Genomic Aromatic Compound Degradation Potential of Novel Paraburkholderia Species: Paraburkholderia domus sp. nov., Paraburkholderia haematera sp. nov. and Paraburkholderia nemoris sp. nov.
Abstract
1. Introduction
2. Results
2.1. Isolation of Paraburkholderia Isolates
2.2. Phylogeny
2.3. Genome Features
2.4. Physiology
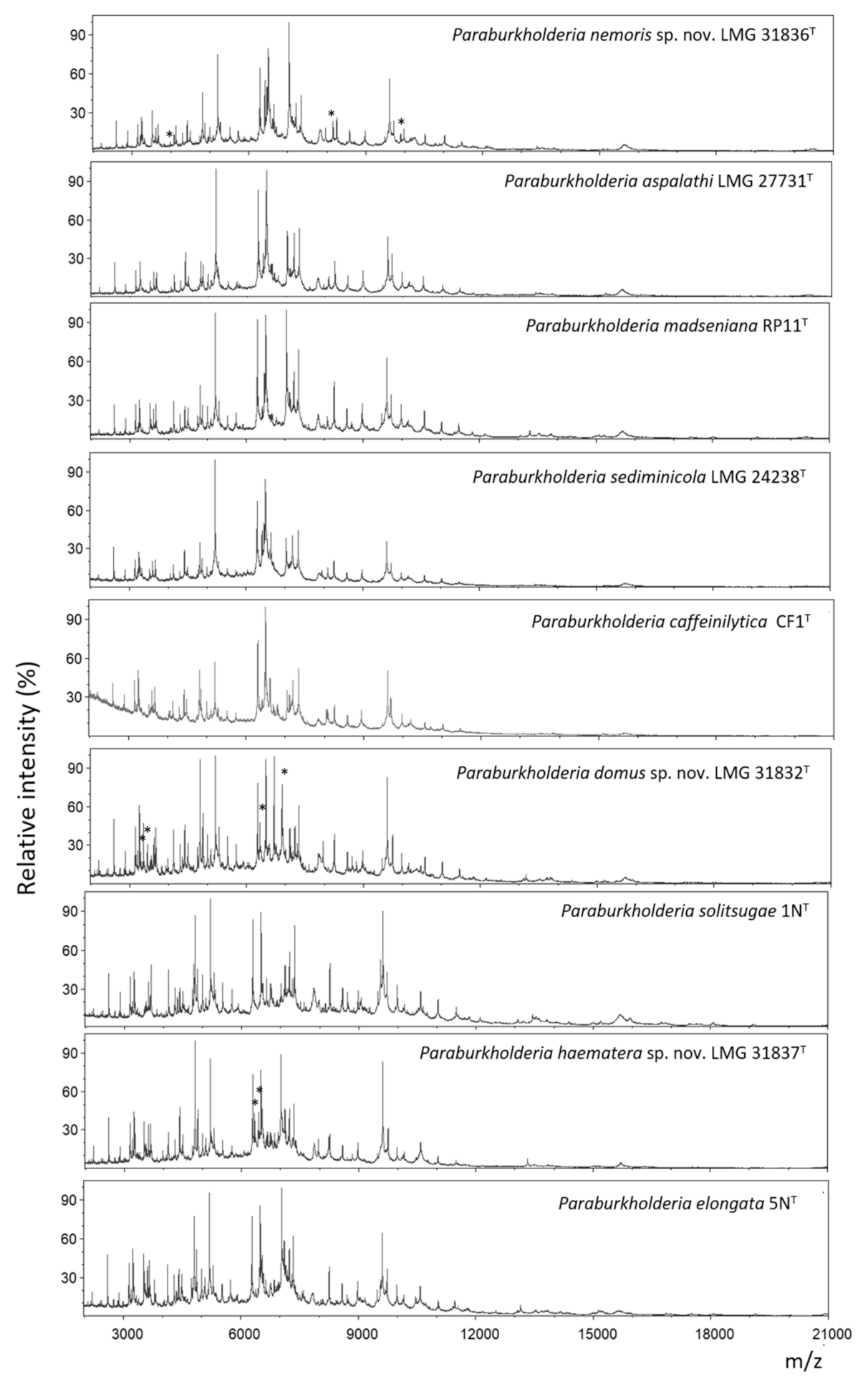
2.5. MALDI-TOF MS Analysis
3. Discussion
4. Materials and Methods
4.1. Isolation of Paraburkholderia Isolates
4.2. Phylogeny
4.3. Functional Genome Annotation
4.4. Physiology
4.5. MALDI-TOF MS Analysis
5. Conclusions
5.1. Description of Paraburkholderia domus sp. nov.
5.2. Description of Paraburkholderia nemoris sp. nov.
5.3. Description of Paraburkholderia haematera sp. nov.
5.4. Emended Description of Paraburkholderia sediminicola Lim et al. 2008
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vandamme, P.; Peeters, C.; De Smet, B.; Price, E.P.; Sarovich, D.S.; Henry, D.A.; Hird, T.J.; Zlosnik, J.E.A.; Mayo, M.; Warner, J.; et al. Comparative Genomics of Burkholderia singularis sp. nov., a Low GCC content, Free-Living bacterium that defies taxonomic dissection of the genus Burkholderia. Front. Microbiol. 2017, 8, 1679. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, R.C.; Murphy, S.J.L.; Feriancek, N.M.; Karasz, D.C.; Derito, C.M.; Newman, J.D.; Buckley, D.H. Paraburkholderia madseniana sp. nov., a phenolic acid-degrading bacterium isolated from acidic forest soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 2137–2146. [Google Scholar] [CrossRef] [PubMed]
- Rusch, A.; Islam, S.; Savalia, P.; Amend, J.P. Burkholderia insulsa sp. nov., a facultatively chemolithotrophic bacterium isolated from an arsenic-rich shallow marine hydrothermal system. Int. J. Syst. Evol. Microbiol. 2015, 65, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.Y.; Gao, Z.H.; Lin, Q.H.; Qiu, L.H. Paraburkholderia pallida sp. Nov. And paraburkholderia silviterrae sp. Nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 3777–3785. [Google Scholar] [CrossRef]
- Mavengere, N.R.; Ellis, A.G.; Le Roux, J.J. Burkholderia aspalathi sp. nov., isolated from root nodules of the South African legume Aspalathus abietina Thunb. Int. J. Syst. Evol. Microbiol. 2014, 64, 1906–1912. [Google Scholar] [CrossRef]
- Xiao, S.Y.; Gao, Z.H.; Yang, Z.; Bi, J.Y.; Qiu, L.H. Paraburkholderia telluris sp. nov., isolated from subtropical forest soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1274–1280. [Google Scholar] [CrossRef]
- Choi, G.M.; Im, W.T. Paraburkholderia azotifigens sp. nov., a nitrogen-fixing bacterium isolated from paddy soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 310–316. [Google Scholar] [CrossRef]
- Nally, E.; Groah, S.L.; Pérez-Losada, M.; Caldovic, L.; Ljungberg, I.; Chandel, N.J.; Sprague, B.; Hsieh, M.H.; Pohl, H.G. Identification of Burkholderia fungorum in the urine of an individual with spinal cord injury and augmentation cystoplasty using 16S sequencing: Copathogen or innocent bystander? Spinal Cord Ser. Cases 2018, 4. [Google Scholar] [CrossRef]
- Loong, S.K.; Soh, Y.H.; Mahfodz, N.H.; Johari, J.; Abu Bakar, S. Synovial tissue infection with Burkholderia fungorum. Emerg. Infect. Dis. 2016, 22, 1834–1835. [Google Scholar] [CrossRef]
- Gerrits, G.P.; Klaassen, C.; Coenye, T.; Vandamme, P.; Meis, J.F. Burkholderia fungorum septicemia. Emerg. Infect. Dis. 2005, 11, 1115–1117. [Google Scholar] [CrossRef]
- Deris, Z.Z.; Van Rostenberghe, H.; Habsah, H.; Noraida, R.; Tan, G.C.; Chan, Y.Y.; Rosliza, A.R.; Ravichandran, M. First isolation of Burkholderia tropica from a neonatal patient successfully treated with imipenem. Int. J. Infect. Dis. 2010, 14, e73–e74. [Google Scholar] [CrossRef][Green Version]
- Marks, L.R.; Dodd, H.; Russo, T.A.; Berenson, C.S. Burkholderia ginsengisoli bacteraemia: Emergence of a novel pathogen. BMJ Case Rep. 2016, 2016, bcr2015213584. [Google Scholar] [CrossRef]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef]
- Paulitsch, F.; Dall’Agnol, R.F.; Delamuta, J.R.M.; Ribeiro, R.A.; da Silva Batista, J.S.; Hungria, M. Paraburkholderia guartelaensis sp. nov., a nitrogen-fixing species isolated from nodules of Mimosa gymnas in an ecotone considered as a hotspot of biodiversity in Brazil. Arch. Microbiol. 2019, 201, 1435–1446. [Google Scholar] [CrossRef]
- Reis, V.M.; Estrada-de los Santos, P.; Tenorio-Salgado, S.; Vogel, J.; Stoffels, M.; Guyon, S.; Mavingui, P.; Baldani, V.L.D.; Schmid, M.; Baldani, J.I.; et al. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 2155–2162. [Google Scholar] [CrossRef]
- Huo, Y.; Kang, J.P.; Kim, Y.J.; Yang, D.C. Paraburkholderia panacihumi sp. nov., an isolate from ginseng-cultivated soil, is antagonistic against root rot fungal pathogen. Arch. Microbiol. 2018, 200, 1151–1158. [Google Scholar] [CrossRef]
- Eberl, L.; Vandamme, P. Members of the genus Burkholderia: Good and bad guys. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Cyle, K.T.; Martinez, C.E.; Karasz, D.C.; Newman, J.D.; Buckley, D.H. Paraburkholderia solitsugae sp. nov. and paraburkholderia elongata sp. nov., phenolic acid-degrading bacteria isolated from forest soil and emended description of paraburkholderia madseniana. Int. J. Syst. Evol. Microbiol. 2020, 70, 5093–5105. [Google Scholar] [CrossRef]
- Nguyen, T.C.; Loganathan, P.; Nguyen, T.V.; Vigneswaran, S.; Kandasamy, J.; Slee, D.; Stevenson, G.; Naidu, R. Polycyclic aromatic hydrocarbons in road-deposited sediments, water sediments, and soils in Sydney, Australia: Comparisons of concentration distribution, sources and potential toxicity. Ecotoxicol. Environ. Saf. 2014, 104, 339–348. [Google Scholar] [CrossRef]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef]
- Morya, R.; Salvachúa, D.; Thakur, I.S. Burkholderia: An Untapped but Promising Bacterial Genus for the Conversion of Aromatic Compounds. Trends Biotechnol. 2020, 38, 963–975. [Google Scholar] [CrossRef]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef]
- Morya, R.; Kumar, M.; Singh, S.S.; Thakur, I.S. Genomic analysis of Burkholderia sp. ISTR5 for biofunneling of lignin-derived compounds. Biotechnol. Biofuels 2019, 12, 277. [Google Scholar] [CrossRef]
- Kögel-Knabner, I. The macromolecular organic composition of Plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 2002, 34, 139–162. [Google Scholar] [CrossRef]
- Ikeya, K.; Hikage, T.; Arai, S.; Watanabe, A. Size distribution of condensed aromatic rings in various soil humic acids. Org. Geochem. 2011, 42, 55–61. [Google Scholar] [CrossRef]
- Cepáková, S.; Frouz, J. Changes in chemical composition of litter during decomposition: A review of published 13C NMR spectra. J. Soil Sci. Plant Nutr. 2015, 15, 805–815. [Google Scholar] [CrossRef]
- Steffen, K.T.; Hatakka, A.; Hofrichter, M. Degradation of humic acids by the litter-decomposing basidiomycete Collybia dryophila. Appl. Environ. Microbiol. 2002, 68, 3442–3448. [Google Scholar] [CrossRef]
- Valášková, V.; Šnajdr, J.; Bittner, B.; Cajthaml, T.; Merhautová, V.; Hofrichter, M.; Baldrian, P. Production of lignocellulose-degrading enzymes and degradation of leaf litter by saprotrophic basidiomycetes isolated from a Quercus petraea forest. Soil Biol. Biochem. 2007, 39, 2651–2660. [Google Scholar] [CrossRef]
- Tikhonov, V.V.; Yakushev, A.V.; Zavgorodnyaya, Y.A.; Byzov, B.A.; Demin, V.V. Effects of humic acids on the growth of bacteria. Eurasian Soil Sci. 2010, 43, 305–313. [Google Scholar] [CrossRef]
- Simmons, B.A.; Loqué, D.; Ralph, J. Advances in modifying lignin for enhanced biofuel production. Curr. Opin. Plant Biol. 2010, 13, 312–319. [Google Scholar] [CrossRef]
- Menon, V.; Rao, M. Trends in bioconversion of lignocellulose: Biofuels, platform chemicals & biorefinery concept. Prog. Energy Combust. Sci. 2012, 38, 522–550. [Google Scholar]
- Akita, H.; Kimura, Z.I.; Yusoff, M.Z.M.; Nakashima, N.; Hoshino, T. Isolation and characterization of Burkholderia sp. strain CCA53 exhibiting ligninolytic potential. Springerplus 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-X.; Wang, T.; Gao, L.-N.; Yin, H.-J.; Lü, X. Isolation, identification and characterization of lignin-degrading bacteria from Qinling, China. J. Appl. Microbiol. 2017, 123, 1447–1460. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-F.; Santhanam, N.; Badri, D.V.; Hunter, W.J.; Manter, D.K.; Decker, S.R.; Vivanco, J.M.; Reardon, K.F. Isolation and characterization of lignin-degrading bacteria from rainforest soils. Biotechnol. Bioeng. 2013, 110, 1616–1626. [Google Scholar] [CrossRef]
- Beckham, G.T.; Johnson, C.W.; Karp, E.M.; Salvachúa, D.; Vardon, D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016, 42, 40–53. [Google Scholar] [CrossRef]
- Qi, S.S.; Bogdanov, A.; Cnockaert, M.; Acar, T.; Ranty-Roby, S.; Coenye, T.; Vandamme, P.; König, G.M.; Crüsemann, M.; Carlier, A. Induction of antibiotic specialized metabolism by co-culturing in a collection of phyllosphere bacteria. Environ. Microbiol. 2021, 23, 2132–2151. [Google Scholar] [CrossRef]
- Wilson, M.S.; Herrick, J.B.; Jeon, C.O.; Hinman, D.E.; Madsen, E.L. Horizontal transfer of phnAc dioxygenase genes within one of two phenotypically and genotypically distinctive naphthalene-degrading guilds from adjacent soil environments. Appl. Environ. Microbiol. 2003, 69, 2172–2181. [Google Scholar] [CrossRef]
- Schindowski, A.; Wittich, R.-M.; Fortnagel, P. Catabolism of 3,5-dichlorosalicylate by Pseudomonas species strain JWS. FEMS Microbiol. Lett. 1991, 84, 63–69. [Google Scholar] [CrossRef]
- Vanlaere, E.; Coenye, T.; Samyn, E.; Plas, C.; Govan, J.; Baets, F.; Boeck, K.; Knoop, C.; Vandamme, P. A novel strategy for the isolation and identification of environmental Burkholderia cepacia complex bacteria. FEMS Microbiol. Lett. 2005, 249, 303–307. [Google Scholar] [CrossRef]
- Salles, J.F.; Samyn, E.; Vandamme, P.; Van Veen, J.A.; Van Elsas, J.D. Changes in agricultural management drive the diversity of Burkholderia species isolated from soil on PCAT medium. Soil Biol. Biochem. 2006, 38, 661–673. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- de Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef]
- Masai, E.; Harada, K.; Peng, X.; Kitayama, H.; Katayama, Y.; Fukuda, M. Cloning and characterization of the ferulic acid catabolic genes of Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 2002, 68, 4416–4424. [Google Scholar] [CrossRef]
- Dumolin, C.; Aerts, M.; Verheyde, B.; Schellaert, S.; Vandamme, T.; Van Der Jeugt, F.; De Canck, E.; Cnockaert, M.; Wieme, A.D.; Cleenwerck, I.; et al. Introducing SPeDE: High-throughput dereplication and accurate determination of microbial diversity from matrix-assisted laser desorption–ionization time of flight mass spectrometry data. Msystems 2019, 4, 437–456. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic Sci. 2014, 9. [Google Scholar] [CrossRef]
- Brink, D.P.; Ravi, K.; Lidén, G.; Gorwa-Grauslund, M.F. Mapping the diversity of microbial lignin catabolism: Experiences from the eLignin database. Appl. Microbiol. Biotechnol. 2019, 103, 3979–4002. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczyński, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- De Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Kamimura, N.; Takahashi, K.; Mori, K.; Araki, T.; Fujita, M.; Higuchi, Y.; Masai, E. Bacterial catabolism of lignin-derived aromatics: New findings in a recent decade: Update on bacterial lignin catabolism. Environ. Microbiol. Rep. 2017, 9, 679–705. [Google Scholar] [CrossRef]
- Stopnisek, N.; Zuhlke, D.; Carlier, A.; Barberan, A.; Fierer, N.; Becher, D.; Riedel, K.; Eberl, L.; Weisskopf, L. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2016, 10, 253–264. [Google Scholar] [CrossRef]
- Jaspers, E.; Overmann, J. Ecological significance of microdiversity: Identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 2004, 70, 4831–4839. [Google Scholar] [CrossRef]
- Larkin, A.A.; Martiny, A.C. Microdiversity shapes the traits, niche space, and biogeography of microbial taxa. Environ. Microbiol. Rep. 2017, 9, 55–70. [Google Scholar] [CrossRef]
- Walle, I.V.; Mussche, S.; Samson, R.; Lust, N.; Lemeur, R. The above- and belowground carbon pools of two mixed deciduous forest stands located in East-Flanders (Belgium). Ann. For. Sci. 2001, 58, 507–517. [Google Scholar] [CrossRef]
- Wang, B.; Blondeel, H.; Baeten, L.; Djukic, I.; De Lombaerde, E.; Verheyen, K. Direct and understorey-mediated indirect effects of human-induced environmental changes on litter decomposition in temperate forest. Soil Biol. Biochem. 2019, 138, 107579. [Google Scholar] [CrossRef]
- Vanhellemont, M.; Baeten, L.; Verheyen, K. Relating changes in understorey diversity to environmental drivers in an ancient forest in northern Belgium. Plant Ecol. Evol. 2014, 147, 22–32. [Google Scholar] [CrossRef]
- Peeters, C.; Depoorter, E.; Praet, J.; Vandamme, P. Extensive cultivation of soil and water samples yields various pathogens in patients with cystic fibrosis but not Burkholderia multivorans. J. Cyst. Fibros. 2016, 15, 769–775. [Google Scholar] [CrossRef][Green Version]
- Gupta, R.; Singal, R.; Shankar, A.; Chander Kuhad, R.; Saxena, R.K. A modified plate assay for screening phosphate solubilizing microorganisms. J. Gen. Appl. Microbiol. 1994, 40, 255–260. [Google Scholar] [CrossRef]
- Wieme, A.D.; Spitaels, F.; Aerts, M.; De Bruyne, K.; Van Landschoot, A.; Vandamme, P. Identification of beer-spoilage bacteria using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Int. J. Food Microbiol. 2014, 185, 41–50. [Google Scholar] [CrossRef]
- Spilker, T.; Baldwin, A.; Bumford, A.; Dowson, C.G.; Mahenthiralingam, E.; Lipuma, J.J. Expanded Multilocus Sequence Typing for Burkholderia Species. J. Clin. Microbiol. 2009, 47, 2607–2610. [Google Scholar] [CrossRef]
- Peeters, C.; Zlosnik, J.E.A.; Spilker, T.; Hird, T.J.; LiPuma, J.J.; Vandamme, P. Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst. Appl. Microbiol. 2013, 36, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp, version 0.20.0, Fastp: An ultra-fast all-in-one FASTQ preprocessor. In Proceedings of the Bioinformatics; Oxford University Press: Oxford, UK, 2018; Volume 34, pp. i884–i890. [Google Scholar]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Li, H. BWA-MEM, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM, version 1.1.2, CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Seemann, T. Prokka, version 1.14.5, Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Ankenbrand, M.J.; Keller, A. BcgTree: Automatized phylogenetic tree building from bacterial core genomes. Genome 2016, 59, 783–791. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef]
- Seemann, T. barrnap 0.9: Rapid Ribosomal RNA Prediction. Available online: https://github.com/tseemann/barrnap (accessed on 18 December 2020).
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 1–6. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- MacFaddin, J. Biochemical Tests for Identification of Medical Bacteria, 3rd ed.; Williams Wilkins: Philadelphia, PA, USA, 2000. [Google Scholar]
- Sierra, G. A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 1957, 23, 15–22. [Google Scholar] [CrossRef]
- Strohalm, M.; Kavan, D.; Novák, P.; Volný, M.; Havlíček, V. mMass 3: A cross-platform software environment for precise analysis of mass spectrometric data. Anal. Chem. 2010, 82, 4648–4651. [Google Scholar] [CrossRef]
- Lim, J.H.; Baek, S.-H.; Lee, S.-T. Burkholderia sediminicola sp. nov., isolated from freshwater sediment. Int. J. Syst. Evol. Microbiol. 2008, 58, 565–569. [Google Scholar] [CrossRef]


| Strain | Source | Country | Year | Isolation Medium | Reference |
|---|---|---|---|---|---|
| Paraburkholderia aspalathi | |||||
| R-20943 (=Hg8) * | Coal tar-contaminated hillside soil | United States | Unknown | / | Wilson et al. [37] |
| R-69658 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69746 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69781 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69848 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69955 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-70036 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-75464 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75465 * | Forest topsoil | Belgium | 2018 | BCEM | This study |
| Paraburkholderia nemoris sp. nov. | |||||
| LMG 31836T * | Forest topsoil | Belgium | 2018 | BCEM | This study |
| LMG 22931 (=G47-12) * | Grassland soil | The Netherlands | 2003 | / | Salles et al. [40] |
| LMG 31840 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69608 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69616 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69643 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69776 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69861 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-70025 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-74083 | Forest topsoil | Belgium | 2018 | Pikovskaya | This study |
| R-74093 | Forest topsoil | Belgium | 2018 | Pikovskaya | This study |
| R-75460 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75463 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75466 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75776 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75777 * | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75793 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75795 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75796 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75797 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75798 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75799 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75800 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75801 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75803 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| Paraburkholderia domus sp. nov. | |||||
| LMG 31832T * | Forest topsoil | Belgium | 2018 | BCEM | This study |
| LMG 19510 (=DSM 6431) | Industrial waste deposit soil | Germany | Unknown | / | Schindowski et al. [38] |
| R-23782 | Succulent soil (porch) | Belgium | 2003 | / | Vanlaere et al. [39] |
| R-69749 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69927 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69973 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69977 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69980 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-69981 | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-70006 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-70199 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-70211 * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-75306 | Forest topsoil | Belgium | 2018 | Pikovskaya | This study |
| R-75471 * | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75473 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75474 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75475 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75476 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75477 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75478 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75479 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75480 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75481 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75482 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75484 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75485 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75486 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75487 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75488 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75589 | Forest topsoil | Belgium | 2018 | BCEM | This study |
| R-75773 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75774 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75784 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75788 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75789 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| R-75804 | Forest topsoil | Belgium | 2017 | PCAT | This study |
| Paraburkholderia haematera sp. nov. | |||||
| LMG 31837T * | Forest topsoil | Belgium | 2016 | DNG | This study |
| R-20943 | R-69658 | R-69746 | R-75465 | LMG 31836T | LMG 22931 | LMG 31840 | R-69608 | R-69776 | R-75777 | LMG 31832T | R-69749 | R-69927 | R-70006 | R-70199 | R-70211 | R-75471 | LMG 31837T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aspalathi LMG 27731T | 84.5 | 82.9 | 85.0 | 85.2 | 62.6 | 63.0 | 63.0 | 63.0 | 62.8 | 62.8 | 40.6 | 40.6 | 40.6 | 40.5 | 40.6 | 40.7 | 40.5 | 39.3 |
| P. madseniana RP11T | 57.4 | 57.3 | 57.1 | 56.7 | 57.3 | 57.6 | 57.5 | 57.5 | 57.0 | 57.4 | 41.9 | 42.0 | 41.9 | 41.9 | 41.9 | 41.9 | 41.9 | 41.0 |
| P. sediminicola LMG 24238T | 55.7 | 55.8 | 55.7 | 55.7 | 55.5 | 55.6 | 55.5 | 55.4 | 55.4 | 55.5 | 39.9 | 40.1 | 40.0 | 40.0 | 39.9 | 40.0 | 40.0 | 38.9 |
| P. solitsugae 1NT | 41.4 | 41.4 | 41.3 | 41.3 | 41.2 | 41.4 | 41.4 | 41.4 | 41.2 | 41.4 | 46.3 | 46.3 | 46.4 | 46.4 | 46.4 | 46.4 | 46.5 | 41.0 |
| P. elongata 5NT | 42.3 | 42.6 | 42.6 | 42.6 | 42.7 | 43.0 | 42.9 | 42.9 | 42.6 | 42.8 | 43.8 | 43.8 | 43.7 | 43.7 | 43.8 | 43.8 | 43.8 | 44.8 |
| P. caffeinilytica CF1T | 43.2 | 43.2 | 43.3 | 43.4 | 44.0 | 44.0 | 43.8 | 43.9 | 43.7 | 43.9 | 39.8 | 39.8 | 39.7 | 39.7 | 39.8 | 39.8 | 39.8 | 39.1 |
| R-20943 | / | 83.4 | 84.9 | 83.9 | 63.2 | 63.4 | 63.2 | 63.3 | 63.3 | 63.1 | 40.4 | 40.6 | 40.4 | 40.4 | 40.4 | 40.5 | 40.4 | 39.3 |
| R-69658 | 83.4 | / | 83.0 | 82.9 | 63.9 | 64.9 | 65.3 | 64.6 | 65.1 | 65.0 | 40.7 | 40.8 | 40.7 | 40.7 | 40.9 | 40.7 | 40.8 | 39.4 |
| R-69746 | 84.9 | 83.0 | / | 85.3 | 62.7 | 63.0 | 63.2 | 63.3 | 63.0 | 63.1 | 40.8 | 41.0 | 40.9 | 40.8 | 40.8 | 40.6 | 40.8 | 39.6 |
| R-75465 | 83.9 | 82.9 | 85.3 | / | 62.9 | 63.0 | 63.2 | 62.8 | 62.8 | 62.9 | 40.6 | 40.9 | 40.7 | 40.7 | 40.7 | 40.6 | 40.6 | 39.3 |
| LMG 31836T | 63.2 | 63.9 | 62.7 | 62.9 | / | 90.4 | 89.2 | 90.6 | 89.5 | 90.1 | 40.7 | 41.0 | 40.6 | 40.6 | 40.7 | 40.7 | 40.6 | 39.5 |
| LMG 22931 | 63.4 | 64.9 | 63.0 | 63.0 | 90.4 | / | 89.4 | 89.6 | 89.7 | 90.1 | 40.9 | 41.3 | 40.8 | 40.9 | 40.9 | 40.8 | 40.9 | 39.7 |
| LMG 31840 | 63.2 | 65.3 | 63.2 | 63.2 | 89.2 | 89.4 | / | 91.4 | 91.2 | 91.6 | 41.2 | 43.7 | 41.1 | 41.1 | 41.2 | 40.8 | 41.1 | 39.7 |
| R-69608 | 63.3 | 64.6 | 63.3 | 62.8 | 90.6 | 89.6 | 91.4 | / | 91.9 | 91.6 | 41.2 | 43.5 | 41.1 | 41.1 | 41.2 | 40.9 | 41.1 | 39.8 |
| R-69776 | 63.3 | 65.1 | 63.0 | 62.8 | 89.5 | 89.7 | 91.2 | 91.9 | / | 98.0 | 41.1 | 43.9 | 41.1 | 41.1 | 41.1 | 40.7 | 41.1 | 39.7 |
| R-75777 | 63.1 | 65.0 | 63.1 | 62.9 | 90.1 | 90.1 | 91.6 | 91.6 | 98.0 | / | 40.9 | 43.5 | 40.9 | 40.9 | 40.9 | 40.8 | 40.9 | 39.8 |
| LMG 31832T | 40.4 | 40.7 | 40.8 | 40.6 | 40.7 | 40.9 | 41.2 | 41.2 | 41.1 | 40.9 | / | 95.1 | 95.2 | 96.1 | 96.2 | 96.1 | 96.6 | 40.7 |
| R-69749 | 40.6 | 40.8 | 41.0 | 40.9 | 41.0 | 41.3 | 43.7 | 43.5 | 43.9 | 43.5 | 95.1 | / | 96.6 | 97.0 | 97.6 | 97.5 | 97.5 | 40.8 |
| R-69927 | 40.4 | 40.7 | 40.9 | 40.7 | 40.6 | 40.8 | 41.1 | 41.1 | 41.1 | 40.9 | 95.2 | 96.6 | / | 97.1 | 97.8 | 97.8 | 98.1 | 40.6 |
| R-70006 | 40.4 | 40.7 | 40.8 | 40.7 | 40.6 | 40.9 | 41.1 | 41.1 | 41.1 | 40.9 | 96.1 | 97.0 | 97.1 | / | 98.1 | 97.7 | 98.4 | 40.6 |
| R-70199 | 40.4 | 40.9 | 40.8 | 40.7 | 40.7 | 40.9 | 41.2 | 41.2 | 41.1 | 40.9 | 96.2 | 97.6 | 97.8 | 98.1 | / | 98.4 | 98.5 | 40.5 |
| R-70211 | 40.5 | 40.7 | 40.6 | 40.6 | 40.7 | 40.8 | 40.8 | 40.9 | 40.7 | 40.8 | 96.1 | 97.5 | 97.8 | 97.7 | 98.4 | / | 98.5 | 40.4 |
| R-75471 | 40.4 | 40.8 | 40.8 | 40.6 | 40.6 | 40.9 | 41.1 | 41.1 | 41.1 | 40.9 | 96.6 | 97.5 | 98.1 | 98.4 | 98.5 | 98.5 | / | 40.5 |
| LMG 31837T | 39.3 | 39.4 | 39.6 | 39.3 | 39.5 | 39.7 | 39.7 | 39.8 | 39.7 | 39.8 | 40.7 | 40.8 | 40.6 | 40.6 | 40.5 | 40.4 | 40.5 | / |
| R-20943 | R-69658 | R-69746 | R-75465 | LMG 31836T | LMG 22931 | LMG 31840 | R-69608 | R-69776 | R-75777 | LMG 31832T | R-69749 | R-69927 | R-70006 | R-70199 | R-70211 | R-75471 | LMG 31837T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aspalathi LMG 27731T | 98.2 | 98.0 | 98.3 | 98.3 | 95.1 | 95.2 | 95.2 | 95.3 | 95.2 | 95.2 | 89.9 | 89.8 | 90.0 | 89.8 | 89.8 | 89.9 | 89.9 | 89.7 |
| P. madseniana RP11T | 94.2 | 94.2 | 94.2 | 94.1 | 94.1 | 94.3 | 94.2 | 94.3 | 94.2 | 94.3 | 90.2 | 90.3 | 90.2 | 90.2 | 90.2 | 90.2 | 90.3 | 90.1 |
| P. sediminicola LMG 24238T | 94.0 | 94.0 | 94.0 | 93.9 | 93.8 | 93.9 | 94.0 | 93.9 | 93.9 | 93.9 | 89.6 | 89.7 | 89.7 | 89.7 | 89.6 | 89.6 | 89.6 | 89.3 |
| P. solitsugae 1NT | 90.1 | 90.0 | 90.0 | 90.0 | 90.1 | 90.2 | 90.1 | 90.3 | 90.1 | 90.3 | 91.5 | 91.5 | 91.5 | 91.6 | 91.5 | 91.6 | 91.6 | 90.0 |
| P. elongata 5NT | 90.4 | 90.5 | 90.5 | 90.5 | 90.7 | 90.8 | 90.6 | 90.6 | 90.6 | 90.6 | 90.8 | 90.7 | 90.8 | 90.7 | 90.7 | 90.8 | 90.8 | 91.2 |
| P. caffeinilytica CF1T | 90.7 | 90.8 | 90.7 | 90.8 | 90.9 | 91.0 | 90.9 | 90.9 | 90.9 | 91.0 | 89.6 | 89.6 | 89.6 | 89.5 | 89.6 | 89.5 | 89.6 | 89.4 |
| R-20943 | / | 98.0 | 98.2 | 98.1 | 95.2 | 95.4 | 95.3 | 95.3 | 95.3 | 95.3 | 89.8 | 89.8 | 89.9 | 89.7 | 89.8 | 89.9 | 89.8 | 89.5 |
| R-69658 | 98.0 | / | 98.0 | 98.1 | 95.4 | 95.6 | 95.6 | 95.5 | 95.6 | 95.6 | 89.9 | 89.8 | 89.9 | 89.9 | 89.9 | 89.8 | 89.9 | 89.5 |
| R-69746 | 98.2 | 98.0 | / | 98.3 | 95.1 | 95.2 | 95.3 | 95.3 | 95.3 | 95.3 | 89.8 | 89.8 | 89.8 | 89.8 | 89.8 | 89.8 | 89.9 | 89.7 |
| R-75465 | 98.1 | 98.1 | 98.3 | / | 95.2 | 95.3 | 95.3 | 95.2 | 95.2 | 95.3 | 89.9 | 90.0 | 89.9 | 89.8 | 89.9 | 89.9 | 89.9 | 89.5 |
| LMG 31836T | 95.2 | 95.4 | 95.1 | 95.2 | / | 98.9 | 98.7 | 98.9 | 98.7 | 98.8 | 89.9 | 89.9 | 89.7 | 89.9 | 89.7 | 89.9 | 89.8 | 89.6 |
| LMG 22931 | 95.4 | 95.6 | 95.2 | 95.3 | 98.9 | / | 98.7 | 98.8 | 98.7 | 98.8 | 89.9 | 90.1 | 90.0 | 90.0 | 90.1 | 89.9 | 89.9 | 89.7 |
| LMG 31840 | 95.3 | 95.6 | 95.3 | 95.3 | 98.7 | 98.7 | / | 99.0 | 98.9 | 99.0 | 90.1 | 90.9 | 90.0 | 90.0 | 90.1 | 89.9 | 90.1 | 89.7 |
| R-69608 | 95.3 | 95.5 | 95.3 | 95.2 | 98.9 | 98.8 | 99.0 | / | 99.0 | 99.0 | 90.0 | 90.8 | 90.0 | 90.0 | 90.0 | 89.9 | 90.1 | 89.8 |
| R-69776 | 95.3 | 95.6 | 95.3 | 95.2 | 98.7 | 98.7 | 98.9 | 99.0 | / | 99.7 | 90.0 | 90.9 | 90.0 | 89.9 | 90.0 | 90.0 | 90.1 | 89.6 |
| R-75777 | 95.3 | 95.6 | 95.3 | 95.3 | 98.8 | 98.8 | 99.0 | 99.0 | 99.7 | / | 89.9 | 90.8 | 89.9 | 89.9 | 89.9 | 89.9 | 90.0 | 89.8 |
| LMG 31832T | 89.8 | 89.9 | 89.8 | 89.9 | 89.9 | 89.9 | 90.1 | 90.0 | 90.0 | 89.9 | / | 99.4 | 99.4 | 99.5 | 99.5 | 99.5 | 99.5 | 90.0 |
| R-69749 | 89.8 | 89.8 | 89.8 | 90.0 | 89.9 | 90.1 | 90.9 | 90.8 | 90.9 | 90.8 | 99.4 | / | 99.6 | 99.6 | 99.6 | 99.6 | 99.7 | 90.1 |
| R-69927 | 89.9 | 89.9 | 89.8 | 89.9 | 89.7 | 90.0 | 90.0 | 90.0 | 90.0 | 89.9 | 99.4 | 99.6 | / | 99.5 | 99.6 | 99.6 | 99.7 | 90.1 |
| R-70006 | 89.7 | 89.9 | 89.8 | 89.8 | 89.9 | 90.0 | 90.0 | 90.0 | 89.9 | 89.9 | 99.5 | 99.6 | 99.5 | / | 99.7 | 99.7 | 99.8 | 90.1 |
| R-70199 | 89.8 | 89.9 | 89.8 | 89.9 | 89.7 | 90.1 | 90.1 | 90.0 | 90.0 | 89.9 | 99.5 | 99.6 | 99.6 | 99.7 | / | 99.7 | 99.8 | 90.1 |
| R-70211 | 89.9 | 89.8 | 89.8 | 89.9 | 89.9 | 89.9 | 89.9 | 89.9 | 90.0 | 89.9 | 99.5 | 99.6 | 99.6 | 99.7 | 99.7 | / | 99.8 | 90.0 |
| R-75471 | 89.8 | 89.9 | 89.9 | 89.9 | 89.8 | 89.9 | 90.1 | 90.1 | 90.1 | 90.0 | 99.5 | 99.7 | 99.7 | 99.8 | 99.8 | 99.8 | / | 90.1 |
| LMG 31837T | 89.5 | 89.5 | 89.7 | 89.5 | 89.6 | 89.7 | 89.7 | 89.8 | 89.6 | 89.8 | 90.0 | 90.1 | 90.1 | 90.1 | 90.1 | 90.0 | 90.1 | / |
| Genome Size (Mb) | Mol % G+C | No. of Contigs | N50 (kb) | No. of CDS | No. of tRNA Genes | No. of rRNA Genes | Accession Number | |
|---|---|---|---|---|---|---|---|---|
| LMG 24238T | 8.64 | 61.40 | 40 | 513.2 | 7690 | 60 | 2 | CADIKC000000000 |
| R-20943 | 8.74 | 61.34 | 39 | 857.6 | 7831 | 60 | 3 | CAJNBA000000000 |
| R-69658 | 9.05 | 61.43 | 380 | 70.9 | 8212 | 60 | 2 | CAJNAU000000000 |
| R-69746 | 9.66 | 61.25 | 520 | 61.7 | 8837 | 61 | 3 | CAJNBE000000000 |
| R-75465 | 9.50 | 61.28 | 387 | 92.4 | 8623 | 60 | 3 | CAJNAX000000000 |
| LMG 31836T | 9.27 | 61.59 | 128 | 261.5 | 8438 | 64 | 2 | CAJNBO000000000 |
| LMG 22931 | 8.64 | 61.47 | 68 | 263.0 | 7692 | 62 | 3 | CAJNBC000000000 |
| LMG 31840 | 8.85 | 61.40 | 152 | 170.6 | 7822 | 60 | 4 | CAJNBI000000000 |
| R-69608 | 8.91 | 61.40 | 116 | 252.5 | 7898 | 61 | 3 | CAJNAW000000000 |
| R-69776 | 9.28 | 61.30 | 113 | 288.6 | 8167 | 62 | 2 | CAJNBH000000000 |
| R-75777 | 9.08 | 61.33 | 98 | 320.9 | 8045 | 61 | 3 | CAJNBN000000000 |
| LMG 31832T | 8.75 | 61.32 | 99 | 229.8 | 7830 | 57 | 3 | CAJNAT000000000 |
| R-69749 | 9.46 | 61.14 | 147 | 210.7 | 8462 | 57 | 2 | CAJNAY000000000 |
| R-69927 | 8.71 | 61.35 | 133 | 205.4 | 7784 | 59 | 3 | CAJNAV000000000 |
| R-70006 | 9.16 | 61.19 | 115 | 208.5 | 8250 | 59 | 2 | CAJNBD000000000 |
| R-70199 | 9.06 | 61.24 | 124 | 265.3 | 8195 | 62 | 3 | CAJNBB000000000 |
| R-70211 | 8.63 | 61.28 | 91 | 323.0 | 7680 | 59 | 2 | CAJNAS000000000 |
| R-75471 | 8.19 | 61.42 | 83 | 359.9 | 7292 | 56 | 2 | CAJNAZ000000000 |
| LMG 31837T | 7.82 | 61.71 | 76 | 270.9 | 6967 | 58 | 2 | CAJNBK000000000 |
| LMG 31836T (cluster B) | LMG 22931 (cluster B) | LMG 31840 (cluster B) | R-69608 (cluster B) | R-69776 (cluster B) | R-75777 (cluster B) | LMG 31832T (cluster C) | R-69749 (cluster C) | R-69927 (cluster C) | R-70006 (cluster C) | R-70199 (cluster C) | R-70211 (cluster C) | R-75471 (cluster C) | LMG 31837T | P. aspalathi LMG 27731T | P. aspalathi R-20943 | P. aspalathi R-69658 | P. aspalathi R-69746 | P. aspalathi R-75465 | P. madseniana RP11T | P. sediminicola LMG 24238T | P. solitsugae 1NT | P. elongata 5NT | P. caffeinilytica CF1T | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
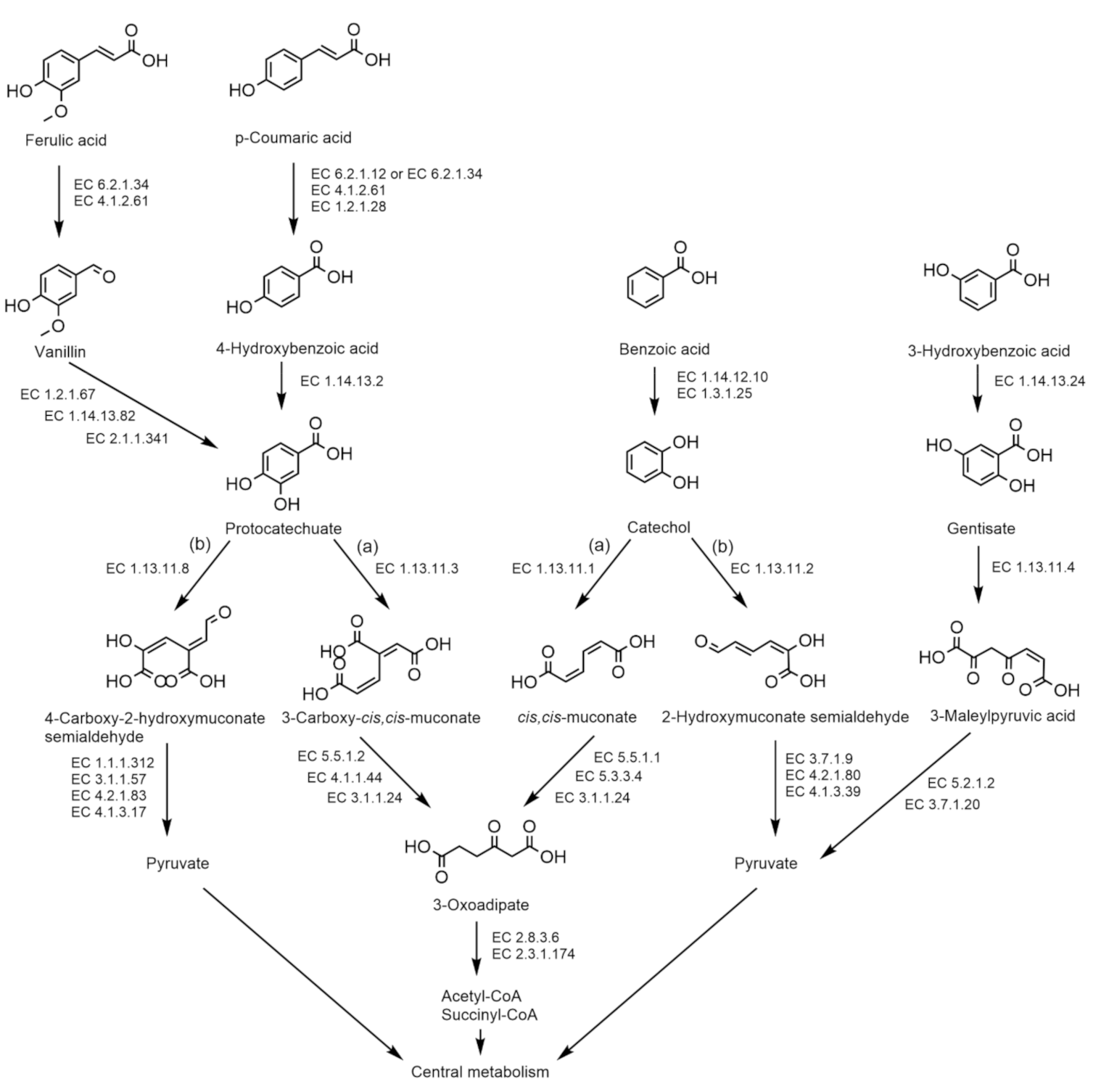
| Catechol (meta-cleavage pathway) | ||||||||||||||||||||||||
| Catechol 2,3 dioxygenase (EC 1.13.11.2) | − | − | + | + | + | + | + | + | + | + | + | − | + | − | − | + | − | − | − | − | − | − | + | − |
| 2-Hydroxymuconic semialdehyde hydrolase (EC 3.7.1.9) | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| 2-Oxopent-4-enoate hydratase (EC 4.2.1.80) | − | − | − | + | − | − | + | + | + | + | + | + | + | − | − | − | − | − | − | + | + | + | + | − |
| 4-Hydroxy-2-oxovalerate aldolase (EC 4.1.3.39) | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | + | + | + |
| Catechol (ortho-cleavage pathway) | ||||||||||||||||||||||||
| Catechol 1,2 dioxygenase (EC 1.13.11.1) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Muconate cycloisomerase (EC 5.5.1.1) | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + |
| Muconolactone isomerase (EC 5.3.3.4) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 3-Oxoadipate enol-lactonase (EC 3.1.1.24) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ferulic acid to vanillin | ||||||||||||||||||||||||
| Feruloyl-CoA synthase (EC 6.2.1.34) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| Feruloyl-CoA hydratase/lyase (EC 4.1.2.61) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| Vanillin to protocatechuate | ||||||||||||||||||||||||
| Vanillin dehydrogenase (EC 1.2.1.67) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | − |
| Vanillate monooxygenase (EC 1.14.13.82) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Vanillate/3-O-methylgallate O-demethylase (EC 2.1.1.341) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| p-Coumaric acid to protocatechuate | ||||||||||||||||||||||||
| 4-Coumarate-CoA ligase (EC 6.2.1.12) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Feruloyl-CoA hydratase/lyase (EC 4.1.2.61) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| Benzaldehyde dehydrogenase (EC 1.2.1.28) | + | + | + | + | + | + | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + | + | + | + |
| 4-Hydroxybenzoate 3-monooxygenase (EC 1.14.13.2) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Protocatechuate (meta-cleavage pathway) | ||||||||||||||||||||||||
| Protocatechuate 4,5-dioxygenase (EC 1.13.11.8) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| 2-Hydroxy-4-carboxymuconate 6-semialdehyde dehydrogenase (EC 1.1.1.312) | − | + | − | − | − | − | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | + | − | − |
| 2-Pyrone-4,6-dicarboxylate hydrolase (EC 3.1.1.57) | − | + | − | − | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | + | − | − |
| 4-Oxalomesaconate hydratase (EC 4.2.1.83) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 4-Carboxy-4-hydroxy-2-oxoadipate aldolase (EC 4.1.3.17) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| 3-Hydroxybenzoic acid | ||||||||||||||||||||||||
| 3-Hydroxybenzoate 6-monooxygenase (EC 1.14.13.24) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − |
| Gentisate 1,2-dioxygenase (EC 1.13.11.4) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | + | + | + | − | + | + | − |
| Maleylacetoacetate isomerase (EC 5.2.1.2) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 3-Fumarylpyruvate hydrolase (EC 3.7.1.20) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Characteristic | LMG 31836T | LMG 31832T | LMG 31837T | P. aspalathi LMG 27731T | P. madseniana RP11T | P. sediminicola LMG 24238T | P. solitsugae 1NT | P. elongata 5NT | P. caffeinilytica CF1T |
|---|---|---|---|---|---|---|---|---|---|
| Hemolysis on horse blood agar | w/+ | w/+ | w/+ | −/+ | −/+ | −/+ | −/w | −/− | −/+ |
| Growth in the presence of 1% NaCl | +/+ | +/+ | +/+ | +/+ | +/+ | −/− | −/− | −/− | −/+ |
| Oxidase activity | + | + | + | + | + | + | w (d) | w (d) | + (d) |
| Catalase activity | w (d) | w | w | + | w | + | + | w (d) | w |
| Lipase activity on: | |||||||||
| Tween 20 | w/+ | w/+ | w/+ | +/+ | +/+ | −/+ | +/+ | +/+ | −/+ |
| Tween 40 | +/+ | w/+ | w/+ | −/+ | −/+ | −/+ | +/+ | −/− | −/+ |
| Tween 60 | +/+ | −/+ | −/+ | −/+ | +/+ | +/+ | +/+ | w/w | +/+ |
| Tween 80 | w/+ | +/+ | +/+ | +/+ | +/+ | −/+ | +/+ | w/w | +/+ |
| API 20NE: | |||||||||
| Nitrate reduction | + | + | − | − | − | − | − | − | − |
| Beta-galactosidase (PNPG) | − | − | − | w | − | w | − | − | − |
| Assimilation of (API 20NE): | |||||||||
| Arabinose | w | w | w | + | w | + | + | + | + |
| N-acetyl-glucosamine | + | − | + | + | + | + | + | + | + |
| Maltose | − | − | − | − | − | − | − | − | w |
| Adipic acid | + | w | w | + | + | + | + | + | w |
| Trisodium citrate | + | − | + | + | + | + | − | w | + |
| Phenylacetic acid | + | − | + | + | + | + | + | + | + |
| API ZYM: | |||||||||
| Alkaline phosphatase | w | w | w | + | w | + | w | w | + |
| Phosphoamidase | w | + | + | + | w | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanwijnsberghe, S.; Peeters, C.; De Ridder, E.; Dumolin, C.; Wieme, A.D.; Boon, N.; Vandamme, P. Genomic Aromatic Compound Degradation Potential of Novel Paraburkholderia Species: Paraburkholderia domus sp. nov., Paraburkholderia haematera sp. nov. and Paraburkholderia nemoris sp. nov. Int. J. Mol. Sci. 2021, 22, 7003. https://doi.org/10.3390/ijms22137003
Vanwijnsberghe S, Peeters C, De Ridder E, Dumolin C, Wieme AD, Boon N, Vandamme P. Genomic Aromatic Compound Degradation Potential of Novel Paraburkholderia Species: Paraburkholderia domus sp. nov., Paraburkholderia haematera sp. nov. and Paraburkholderia nemoris sp. nov. International Journal of Molecular Sciences. 2021; 22(13):7003. https://doi.org/10.3390/ijms22137003
Chicago/Turabian StyleVanwijnsberghe, Sarah, Charlotte Peeters, Emmelie De Ridder, Charles Dumolin, Anneleen D. Wieme, Nico Boon, and Peter Vandamme. 2021. "Genomic Aromatic Compound Degradation Potential of Novel Paraburkholderia Species: Paraburkholderia domus sp. nov., Paraburkholderia haematera sp. nov. and Paraburkholderia nemoris sp. nov." International Journal of Molecular Sciences 22, no. 13: 7003. https://doi.org/10.3390/ijms22137003
APA StyleVanwijnsberghe, S., Peeters, C., De Ridder, E., Dumolin, C., Wieme, A. D., Boon, N., & Vandamme, P. (2021). Genomic Aromatic Compound Degradation Potential of Novel Paraburkholderia Species: Paraburkholderia domus sp. nov., Paraburkholderia haematera sp. nov. and Paraburkholderia nemoris sp. nov. International Journal of Molecular Sciences, 22(13), 7003. https://doi.org/10.3390/ijms22137003






