Concentrated Raw Fibers Enhance the Fiber-Degrading Capacity of a Synthetic Human Gut Microbiome
Abstract
:1. Introduction
2. Results
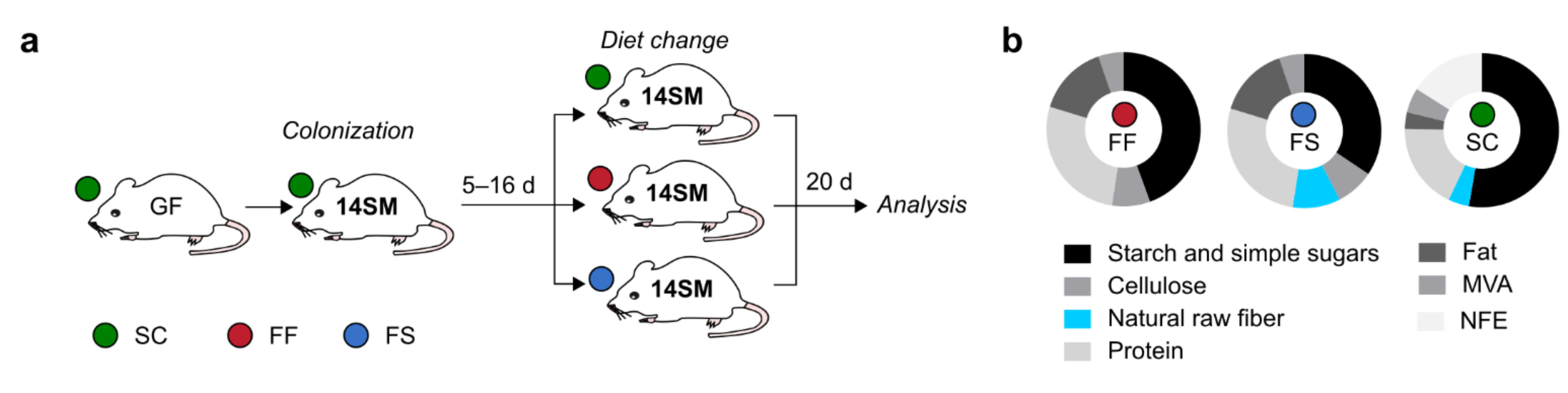
2.1. Experimental Setup to Study the Specific Effects of Concentrated Raw Fibers on Composition and Function of A 14-Member Microbial Community in Mice
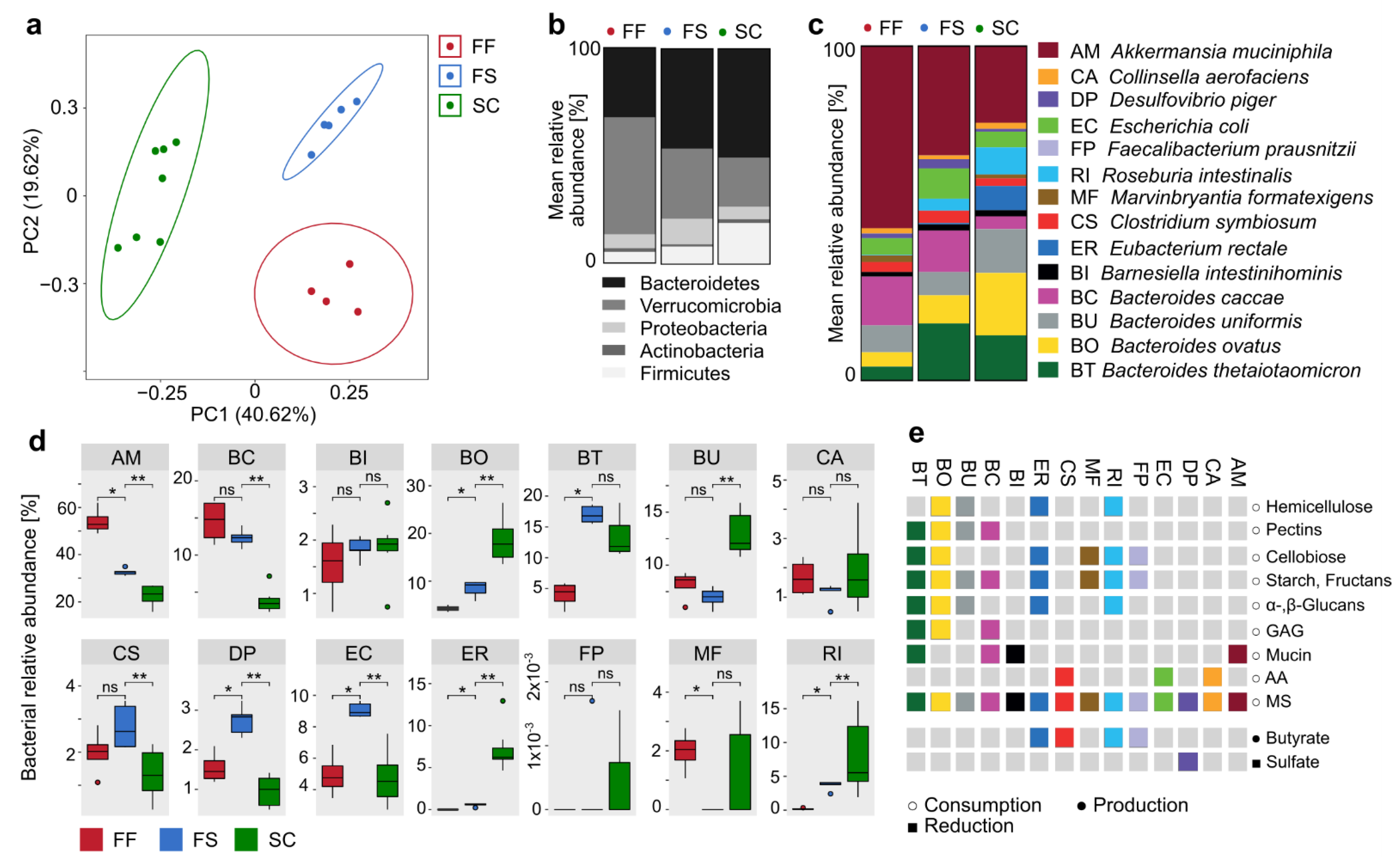
2.2. Increase in Relative Abundance of Certain Fiber-Degrading Commensals in Response to Dietary Fiber Supplementation
2.3. Inter-Bacterial Relations in Relative Abundance within the 14-Member Microbial Community
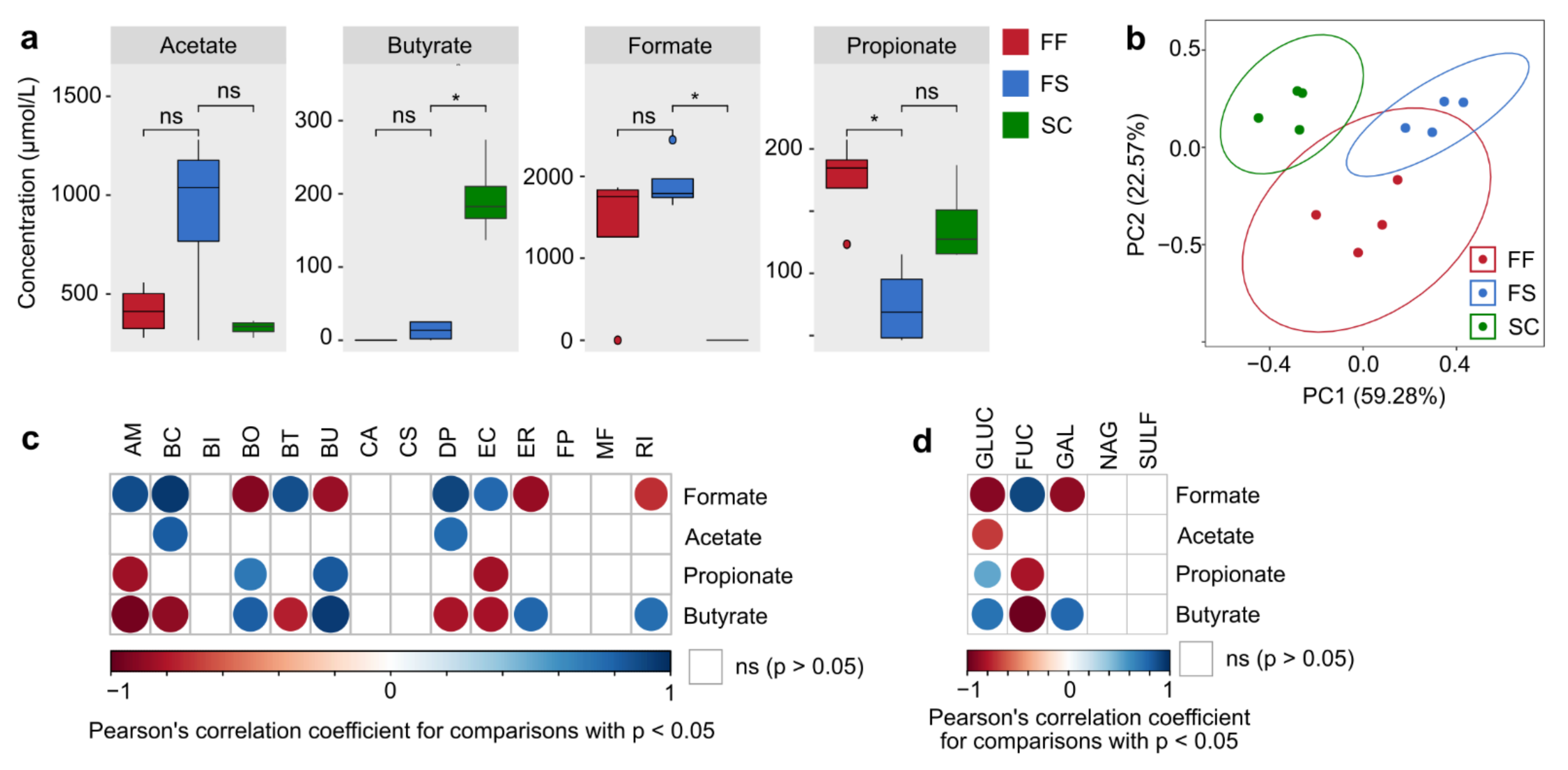
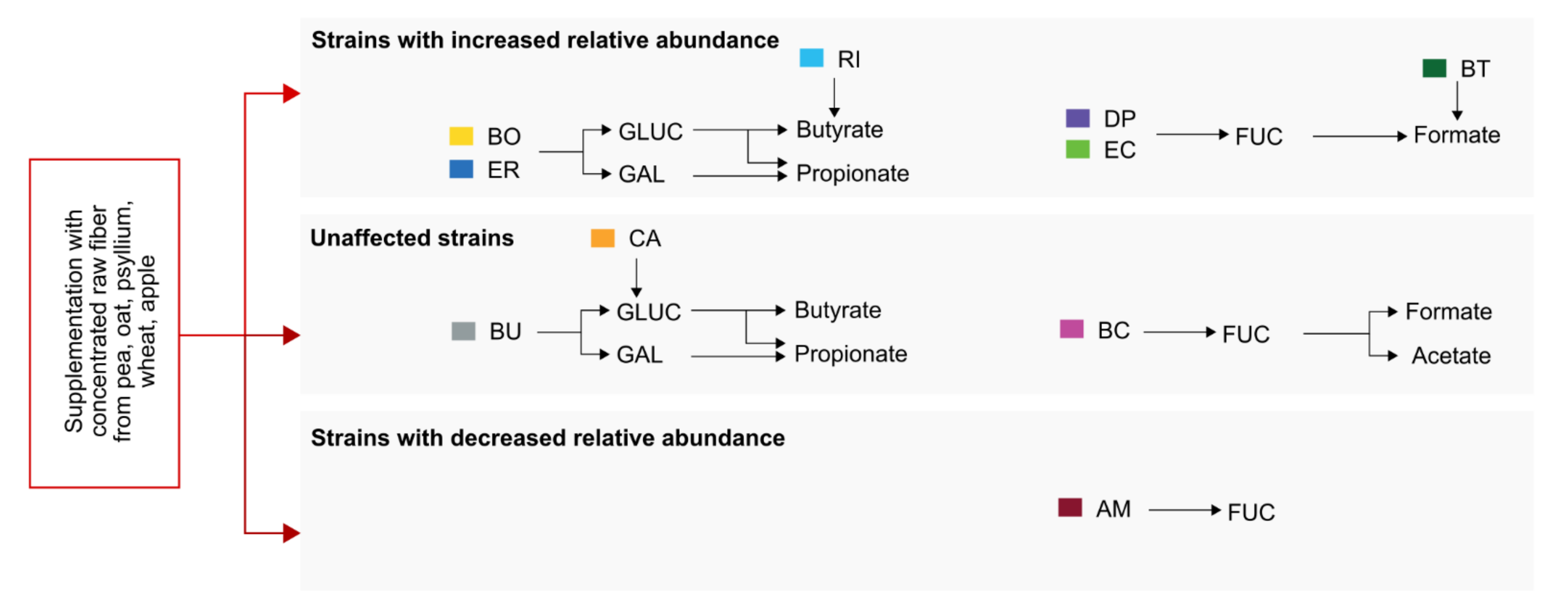
2.4. Concentrated Raw Fiber Supplementation Is Associated with Changes in Fecal Bacterial Glycan-Degrading Enzymes
2.5. Changes of Bacterial Glycan-Degrading Enzyme Activities Are Interlinked with a Specific Short-Chain Fatty Acid Production Profile
3. Discussion
4. Materials and Methods
4.1. Mouse Experiments
4.2. Animal Diets
4.3. Culturing and Colonization of Germ-Free Mice with Synthetic Microbiota
4.4. Isolation of Bacterial DNA from Mouse Fecal Samples
4.5. Illumina 16S rRNA Gene Sequencing and Data Analysis
4.6. Intestinal Fatty Acid Analysis
4.7. Detection of Glycan-Degrading Enzyme Activities in Fecal Samples
4.8. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | Amino acids |
| AM | Akkermansia muciniphila |
| BC | Bacteroides caccae |
| BI | Barnesiella intestinihominis |
| BO | Bacteroides ovatus |
| BT | Bacteroides thetaiotaomicron |
| BU | Bacteroides uniformis |
| CA | Collinsella aerofaciens |
| CRF | Concentrated raw fiber |
| CS | Clostridium symbiosum |
| DP | Desulfovibrio piger |
| EC | Escherichia coli |
| ER | Eubacterium rectale |
| FF | Fiber-free |
| FP | Faecalibacterium prausnitzii |
| FS | Fiber-supplemented |
| FUC | α-Fucosidase |
| GAG | Glucosaminoglycans |
| GAL | α-Galactosidase |
| GF | Germ-free |
| GLUC | β-Glucosidase |
| HPLC | High-performance liquid chromatography |
| I/S-ratio | Insoluble-to-soluble fibers |
| MF | Marvinbryantia formatexigens |
| MS | Mono-saccharides |
| MVA | Minerals, vitamins, and ash |
| NAG | β-N-Acetylglucosaminidase |
| NFE | Nitrogen-free extracts |
| PCA | Principal components cnalysis |
| RI | Roseburia intestinalis |
| SC | Standard mouse chow |
| SCFA | Short-chain fatty acid |
| SM | Synthetic microbiome |
| SULF | Sulfatase |
References
- Statovci, D.; Aguilera, M.; Mac Sharry, J.; Melgar, S. The Impact of Western Diet and Nutrients on the Microbiota and Immune Response at Mucosal Interfaces. Front. Immunol. 2017, 8, 838. [Google Scholar] [CrossRef] [Green Version]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Scott, K.P.; Antoine, J.-M.; Midtvedt, T.; Van Hemert, S. Manipulating the gut microbiota to maintain health and treat disease. Microb. Ecol. Health Dis. 2015, 26, 25877. [Google Scholar] [CrossRef]
- Rezende, E.S.V.; Lima, G.C.; Naves, M.M.V. Dietary fibers as beneficial microbiota modulators: A proposal classification by prebiotic categories. Nutrition 2021, 89, 111217. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brüssow, H. Probiotics and prebiotics in clinical tests: An update. F1000Research 2019, 8, 1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Ortigoza, V.; Garcia-Amezquita, L.E.; Serna-Saldívar, S.O.; Welti-Chanes, J. Advances in the Functional Characterization and Extraction Processes of Dietary Fiber. Food Eng. Rev. 2016, 8, 251–271. [Google Scholar] [CrossRef]
- De Wit, N.; Esser, D.; Siebelink, E.; Fischer, A.; Sieg, J.; Mes, J.J. Extrinsic wheat fibre consumption enhances faecal bulk and stool frequency; a randomized controlled trial. Food Funct. 2019, 10, 646–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamargo, A.; Cueva, C.; Alvarez, M.D.; Herranz, B.; Moreno-Arribas, M.V.; Laguna, L. Physical effects of dietary fibre on simulated luminal flow, studied byin vitrodynamic gastrointestinal digestion and fermentation. Food Funct. 2019, 10, 3452–3465. [Google Scholar] [CrossRef] [Green Version]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [Green Version]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; Von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I.; Consortium, N.-P.D. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Stoll, M.L.; Kumar, R.; Morrow, C.D.; Lefkowitz, E.J.; Cui, X.; Genin, A.; Cron, R.Q.; Elson, C. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res. Ther. 2014, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Earley, H.; Lennon, G.; Balfe, Á.; Coffey, J.C.; Winter, D.C.; O’Connell, P.R. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vetvicka, V.; Vannucci, L.; Sima, P.; Richter, J. Beta Glucan: Supplement or Drug? From Laboratory to Clinical Trials. Molecules 2019, 24, 1251. [Google Scholar] [CrossRef] [Green Version]
- Stams, A.J.M.; Plugge, C.M. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Genet. 2009, 7, 568–577. [Google Scholar] [CrossRef]
- Wegmann, U.; Palop, C.N.; Mayer, M.J.; Crost, E.; Narbad, A. Complete Genome Sequence of Desulfovibrio piger FI11049. Genome Announc. 2017, 5, e01528-16. [Google Scholar] [CrossRef] [Green Version]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Genet. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Lin, L.; Zhang, J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietzke, M.; Meiser, J.; Vazquez, A. Formate metabolism in health and disease. Mol. Metab. 2020, 33, 23–37. [Google Scholar] [CrossRef]
- Hughes, E.R.; Winter, M.G.; Duerkop, B.A.; Spiga, L.; De Carvalho, T.F.; Zhu, W.; Gillis, C.C.; Büttner, L.; Smoot, M.P.; Behrendt, C.L.; et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe 2017, 21, 208–219. [Google Scholar] [CrossRef] [Green Version]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nat. Cell Biol. 2016, 529, 212–215. [Google Scholar] [CrossRef] [Green Version]
- Russell, W.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.; Wallace, R.J.; et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Buyken, A.; Goletzke, J.; Joslowski, G.; Felbick, A.; Cheng, G.; Herder, C.; Brand-Miller, J.C. Association between carbohydrate quality and inflammatory markers: Systematic review of observational and interventional studies. Am. J. Clin. Nutr. 2014, 99, 813–833. [Google Scholar] [CrossRef] [Green Version]
- Dewulf, E.M.; Cani, P.D.; Neyrinck, A.; Possemiers, S.; Van Holle, A.; Muccioli, G.; Deldicque, L.; Bindels, L.B.; Pachikian, B.D.; Sohet, F.M.; et al. Inulin-type fructans with prebiotic properties counteract GPR43 overexpression and PPARγ-related adipogenesis in the white adipose tissue of high-fat diet-fed mice. J. Nutr. Biochem. 2011, 22, 712–722. [Google Scholar] [CrossRef]
- Deehan, E.C.; Yang, C.; Perez-Muñoz, M.E.; Nguyen, N.K.; Cheng, C.C.; Triador, L.; Zhang, Z.; Bakal, J.A.; Walter, J. Precision Microbiome Modulation with Discrete Dietary Fiber Structures Directs Short-Chain Fatty Acid Production. Cell Host Microbe 2020, 27, 389–404.e6. [Google Scholar] [CrossRef]
- Volkova, A.; Ruggles, K.V. Predictive Metagenomic Analysis of Autoimmune Disease Identifies Robust Autoimmunity and Disease Specific Microbial Signatures. Front. Microbiol. 2021, 12, 621310. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, Y.; Yang, Y.; Gong, W.; Sun, X.; Yang, L.; Zhang, Q.; Jin, M. Akkermansia muciniphila Improves Host Defense Against Influenza Virus Infection. Front. Microbiol. 2021, 11, 586476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J. Funct. Foods 2017, 33, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Becken, B.; Davey, L.; Middleton, D.R.; Mueller, K.D.; Sharma, A.; Holmes, Z.C.; Dallow, E.; Remick, B.; Barton, G.M.; David, L.A.; et al. Genotypic and Phenotypic Diversity among Human Isolates of Akkermansia muciniphila. mBio 2021, 12. [Google Scholar] [CrossRef]
- Swidsinski, A.; Loening-Baucke, V.; Theissig, F.; Engelhardt, H.; Bengmark, S.; Koch, S.; Lochs, H.; Dörffel, Y. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 2007, 56, 343–350. [Google Scholar] [CrossRef]
- Van der Post, S.; Jabbar, K.; Birchenough, G.; Arike, L.; Akhtar, N.; Sjovall, H.; Johansson, M.E.V.; Hansson, G.C. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 2019, 68, 2142–2151. [Google Scholar] [CrossRef] [Green Version]
- Odenwald, M.A.; Turner, M.A.O.J.R. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 9–21. [Google Scholar] [CrossRef]
- Steimle, A.; De Sciscio, A.; Neumann, M.; Grant, E.T.; Pereira, G.V.; Ohno, H.; Martens, E.C.; Desai, M.S. Constructing a gnotobiotic mouse model with a synthetic human gut microbiome to study host–microbe cross talk. STAR Protocols 2021, 2, 100607. [Google Scholar] [CrossRef]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2016, 533, 543–546. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D. MiSeq SOP. Available online: https://mothur.org/wiki/miseq_sop/ (accessed on 23 October 2019).
- Greenhalgh, K.; Ramiro-Garcia, J.; Heinken, A.; Ullmann, P.; Bintener, T.; Pacheco, M.P.; Baginska, J.; Shah, P.; Frachet, A.; Halder, R.; et al. Integrated In Vitro and In Silico Modeling Delineates the Molecular Effects of a Synbiotic Regimen on Colorectal-Cancer-Derived Cells. Cell Rep. 2019, 27, 1621–1632.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steimle, A.; Grant, E.T.; Desai, M.S. Quantitative assay to detect bacterial glycan-degrading enzyme activities in mouse and human fecal samples. STAR Protoc. 2021, 2, 100326. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Bryan, J. Read Excel Files. R Package Version 1.3.1. Available online: https://CRAN.R-project.org/package=readxl (accessed on 3 June 2021).
- Wickham, H. Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kassambara, A. ‘ggplot2’ Based Publication Ready Plots. R Package Version 0.4.0. Available online: https://CRAN.R-project.org/package=ggpubr (accessed on 3 June 2021).
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Results of Popular R Packages. R J. 2016, 8, 474–485. [Google Scholar] [CrossRef] [Green Version]
- Harrell, F.E.J. Hmisc: Harrell Miscellaneous. R Package Version 4.5-0. Available online: https://CRAN.R-project.org/package=Hmisc (accessed on 3 June 2021).
- Xie, Y. xfun: Supporting Functions for Packages Maintained by ‘Yihui Xie’. R Package Version 0.23. Available online: https://CRAN.R-project.org/package=xfun (accessed on 3 June 2021).
- Wei, T.; Simko, V. Visualization of a Correlation Matrix (Version 0.84). Available online: https://github.com/taiyun/corrplot (accessed on 3 June 2021).
- Kuhn, M.; Jackson, S.; Cimentada, J. Correlations in R. R Package Version 0.4.3. Available online: https://CRAN.R-project.org/package=corrr (accessed on 3 June 2021).


| Origin of Fiber Preparation | Total Dietary Fiber Content | I/S Ratio | pH Value (10% Solution) | Pectin Content | Oxide Ash | Bulk Density | Average Fiber Length |
|---|---|---|---|---|---|---|---|
| Pea | ~70% | ~34 | 4.0–7.0 | N/A | max. 5% | 300–620 g/L | N/A |
| Oat | ~90% | N/A | 5.5–7.5 | N/A | max. 5% | 260–385 g/L | 75 µm |
| Psyllium | ~80% | ~0.2 | 5.0–7.0 | N/A | max. 3% | 350–700 g/L | N/A |
| Wheat | ~97% | N/A | 5.0–8.0 | N/A | max. 3% | 260–355 g/L | 50 µm |
| Apple | ~55% | ~4.5 | 3.0–5.0 | ~9% | max. 3% | 255–345 g/L | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steimle, A.; Neumann, M.; Grant, E.T.; Turner, J.D.; Desai, M.S. Concentrated Raw Fibers Enhance the Fiber-Degrading Capacity of a Synthetic Human Gut Microbiome. Int. J. Mol. Sci. 2021, 22, 6855. https://doi.org/10.3390/ijms22136855
Steimle A, Neumann M, Grant ET, Turner JD, Desai MS. Concentrated Raw Fibers Enhance the Fiber-Degrading Capacity of a Synthetic Human Gut Microbiome. International Journal of Molecular Sciences. 2021; 22(13):6855. https://doi.org/10.3390/ijms22136855
Chicago/Turabian StyleSteimle, Alex, Mareike Neumann, Erica T. Grant, Jonathan D. Turner, and Mahesh S. Desai. 2021. "Concentrated Raw Fibers Enhance the Fiber-Degrading Capacity of a Synthetic Human Gut Microbiome" International Journal of Molecular Sciences 22, no. 13: 6855. https://doi.org/10.3390/ijms22136855
APA StyleSteimle, A., Neumann, M., Grant, E. T., Turner, J. D., & Desai, M. S. (2021). Concentrated Raw Fibers Enhance the Fiber-Degrading Capacity of a Synthetic Human Gut Microbiome. International Journal of Molecular Sciences, 22(13), 6855. https://doi.org/10.3390/ijms22136855







