, T.; Mulet, C.; De Castro, C.; Molinaro, A.; Saffarian, A.; Nigro, G.; Bérard, M.; Clerc, M.; Pedersen, A.B.; Sansonetti, P.J.; et al. Lipopolysaccharide from Crypt-Specific Core Microbiota Modulates the Colonic Epithelial Proliferation-to-Differentiation Balance. mBio 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, Y.; Yin, F.; Yu, J.; Silverman, N.; Pan, D. Toll Receptor-Mediated Hippo Signaling Controls Innate Immunity in Drosophila. Cell 2016, 164, 406–419. [Google Scholar] [CrossRef] [Green Version]
- Levy, A.; Stedman, A.; Deutsch, E.; Donnadieu, F.; Virgin, H.W.; Sansonetti, P.J.; Nigro, G. Innate immune receptor NOD2 mediates LGR5+ intestinal stem cell protection against ROS cytotoxicity via mitophagy stimulation. Proc. Natl. Acad. Sci. USA 2020, 117, 1994–2003. [Google Scholar] [CrossRef] [Green Version]
- Nigro, G.; Rossi, R.; Commere, P.-H.; Jay, P.; Sansonetti, P.J. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 2014, 15, 792–798. [Google Scholar] [CrossRef] [Green Version]
- Kaiko, G.E.; Ryu, S.H.; Koues, O.I.; Collins, P.L.; Solnica-Krezel, L.; Pearce, E.J.; Pearce, E.L.; Oltz, E.M.; Stappenbeck, T.S. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 2016, 165, 1708–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El Alam, G.; Bou Sleiman, M.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 2020, 159, 956–968. [Google Scholar] [CrossRef]
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, W.Y.; Blutt, S.E.; Zeng, X.-L.; Chen, M.-S.; Lo, Y.-H.; Castillo-Azofeifa, D.; Klein, O.D.; Shroyer, N.F.; Donowitz, M.; Estes, M.K. Epithelial WNT Ligands Are Essential Drivers of Intestinal Stem Cell Activation. Cell Rep. 2018, 22, 1003–1015. [Google Scholar] [CrossRef] [Green Version]
- Cliffe, L.J.; Humphreys, N.E.; Lane, T.E.; Potten, C.S.; Booth, C.; Grencis, R.K. Accelerated intestinal epithelial cell turnover: A new mechanism of parasite expulsion. Science 2005, 308, 1463–1465. [Google Scholar] [CrossRef] [PubMed]
- Nusse, Y.M.; Savage, A.K.; Marangoni, P.; Rosendahl-Huber, A.K.M.; Landman, T.A.; De Sauvage, F.J.; Locksley, R.M.; Klein, O.D. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 2018, 559, 109–113. [Google Scholar] [CrossRef]
- Pocaterra, A.; Romani, P.; Dupont, S. YAP/TAZ functions and their regulation at a glance. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Wu, L.-W.; Grivennikov, S.I.; de Jong, P.R.; Lian, I.; Yu, F.-X.; Wang, K.; Ho, S.B.; Boland, B.S.; Chang, J.T.; et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 2015, 519, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romera-Hernández, M.; Aparicio-Domingo, P.; Papazian, N.; Karrich, J.J.; Cornelissen, F.; Hoogenboezem, R.M.; Samsom, J.N.; Cupedo, T. Yap1-Driven Intestinal Repair Is Controlled by Group 3 Innate Lymphoid Cells. Cell Rep. 2020, 30, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Roulis, M.; Kaklamanos, A.; Schernthanner, M.; Bielecki, P.; Zhao, J.; Kaffe, E.; Frommelt, L.-S.; Qu, R.; Knapp, M.S.; Henriques, A.; et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature 2020, 580, 524–529. [Google Scholar] [CrossRef]
- Brown, S.L.; Riehl, T.E.; Walker, M.R.; Geske, M.J.; Doherty, J.M.; Stenson, W.F.; Stappenbeck, T.S. Myd88-dependent positioning of Ptgs2- expressing stromal cells maintains colonic epithelial proliferation during injury. J. Clin. Investig. 2007, 117, 258–269. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-B.; Kim, M.; Park, Y.-S.; Park, I.; Kim, T.; Yang, S.-Y.; Cho, C.J.; Hwang, D.; Jung, J.-H.; Markowitz, S.D.; et al. Prostaglandin E2 Activates YAP and a Positive-Signaling Loop to Promote Colon Regeneration After Colitis but Also Carcinogenesis in Mice. Gastroenterology 2017, 152, 616–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, H.; VanDussen, K.L.; Malvin, N.P.; Ryu, S.H.; Wang, Y.; Sonnek, N.M.; Lai, C.; Stappenbeck, T.S. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2017, 36, 5–24. [Google Scholar] [CrossRef] [PubMed]
- De Salvo, C.; Pastorelli, L.; Petersen, C.P.; Buttò, L.F.; Buela, K.-A.; Omenetti, S.; Locovei, S.A.; Ray, S.; Friedman, H.R.; Duijser, J.; et al. Interleukin 33 Triggers Early Eosinophil-Dependent Events Leading to Metaplasia in a Chronic Model of Gastritis-Prone Mice. Gastroenterology 2020, 160, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.R.; Engevik, A.C.; Madorsky, T.; Belmont, E.; Stier, M.T.; Norlander, A.E.; Pilkinton, M.A.; McDonnell, W.J.; Weis, J.A.; Jang, B.; et al. Group 2 innate lymphoid cells coordinate damage response in the stomach. Gastroenterology 2020, 159, 2077–2091. [Google Scholar] [CrossRef]
- Petersen, C.P.; Meyer, A.R.; De Salvo, C.; Choi, E.; Schlegel, C.; Petersen, A.; Engevik, A.C.; Prasad, N.; Levy, S.E.; Peebles, R.S.; et al. A signalling cascade of IL-33 to IL-13 regulates metaplasia in the mouse stomach. Gut 2018, 67, 805–817. [Google Scholar] [CrossRef]
- Yimlamai, D.; Christodoulou, C.; Galli, G.G.; Yanger, K.; Pepe-Mooney, B.; Gurung, B.; Shrestha, K.; Cahan, P.; Stanger, B.Z.; Camargo, F.D. Hippo pathway activity influences liver cell fate. Cell 2014, 157, 1324–1338. [Google Scholar] [CrossRef] [Green Version]
- Totaro, A.; Zhuang, Q.; Panciera, T.; Battilana, G.; Azzolin, L.; Brumana, G.; Gandin, A.; Brusatin, G.; Cordenonsi, M.; Piccolo, S. Cell phenotypic plasticity requires autophagic flux driven by YAP/TAZ mechanotransduction. Proc. Natl. Acad. Sci. USA 2019, 116, 17848–17857. [Google Scholar] [CrossRef] [Green Version]
- Davidson, N.O.; Lewis, M.A.; Blanc, V.; Mills, J.C.; Willet, S.G.; Liu, D.; Wang, Z.; Radyk, M.D.; Cunningham, R.L.; Sibbel, G.; et al. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J. 2018, 37, e98311. [Google Scholar] [CrossRef]
- Miao, Z.F.; Sun, J.X.; Adkins-Threats, M.; Pang, M.J.; Zhao, J.H.; Wang, X.; Tang, K.W.; Wang, Z.N.; Mills, J.C. DDIT4 Licenses Only Healthy Cells to Proliferate During Injury-induced Metaplasia. Gastroenterology 2021, 160, 260–271. [Google Scholar] [CrossRef]
- Tumaneng, K.; Russell, R.C.; Guan, K.-L. Organ size control by Hippo and TOR pathways. Curr. Biol. 2012, 22, R368–R379. [Google Scholar] [CrossRef] [Green Version]
- Rhinn, M.; Dollé, P. Retinoic acid signalling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef] [Green Version]
- Iwafuchi-Doi, M.; Zaret, K.S. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014, 28, 2679–2692. [Google Scholar] [CrossRef] [Green Version]
- Kazakevych, J.; Sayols, S.; Messner, B.; Krienke, C.; Soshnikova, N. Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res. 2017, 45, 5770–5784. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Li, F.; Ferreiro-Neira, I.; Ho, L.-L.; Luyten, A.; Nalapareddy, K.; Long, H.; Verzi, M.; Shivdasani, R.A. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature 2014, 506, 511–515. [Google Scholar] [CrossRef]
- Verzi, M.P.; Shivdasani, R.A. Epigenetic regulation of intestinal stem cell differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 319, G189–G196. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, U.; Saxena, M.; O’Neill, N.K.; Saadatpour, A.; Yuan, G.-C.; Herbert, Z.; Murata, K.; Shivdasani, R.A. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell 2017, 21, 65–77. [Google Scholar] [CrossRef] [Green Version]
- Zwiggelaar, R.T.; Lindholm, H.T.; Fosslie, M.; Terndrup Pedersen, M.; Ohta, Y.; Díez-Sánchez, A.; Martín-Alonso, M.; Ostrop, J.; Matano, M.; Parmar, N.; et al. LSD1 represses a neonatal/reparative gene program in adult intestinal epithelium. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Xiong, L.; Lyu, Y.; Zhang, X.; Shen, J.; Guan, J.; Chai, P.; Lin, Z.; Nie, B.; Li, C.; et al. Establishment of intestinal organoid cultures modeling injury-associated epithelial regeneration. Cell Res. 2021, 31, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Van Es, J.H.; Wiebrands, K.; López-Iglesias, C.; van de Wetering, M.; Zeinstra, L.; van den Born, M.; Korving, J.; Sasaki, N.; Peters, P.J.; van Oudenaarden, A.; et al. Enteroendocrine and tuft cells support Lgr5 stem cells on Paneth cell depletion. Proc. Natl. Acad. Sci. USA 2019, 116, 26599–26605. [Google Scholar] [CrossRef] [PubMed]
- Mosa, M.H.; Michels, B.E.; Menche, C.; Nicolas, A.M.; Darvishi, T.; Greten, F.R.; Farin, H.F. A wnt-induced phenotypic switch in cancer-associated fibroblasts inhibits EMT in colorectal cancer. Cancer Res. 2020, 80, 5569–5582. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouladan, S.; Gregorieff, A. Taking a Step Back: Insights into the Mechanisms Regulating Gut Epithelial Dedifferentiation. Int. J. Mol. Sci. 2021, 22, 7043. https://doi.org/10.3390/ijms22137043
Ouladan S, Gregorieff A. Taking a Step Back: Insights into the Mechanisms Regulating Gut Epithelial Dedifferentiation. International Journal of Molecular Sciences. 2021; 22(13):7043. https://doi.org/10.3390/ijms22137043
Chicago/Turabian StyleOuladan, Shaida, and Alex Gregorieff. 2021. "Taking a Step Back: Insights into the Mechanisms Regulating Gut Epithelial Dedifferentiation" International Journal of Molecular Sciences 22, no. 13: 7043. https://doi.org/10.3390/ijms22137043
APA StyleOuladan, S., & Gregorieff, A. (2021). Taking a Step Back: Insights into the Mechanisms Regulating Gut Epithelial Dedifferentiation. International Journal of Molecular Sciences, 22(13), 7043. https://doi.org/10.3390/ijms22137043




