Multifaceted Regulation of MicroRNA Biogenesis: Essential Roles and Functional Integration in Neuronal and Glial Development
Abstract
1. Introduction
2. Canonical and Non-Canonical miRNA Processing Pathways
2.1. The Canonical miRNA Biogenesis Pathway
2.2. Emerging Roles of Non-Canonical miRNA Biogenesis Pathways in the Central Nervous System
3. MiRNAs Play Key Roles in Governing Neuronal and Glial Development
3.1. The Most Abundant MiRNA in the Brain: MiR-124 and its Anti-Neurogenic Targets
3.2. Convergence of Distinct Pro-Neurogenic miRNAs on Key Inhibitors of Neuronal Differentiation
3.3. MiRNAs Govern Glial Lineage Development and the Functional Interplay between Neurons and Glia
4. Neural miRNA Multigene Families
5. Regulation of miRNAs in Neural Development: Divergence by Biogenesis and Convergence on Key Targets
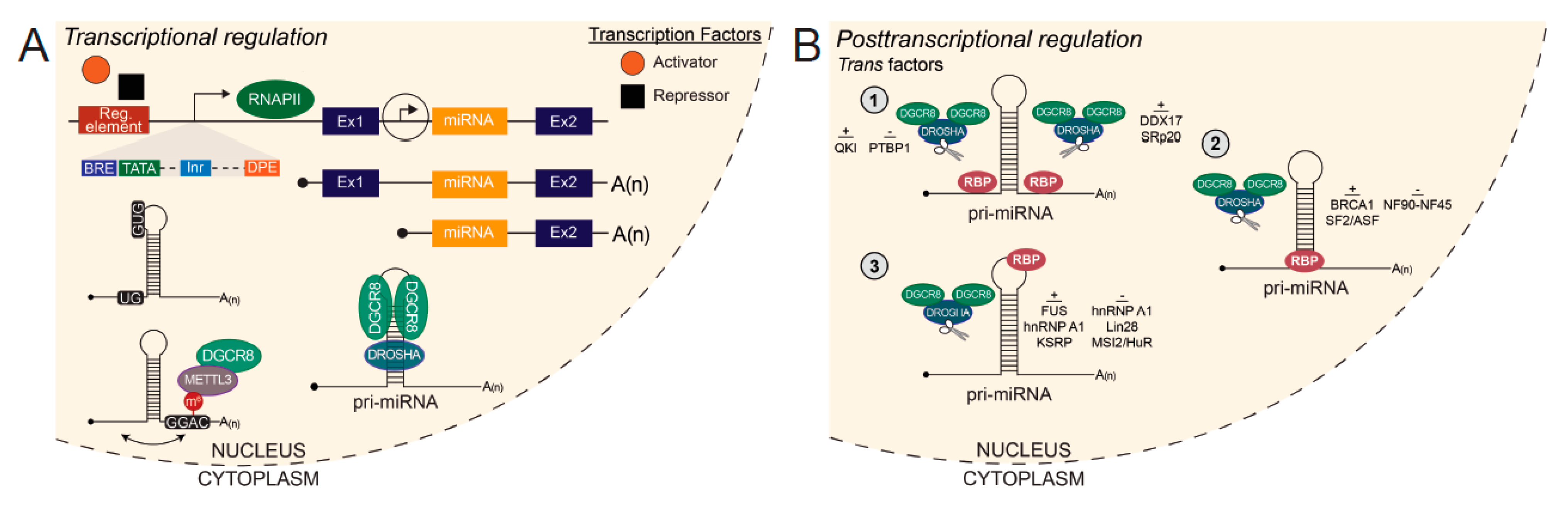
5.1. Transcriptional Regulation of miRNA Genes
5.1.1. Transcriptional Regulation of Neuronal miRNA Genes
5.1.2. Transcriptional Regulation of Oligodendroglial miRNA Genes
5.2. Posttranscriptional Regulation of miRNA Biogenesis
5.3. Posttranscriptional Regulation of miRNA Activity
6. Neural miRNAs in Brain Disorders: Dysregulation, Etiology, and Therapeutic Potential
6.1. Neuronal miRNAs in Brain Diseases
6.2. MiRNAs in Glial Cells and Glia-Neuron Communication Implicated in Brain Diseases
7. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef]
- Pham, J.T.; Gallicano, G.I. Specification of neural cell fate and regulation of neural stem cell proliferation by microRNAs. Am. J. Stem Cells 2012, 1, 182–195. [Google Scholar]
- Loya, C.M.; Van Vactor, D.; Fulga, T.A. Understanding neuronal connectivity through the post-transcriptional toolkit. Genes Dev. 2010, 24, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhang, L.; Pearce, W.J. MicroRNAs in brain development and cerebrovascular pathophysiology. Am. J. Physiol. Cell Physiol. 2019, 317, C3–C19. [Google Scholar] [CrossRef] [PubMed]
- Fiorenza, A.; Barco, A. Role of Dicer and the miRNA system in neuronal plasticity and brain function. Neurobiol. Learn Mem. 2016, 135, 3–12. [Google Scholar] [CrossRef]
- Tiane, A.; Schepers, M.; Rombaut, B.; Hupperts, R.; Prickaerts, J.; Hellings, N.; van den Hove, D.; Vanmierlo, T. From OPC to Oligodendrocyte: An Epigenetic Journey. Cells 2019, 8, 1236. [Google Scholar] [CrossRef] [PubMed]
- Brites, D. Regulatory function of microRNAs in microglia. Glia 2020, 68, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Su, X.; Piao, L.; Jin, Z.; Jin, R. Involvement of Astrocytes and microRNA Dysregulation in Neurodegenerative Diseases: From Pathogenesis to Therapeutic Potential. Front. Mol. Neurosci. 2021, 14, 556215. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Bozhilova, R.; Kaneva, R.; Milanova, V. The Dysregulation of microRNAs and the Role of Stress in the Pathogenesis of Mental Disorders. Curr. Top. Med. Chem. 2018, 18, 1893–1907. [Google Scholar] [CrossRef]
- Gruzdev, S.K.; Yakovlev, A.A.; Druzhkova, T.A.; Guekht, A.B.; Gulyaeva, N.V. The Missing Link: How Exosomes and miRNAs can Help in Bridging Psychiatry and Molecular Biology in the Context of Depression, Bipolar Disorder and Schizophrenia. Cell. Mol. Neurobiol. 2019, 39, 729–750. [Google Scholar] [CrossRef]
- Paul, S.; Reyes, P.R.; Garza, B.S.; Sharma, A. MicroRNAs and Child Neuropsychiatric Disorders: A Brief Review. Neurochem. Res. 2020, 45, 232–240. [Google Scholar] [CrossRef]
- Schulze, M.; Sommer, A.; Plotz, S.; Farrell, M.; Winner, B.; Grosch, J.; Winkler, J.; Riemenschneider, M.J. Sporadic Parkinson’s disease derived neuronal cells show disease-specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol. Commun. 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Rizzuti, M.; Filosa, G.; Melzi, V.; Calandriello, L.; Dioni, L.; Bollati, V.; Bresolin, N.; Comi, G.P.; Barabino, S.; Nizzardo, M.; et al. MicroRNA expression analysis identifies a subset of downregulated miRNAs in ALS motor neuron progenitors. Sci. Rep. 2018, 8, 10105. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002, 16, 720–728. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Biasiolo, M.; Sales, G.; Lionetti, M.; Agnelli, L.; Todoerti, K.; Bisognin, A.; Coppe, A.; Romualdi, C.; Neri, A.; Bortoluzzi, S. Impact of host genes and strand selection on miRNA and miRNA * expression. PLoS ONE 2011, 6, e23854. [Google Scholar] [CrossRef]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bifunctional microRNA miR-9/miR-9 * regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008, 28, 14341–14346. [Google Scholar] [CrossRef]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Knuckles, P.; Vogt, M.A.; Lugert, S.; Milo, M.; Chong, M.M.; Hautbergue, G.M.; Wilson, S.A.; Littman, D.R.; Taylor, V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012, 15, 962–969. [Google Scholar] [CrossRef]
- Nigro, A.; Menon, R.; Bergamaschi, A.; Clovis, Y.M.; Baldi, A.; Ehrmann, M.; Comi, G.; De Pietri Tonelli, D.; Farina, C.; Martino, G.; et al. MiR-30e and miR-181d control radial glia cell proliferation via HtrA1 modulation. Cell Death Dis. 2012, 3, e360. [Google Scholar] [CrossRef][Green Version]
- O’Toole, S.M.; Ferrer, M.M.; Mekonnen, J.; Zhang, H.; Shima, Y.; Ladle, D.R.; Nelson, S.B. Dicer maintains the identity and function of proprioceptive sensory neurons. J. Neurophysiol. 2017, 117, 1057–1069. [Google Scholar] [CrossRef]
- Chmielarz, P.; Konovalova, J.; Najam, S.S.; Alter, H.; Piepponen, T.P.; Erfle, H.; Sonntag, K.C.; Schutz, G.; Vinnikov, I.A.; Domanskyi, A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017, 8, e2813. [Google Scholar] [CrossRef]
- Davis, T.H.; Cuellar, T.L.; Koch, S.M.; Barker, A.J.; Harfe, B.D.; McManus, M.T.; Ullian, E.M. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008, 28, 4322–4330. [Google Scholar] [CrossRef]
- Shin, D.; Shin, J.Y.; McManus, M.T.; Ptácek, L.J.; Fu, Y.H. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol. 2009, 66, 843–857. [Google Scholar] [CrossRef]
- Varol, D.; Mildner, A.; Blank, T.; Shemer, A.; Barashi, N.; Yona, S.; David, E.; Boura-Halfon, S.; Segal-Hayoun, Y.; Chappell-Maor, L.; et al. Dicer Deficiency Differentially Impacts Microglia of the Developing and Adult Brain. Immunity 2017, 46, 1030–1044.e8. [Google Scholar] [CrossRef]
- Sun, C.; Zhu, L.; Ma, R.; Ren, J.; Wang, J.; Gao, S.; Yang, D.; Ning, K.; Ling, B.; Lu, B.; et al. Astrocytic miR-324-5p is essential for synaptic formation by suppressing the secretion of CCL5 from astrocytes. Cell Death Dis. 2019, 10, 141. [Google Scholar] [CrossRef]
- Tao, J.; Wu, H.; Lin, Q.; Wei, W.; Lu, X.H.; Cantle, J.P.; Ao, Y.; Olsen, R.W.; Yang, X.W.; Mody, I.; et al. Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J. Neurosci. 2011, 31, 8306–8319. [Google Scholar] [CrossRef]
- Ristori, E.; Lopez-Ramirez, M.A.; Narayanan, A.; Hill-Teran, G.; Moro, A.; Calvo, C.F.; Thomas, J.L.; Nicoli, S. A Dicer-miR-107 Interaction Regulates Biogenesis of Specific miRNAs Crucial for Neurogenesis. Dev. Cell 2015, 32, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kim, P.; Phay, M.; Yoo, S. Identification of precursor microRNAs within distal axons of sensory neuron. J. Neurochem. 2015, 134, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Lugli, G.; Larson, J.; Martone, M.E.; Jones, Y.; Smalheiser, N.R. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 2005, 94, 896–905. [Google Scholar] [CrossRef]
- Sambandan, S.; Akbalik, G.; Kochen, L.; Rinne, J.; Kahlstatt, J.; Glock, C.; Tushev, G.; Alvarez-Castelao, B.; Heckel, A.; Schuman, E.M. Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 2017, 355, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Lai, E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 2011, 43, 892–903. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Hsu, R.; Melton, C.; Thomas, M.; Ullian, E.M.; Blelloch, R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA 2011, 17, 1489–1501. [Google Scholar] [CrossRef]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Cifuentes, D.; Izumi, N.; Sakaguchi, Y.; Suzuki, T.; Giraldez, A.J.; Tomari, Y. Poly(A)-specific ribonuclease mediates 3’-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013, 5, 715–726. [Google Scholar] [CrossRef]
- Nan, Y.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Yang, Y.; Yue, X.; Pu, P.; Zhong, Y.; Kang, C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010, 1359, 14–21. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, L.; Li, T.; Zhu, M.; Zhang, C.; Chen, L.; Zhao, P.; Zhou, H.; Yu, S.; Yang, X. The role of miR-451 in the switching between proliferation and migration in malignant glioma cells: AMPK signaling, mTOR modulation and Rac1 activation required. Int. J. Oncol. 2017, 50, 1989–1999. [Google Scholar] [CrossRef]
- Fu, C.; Chen, S.; Cai, N.; Liu, Z.; Wang, P.; Zhao, J. Potential Neuroprotective Effect of miR-451 Against Cerebral Ischemia/Reperfusion Injury in Stroke Patients and a Mouse Model. World Neurosurg. 2019, 130, e54–e61. [Google Scholar] [CrossRef]
- Hansen, T.B.; Venø, M.T.; Jensen, T.I.; Schaefer, A.; Damgaard, C.K.; Kjems, J. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016, 7, 11538. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef]
- Kutsche, L.K.; Gysi, D.M.; Fallmann, J.; Lenk, K.; Petri, R.; Swiersy, A.; Klapper, S.D.; Pircs, K.; Khattak, S.; Stadler, P.F.; et al. Combined Experimental and System-Level Analyses Reveal the Complex Regulatory Network of miR-124 during Human Neurogenesis. Cell Syst. 2018, 7, 438–452.e8. [Google Scholar] [CrossRef]
- Hou, Q.; Ruan, H.; Gilbert, J.; Wang, G.; Ma, Q.; Yao, W.D.; Man, H.Y. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat. Commun. 2015, 6, 10045. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Grafe, A.; Seiler, A.; Schumacher, S.; Nitsch, R.; Wulczyn, F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005, 21, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Deo, M.; Yu, J.Y.; Chung, K.H.; Tippens, M.; Turner, D.L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006, 235, 2538–2548. [Google Scholar] [CrossRef] [PubMed]
- Akerblom, M.; Sachdeva, R.; Barde, I.; Verp, S.; Gentner, B.; Trono, D.; Jakobsson, J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 2012, 32, 8879–8889. [Google Scholar] [CrossRef]
- Franke, K.; Otto, W.; Johannes, S.; Baumgart, J.; Nitsch, R.; Schumacher, S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012, 31, 2908–2921. [Google Scholar] [CrossRef]
- Wei, C.; Ren, L.; Li, K.; Lu, Z. The regulation of survival and differentiation of neural stem cells by miR-124 via modulating PAX3. Neurosci. Lett. 2018, 683, 19–26. [Google Scholar] [CrossRef]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef]
- Conaco, C.; Otto, S.; Han, J.J.; Mandel, G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. USA 2006, 103, 2422–2427. [Google Scholar] [CrossRef]
- Yeom, K.H.; Mitchell, S.; Linares, A.J.; Zheng, S.; Lin, C.H.; Wang, X.J.; Hoffmann, A.; Black, D.L. Polypyrimidine tract-binding protein blocks miRNA-124 biogenesis to enforce its neuronal-specific expression in the mouse. Proc. Natl. Acad. Sci. USA 2018, 115, E11061–E11070. [Google Scholar] [CrossRef]
- Xue, Y.; Ouyang, K.; Huang, J.; Zhou, Y.; Ouyang, H.; Li, H.; Wang, G.; Wu, Q.; Wei, C.; Bi, Y.; et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013, 152, 82–96. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; Sonntag, K.C.; Isacson, O.; Kosik, K.S. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 2006, 24, 857–864. [Google Scholar] [CrossRef]
- Neo, W.H.; Yap, K.; Lee, S.H.; Looi, L.S.; Khandelia, P.; Neo, S.X.; Makeyev, E.V.; Su, I.H. MicroRNA miR-124 controls the choice between neuronal and astrocyte differentiation by fine-tuning Ezh2 expression. J. Biol. Chem. 2014, 289, 20788–20801. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Oh, Y.M.; Lu, Y.L.; Kim, W.K.; Yoo, A.S. MicroRNAs Overcome Cell Fate Barrier by Reducing EZH2-Controlled REST Stability during Neuronal Conversion of Human Adult Fibroblasts. Dev. Cell 2018, 46, 73–84.e7. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Santos, M.C.; Tegge, A.N.; Correa, B.R.; Mahesula, S.; Kohnke, L.Q.; Qiao, M.; Ferreira, M.A.; Kokovay, E.; Penalva, L.O. miR-124, -128, and -137 Orchestrate Neural Differentiation by Acting on Overlapping Gene Sets Containing a Highly Connected Transcription Factor Network. Stem Cells 2016, 34, 220–232. [Google Scholar] [CrossRef]
- Mondanizadeh, M.; Arefian, E.; Mosayebi, G.; Saidijam, M.; Khansarinejad, B.; Hashemi, S.M. MicroRNA-124 regulates neuronal differentiation of mesenchymal stem cells by targeting Sp1 mRNA. J. Cell. Biochem. 2015, 116, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Cai, N.; Tao, T.; Zhang, R.; Yan, W.; Li, R.; Zhang, J.; Luo, H.; Shi, Y.; Luan, W.; et al. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS ONE 2014, 9, e98651. [Google Scholar] [CrossRef]
- Dai, Y.; Zhang, W.R.; Wang, Y.M.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. MicroRNA-128 regulates the proliferation and differentiation of bovine skeletal muscle satellite cells by repressing Sp1. Mol. Cell. Biochem. 2016, 414, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xu, Z.; Gu, J.; Huang, H.; Gao, G.; Zhang, X.; Li, J.; Jin, H.; Jiang, G.; Sun, H.; et al. Induction of miR-137 by Isorhapontigenin (ISO) Directly Targets Sp1 Protein Translation and Mediates Its Anticancer Activity Both In Vitro and In Vivo. Mol. Cancer Ther. 2016, 15, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228–231. [Google Scholar] [CrossRef]
- Ballas, N.; Grunseich, C.; Lu, D.D.; Speh, J.C.; Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 2005, 121, 645–657. [Google Scholar] [CrossRef]
- Andres, M.E.; Burger, C.; Peral-Rubio, M.J.; Battaglioli, E.; Anderson, M.E.; Grimes, J.; Dallman, J.; Ballas, N.; Mandel, G. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 1999, 96, 9873–9878. [Google Scholar] [CrossRef]
- Grimes, J.A.; Nielsen, S.J.; Battaglioli, E.; Miska, E.A.; Speh, J.C.; Berry, D.L.; Atouf, F.; Holdener, B.C.; Mandel, G.; Kouzarides, T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 2000, 275, 9461–9467. [Google Scholar] [CrossRef]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005, 307, 596–600. [Google Scholar] [CrossRef]
- Philips, T.; Rothstein, J.D. Oligodendroglia: Metabolic supporters of neurons. J. Clin. Investig. 2017, 127, 3271–3280. [Google Scholar] [CrossRef]
- Emery, B. Transcriptional and post-transcriptional control of CNS myelination. Curr. Opin. Neurobiol. 2010, 20, 601–607. [Google Scholar] [CrossRef]
- Bockbrader, K.; Feng, Y. Essential function, sophisticated regulation and pathological impact of the selective RNA-binding protein QKI in CNS myelin development. Future Neurol. 2008, 3, 655–668. [Google Scholar] [CrossRef]
- Dugas, J.C.; Cuellar, T.L.; Scholze, A.; Ason, B.; Ibrahim, A.; Emery, B.; Zamanian, J.L.; Foo, L.C.; McManus, M.T.; Barres, B.A. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron 2010, 65, 597–611. [Google Scholar] [CrossRef]
- Zhao, X.; He, X.; Han, X.; Yu, Y.; Ye, F.; Chen, Y.; Hoang, T.; Xu, X.; Mi, Q.S.; Xin, M.; et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron 2010, 65, 612–626. [Google Scholar] [CrossRef]
- Wang, H.; Moyano, A.L.; Ma, Z.; Deng, Y.; Lin, Y.; Zhao, C.; Zhang, L.; Jiang, M.; He, X.; Ma, Z.; et al. miR-219 Cooperates with miR-338 in Myelination and Promotes Myelin Repair in the CNS. Dev. Cell 2017, 40, 566–582.e5. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.G.; Lu, M.; Wang, X.; Shang, X.; Elias, S.B.; Chopp, M. MiR-146a promotes remyelination in a cuprizone model of demyelinating injury. Neuroscience 2017, 348, 252–263. [Google Scholar] [CrossRef]
- Liu, X.S.; Chopp, M.; Pan, W.L.; Wang, X.L.; Fan, B.Y.; Zhang, Y.; Kassis, H.; Zhang, R.L.; Zhang, X.M.; Zhang, Z.G. MicroRNA-146a Promotes Oligodendrogenesis in Stroke. Mol. Neurobiol. 2017, 54, 227–237. [Google Scholar] [CrossRef]
- Wang, C.Y.; Deneen, B.; Tzeng, S.F. MicroRNA-212 inhibits oligodendrocytes during maturation by down-regulation of differentiation-associated gene expression. J. Neurochem. 2017, 143, 112–125. [Google Scholar] [CrossRef]
- Buller, B.; Chopp, M.; Ueno, Y.; Zhang, L.; Zhang, R.L.; Morris, D.; Zhang, Y.; Zhang, Z.G. Regulation of serum response factor by miRNA-200 and miRNA-9 modulates oligodendrocyte progenitor cell differentiation. Glia 2012, 60, 1906–1914. [Google Scholar] [CrossRef] [PubMed]
- Marangon, D.; Abbracchio, M.P.; Lecca, D. Pathway-Focused Profiling of Oligodendrocytes Over-Expressing miR-125a-3p Reveals Alteration of Wnt and Cell-to-Cell Signaling. Cell. Mol. Neurobiol. 2021, 41, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi-Kasman, M.; Shojaei, A.; Gol, M.; Moghadamnia, A.A.; Baharvand, H.; Javan, M. miR-302/367-induced neurons reduce behavioral impairment in an experimental model of Alzheimer’s disease. Mol. Cell. Neurosci. 2018, 86, 50–57. [Google Scholar] [CrossRef]
- Ghasemi-Kasman, M.; Zare, L.; Baharvand, H.; Javan, M. In vivo conversion of astrocytes to myelinating cells by miR-302/367 and valproate to enhance myelin repair. J. Tissue Eng. Regen. Med. 2018, 12, e462–e472. [Google Scholar] [CrossRef]
- Ge, X.; Guo, M.; Hu, T.; Li, W.; Huang, S.; Yin, Z.; Li, Y.; Chen, F.; Zhu, L.; Kang, C.; et al. Increased Microglial Exosomal miR-124-3p Alleviates Neurodegeneration and Improves Cognitive Outcome after rmTBI. Mol. Ther. 2020, 28, 503–522. [Google Scholar] [CrossRef]
- Song, Y.; Li, Z.; He, T.; Qu, M.; Jiang, L.; Li, W.; Shi, X.; Pan, J.; Zhang, L.; Wang, Y.; et al. M2 microglia-derived exosomes protect the mouse brain from ischemia-reperfusion injury via exosomal miR-124. Theranostics 2019, 9, 2910–2923. [Google Scholar] [CrossRef]
- Men, Y.; Yelick, J.; Jin, S.; Tian, Y.; Chiang, M.S.R.; Higashimori, H.; Brown, E.; Jarvis, R.; Yang, Y. Exosome reporter mice reveal the involvement of exosomes in mediating neuron to astroglia communication in the CNS. Nat. Commun. 2019, 10, 4136. [Google Scholar] [CrossRef]
- Genoud, C.; Quairiaux, C.; Steiner, P.; Hirling, H.; Welker, E.; Knott, G.W. Plasticity of astrocytic coverage and glutamate transporter expression in adult mouse cortex. PLoS Biol. 2006, 4, e343. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef]
- Willemsen, M.H.; Valles, A.; Kirkels, L.A.; Mastebroek, M.; Olde Loohuis, N.; Kos, A.; Wissink-Lindhout, W.M.; de Brouwer, A.P.; Nillesen, W.M.; Pfundt, R.; et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J. Med. Genet. 2011, 48, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Bofill-De Ros, X.; Kasprzak, W.K.; Bhandari, Y.; Fan, L.; Cavanaugh, Q.; Jiang, M.; Dai, L.; Yang, A.; Shao, T.J.; Shapiro, B.A.; et al. Structural Differences between Pri-miRNA Paralogs Promote Alternative Drosha Cleavage and Expand Target Repertoires. Cell Rep. 2019, 26, 447–459.e4. [Google Scholar] [CrossRef] [PubMed]
- Dhir, A.; Dhir, S.; Proudfoot, N.J.; Jopling, C.L. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat. Struct. Mol. Biol. 2015, 22, 319–327. [Google Scholar] [CrossRef]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef]
- Kim, B.; Jeong, K.; Kim, V.N. Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Mol. Cell 2017, 66, 258–269.e5. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Sanuki, R.; Onishi, A.; Koike, C.; Muramatsu, R.; Watanabe, S.; Muranishi, Y.; Irie, S.; Uneo, S.; Koyasu, T.; Matsui, R.; et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 2011, 14, 1125–1134. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef]
- Yin, S.; Yu, Y.; Reed, R. Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Sci. Rep. 2015, 5, 11992. [Google Scholar] [CrossRef]
- Pawlicki, J.M.; Steitz, J.A. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008, 182, 61–76. [Google Scholar] [CrossRef]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Shen, C.; Huang, Y.W.; Huang, K.; Huang, T.H.; Nephew, K.P.; Li, L.; Liu, Y. RNA polymerase II binding patterns reveal genomic regions involved in microRNA gene regulation. PLoS ONE 2010, 5, e13798. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Gerstein, M. Modeling the relative relationship of transcription factor binding and histone modifications to gene expression levels in mouse embryonic stem cells. Nucleic Acids Res. 2012, 40, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Marsico, A.; Huska, M.R.; Lasserre, J.; Hu, H.; Vucicevic, D.; Musahl, A.; Orom, U.; Vingron, M. PROmiRNA: A new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol. 2013, 14, R84. [Google Scholar] [CrossRef] [PubMed]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Astrom, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Conrad, T.; Marsico, A.; Gehre, M.; Orom, U.A. Microprocessor activity controls differential miRNA biogenesis In Vivo. Cell Rep. 2014, 9, 542–554. [Google Scholar] [CrossRef]
- He, C.; Li, Z.; Chen, P.; Huang, H.; Hurst, L.D.; Chen, J. Young intragenic miRNAs are less coexpressed with host genes than old ones: Implications of miRNA-host gene coevolution. Nucleic Acids Res. 2012, 40, 4002–4012. [Google Scholar] [CrossRef]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010, 189, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Stanley, P.; Papalopulu, N. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012, 2, 10–18. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef]
- Reiprich, S.; Cantone, M.; Weider, M.; Baroti, T.; Wittstatt, J.; Schmitt, C.; Kuspert, M.; Vera, J.; Wegner, M. Transcription factor Sox10 regulates oligodendroglial Sox9 levels via microRNAs. Glia 2017, 65, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Roden, C.; Gaillard, J.; Kanoria, S.; Rennie, W.; Barish, S.; Cheng, J.; Pan, W.; Liu, J.; Cotsapas, C.; Ding, Y.; et al. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017, 27, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Auyeung, V.C.; Ulitsky, I.; McGeary, S.E.; Bartel, D.P. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013, 152, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Wang, F.; Song, W.; Zhao, H.; Ma, Y.; Li, Y.; Zhai, D.; Pi, J.; Si, Y.; Xu, J.; Dong, L.; et al. The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 2017, 27, 416–439. [Google Scholar] [CrossRef]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Caceres, J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007, 14, 591–596. [Google Scholar] [CrossRef]
- Michlewski, G.; Guil, S.; Semple, C.A.; Caceres, J.F. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell 2008, 32, 383–393. [Google Scholar] [CrossRef]
- Kawai, S.; Amano, A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 2012, 197, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Morlando, M.; Dini Modigliani, S.; Torrelli, G.; Rosa, A.; Di Carlo, V.; Caffarelli, E.; Bozzoni, I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012, 31, 4502–4510. [Google Scholar] [CrossRef]
- Ruggiero, T.; Trabucchi, M.; De Santa, F.; Zupo, S.; Harfe, B.D.; McManus, M.T.; Rosenfeld, M.G.; Briata, P.; Gherzi, R. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB J. 2009, 23, 2898–2908. [Google Scholar] [CrossRef]
- Wu, H.; Sun, S.; Tu, K.; Gao, Y.; Xie, B.; Krainer, A.R.; Zhu, J. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell 2010, 38, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Aoki, K.; Higuchi, T.; Todaka, H.; Morisawa, K.; Tamaki, N.; Hatano, E.; Fukushima, A.; Taniguchi, T.; Agata, Y. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 2009, 29, 3754–3769. [Google Scholar] [CrossRef]
- Michlewski, G.; Caceres, J.F. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010, 17, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, N.R.; de Lima Alves, F.; de Andres-Aguayo, L.; Graf, T.; Caceres, J.F.; Rappsilber, J.; Michlewski, G. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013, 27, 24–38. [Google Scholar] [CrossRef]
- Piskounova, E.; Polytarchou, C.; Thornton, J.E.; LaPierre, R.J.; Pothoulakis, C.; Hagan, J.P.; Iliopoulos, D.; Gregory, R.I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079. [Google Scholar] [CrossRef]
- Takeuchi, A.; Takahashi, Y.; Iida, K.; Hosokawa, M.; Irie, K.; Ito, M.; Brown, J.B.; Ohno, K.; Nakashima, K.; Hagiwara, M. Identification of Qk as a Glial Precursor Cell Marker that Governs the Fate Specification of Neural Stem Cells to a Glial Cell Lineage. Stem Cell Rep. 2020, 15, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Dawson, M.R.; Reynolds, R.; Hardy, R.J. Expression of QKI proteins and MAP1B identifies actively myelinating oligodendrocytes in adult rat brain. Mol. Cell. Neurosci. 2001, 17, 292–302. [Google Scholar] [CrossRef]
- Hardy, R.J.; Loushin, C.L.; Friedrich, V.L., Jr.; Chen, Q.; Ebersole, T.A.; Lazzarini, R.A.; Artzt, K. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice. J. Neurosci. 1996, 16, 7941–7949. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.P.; Terrone, S.; Giraud, G.; Benoit-Pilven, C.; Cluet, D.; Combaret, V.; Mortreux, F.; Auboeuf, D.; Bourgeois, C.F. The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res. 2018, 46, 7686–7700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357. [Google Scholar] [CrossRef]
- Mahmoudi, E.; Fitzsimmons, C.; Geaghan, M.P.; Shannon Weickert, C.; Atkins, J.R.; Wang, X.; Cairns, M.J. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacology 2019, 44, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Dube, U.; Del-Aguila, J.L.; Li, Z.; Budde, J.P.; Jiang, S.; Hsu, S.; Ibanez, L.; Fernandez, M.V.; Farias, F.; Norton, J.; et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019, 22, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Hoffmann, A.; Ziller, M.; Spengler, D. Progress in iPSC-Based Modeling of Psychiatric Disorders. Int. J. Mol. Sci. 2019, 20, 4896. [Google Scholar] [CrossRef]
- Pan, Y.H.; Wu, N.; Yuan, X.B. Toward a Better Understanding of Neuronal Migration Deficits in Autism Spectrum Disorders. Front. Cell Dev. Biol. 2019, 7, 205. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B., Jr.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Primers 2017, 3, 17065. [Google Scholar] [CrossRef] [PubMed]
- Juzwik, C.A.; Drake, S.S.; Zhang, Y.; Paradis-Isler, N.; Sylvester, A.; Amar-Zifkin, A.; Douglas, C.; Morquette, B.; Moore, C.S.; Fournier, A.E. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog. Neurobiol. 2019, 182, 101664. [Google Scholar] [CrossRef] [PubMed]
- Muddashetty, R.S.; Nalavadi, V.C.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.T.; Bassell, G.J. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 2011, 42, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, E.; Paudel, Y.N.; Piperi, C. miR-124 and Parkinson’s disease: A biomarker with therapeutic potential. Pharmacol. Res. 2019, 150, 104515. [Google Scholar] [CrossRef]
- He, S.; Liu, X.; Jiang, K.; Peng, D.; Hong, W.; Fang, Y.; Qian, Y.; Yu, S.; Li, H. Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J. Psychiatr. Res. 2016, 78, 65–71. [Google Scholar] [CrossRef]
- Le Grand, J.N.; Gonzalez-Cano, L.; Pavlou, M.A.; Schwamborn, J.C. Neural stem cells in Parkinson’s disease: A role for neurogenesis defects in onset and progression. Cell. Mol. Life Sci. 2015, 72, 773–797. [Google Scholar] [CrossRef]
- Zhao, Y.; Jaber, V.; Alexandrov, P.N.; Vergallo, A.; Lista, S.; Hampel, H.; Lukiw, W.J. microRNA-Based Biomarkers in Alzheimer’s Disease (AD). Front. Neurosci. 2020, 14, 585432. [Google Scholar] [CrossRef]
- Aloizou, A.M.; Siokas, V.; Sapouni, E.M.; Sita, N.; Liampas, I.; Brotis, A.G.; Rakitskii, V.N.; Burykina, T.I.; Aschner, M.; Bogdanos, D.P.; et al. Parkinson’s disease and pesticides: Are microRNAs the missing link? Sci. Total Environ. 2020, 744, 140591. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L. MicroRNAs in amyotrophic lateral sclerosis: From pathogenetic involvement to diagnostic biomarker and therapeutic agent development. Neurol. Sci. 2020, 41, 3569–3577. [Google Scholar] [CrossRef]
- Tolosa, E.; Botta-Orfila, T.; Morato, X.; Calatayud, C.; Ferrer-Lorente, R.; Marti, M.J.; Fernandez, M.; Gaig, C.; Raya, A.; Consiglio, A.; et al. MicroRNA alterations in iPSC-derived dopaminergic neurons from Parkinson disease patients. Neurobiol. Aging 2018, 69, 283–291. [Google Scholar] [CrossRef]
- Church, V.A.; Cates, K.; Capano, L.; Aryal, S.; Kim, W.K.; Yoo, A.S. Generation of Human Neurons by microRNA-Mediated Direct Conversion of Dermal Fibroblasts. Methods Mol. Biol. 2021, 2239, 77–100. [Google Scholar]
- Ghafouri-Fard, S.; Shoorei, H.; Bahroudi, Z.; Abak, A.; Majidpoor, J.; Taheri, M. An update on the role of miR-124 in the pathogenesis of human disorders. Biomed. Pharmacother. 2021, 135, 111198. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.S.; Costain, G.; Fung, W.L.; Russell, K.J.; Pierce, L.; Kapadia, R.; Carter, R.F.; Chow, E.W.; Forsythe, P.J. Clinically detectable copy number variations in a Canadian catchment population of schizophrenia. J. Psychiatr. Res. 2010, 44, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Faraone, S.V.; Lasky-Su, J.; Tsuang, M.T. Genome-wide scan of homogeneous subtypes of NIMH genetics initiative schizophrenia families. Psychiatry Res. 2005, 133, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Baulac, S.; Gourfinkel-An, I.; Couarch, P.; Depienne, C.; Kaminska, A.; Dulac, O.; Baulac, M.; LeGuern, E.; Nabbout, R. A novel locus for generalized epilepsy with febrile seizures plus in French families. Arch. Neurol. 2008, 65, 943–951. [Google Scholar] [CrossRef]
- Claeys, I.; Holvoet, M.; Eyskens, B.; Adriaensens, P.; Gewillig, M.; Fryns, J.P.; Devriendt, K. A recognisable behavioural phenotype associated with terminal deletions of the short arm of chromosome 8. Am. J. Med. Genet. 1997, 74, 515–520. [Google Scholar] [CrossRef]
- Roy, B.; Dunbar, M.; Shelton, R.C.; Dwivedi, Y. Identification of MicroRNA-124-3p as a Putative Epigenetic Signature of Major Depressive Disorder. Neuropsychopharmacology 2017, 42, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, D.; Huang, H.Z.; Wang, Z.H.; Hou, T.Y.; Yang, X.; Pang, P.; Wei, N.; Zhou, Y.F.; Dupras, M.J.; et al. A Novel MicroRNA-124/PTPN1 Signal Pathway Mediates Synaptic and Memory Deficits in Alzheimer’s Disease. Biol. Psychiatry 2018, 83, 395–405. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Gong, G.; Wang, Y.; Bian, M.; Yu, L.; Wei, C. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 2017, 8, 114065–114071. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Huo, X.; Yang, Y.; Hu, Z.; Botchway, B.O.A.; Jiang, Y.; Fang, M. miR-124 downregulates BACE 1 and alters autophagy in APP/PS1 transgenic mice. Toxicol. Lett. 2017, 280, 195–205. [Google Scholar] [CrossRef]
- Gascon, E.; Lynch, K.; Ruan, H.; Almeida, S.; Verheyden, J.M.; Seeley, W.W.; Dickson, D.W.; Petrucelli, L.; Sun, D.; Jiao, J.; et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med. 2014, 20, 1444–1451. [Google Scholar] [CrossRef]
- Johnson, R.; Zuccato, C.; Belyaev, N.D.; Guest, D.J.; Cattaneo, E.; Buckley, N.J. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 2008, 29, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Zhu, Z.; Wu, J.; Zhang, Y.; Zhang, H.; Sun, X.; Qian, C.; Wang, B.; Xie, L.; Zhang, S.; et al. MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson’s disease. FASEB J. 2019, 33, 8648–8665. [Google Scholar] [CrossRef]
- Geng, L.; Liu, W.; Chen, Y. miR-124-3p attenuates MPP(+)-induced neuronal injury by targeting STAT3 in SH-SY5Y cells. Exp. Biol. Med. 2017, 242, 1757–1764. [Google Scholar] [CrossRef]
- Gong, X.; Wang, H.; Ye, Y.; Shu, Y.; Deng, Y.; He, X.; Lu, G.; Zhang, S. miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am. J. Transl. Res. 2016, 8, 2127–2137. [Google Scholar]
- Han, D.; Dong, X.; Zheng, D.; Nao, J. MiR-124 and the Underlying Therapeutic Promise of Neurodegenerative Disorders. Front. Pharmacol. 2019, 10, 1555. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, E.; Cairns, M.J. MiR-137: An important player in neural development and neoplastic transformation. Mol. Psychiatry 2017, 22, 44–55. [Google Scholar] [CrossRef]
- Guella, I.; Sequeira, A.; Rollins, B.; Morgan, L.; Torri, F.; van Erp, T.G.; Myers, R.M.; Barchas, J.D.; Schatzberg, A.F.; Watson, S.J.; et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J. Psychiatr. Res. 2013, 47, 1215–1221. [Google Scholar] [CrossRef]
- Pacheco, A.; Berger, R.; Freedman, R.; Law, A.J. A VNTR Regulates miR-137 Expression Through Novel Alternative Splicing and Contributes to Risk for Schizophrenia. Sci. Rep. 2019, 9, 11793. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Z.M.; Tan, W.; Wang, X.; Li, Y.; Bai, B.; Li, Y.; Zhang, S.F.; Yan, H.L.; Chen, Z.L.; et al. Partial loss of psychiatric risk gene Mir137 in mice causes repetitive behavior and impairs sociability and learning via increased Pde10a. Nat. Neurosci. 2018, 21, 1689–1703. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.T.; Anderson, B.R.; Shah, N.; Zimmer, S.E.; Hawkins, D.; Valdez, A.N.; Gu, Q.; Bassell, G.J. Inhibition of the Schizophrenia-Associated MicroRNA miR-137 Disrupts Nrg1alpha Neurodevelopmental Signal Transduction. Cell Rep. 2017, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.T.; Gross, C.; Bassell, G.J. microRNAs Sculpt Neuronal Communication in a Tight Balance That Is Lost in Neurological Disease. Front. Mol. Neurosci. 2018, 11, 455. [Google Scholar] [CrossRef]
- Raabe, F.J.; Slapakova, L.; Rossner, M.J.; Cantuti-Castelvetri, L.; Simons, M.; Falkai, P.G.; Schmitt, A. Oligodendrocytes as A New Therapeutic Target in Schizophrenia: From Histopathological Findings to Neuron-Oligodendrocyte Interaction. Cells 2019, 8, 1496. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Song, X.; Luo, H.; Sun, J.; Han, C.; Gu, X.; Li, J.; Cai, G.; Zhu, Y.; et al. Modulation of Stem Cells as Therapeutics for Severe Mental Disorders and Cognitive Impairments. Front. Psychiatry 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.J.; Yu, Y.; Zhu, G.C.; Sun, Z.H.; Xu, J.; Cao, J.H.; Ge, J.X. Association between single nucleotide polymorphisms in MiR219-1 and MiR137 and susceptibility to schizophrenia in a Chinese population. FEBS Open Bio 2015, 5, 774–778. [Google Scholar] [CrossRef][Green Version]
- Cserep, C.; Posfai, B.; Denes, A. Shaping Neuronal Fate: Functional Heterogeneity of Direct Microglia-Neuron Interactions. Neuron 2021, 109, 222–240. [Google Scholar] [CrossRef]
- Tan, C.X.; Burrus Lane, C.J.; Eroglu, C. Role of astrocytes in synapse formation and maturation. Curr. Top. Dev. Biol. 2021, 142, 371–407. [Google Scholar] [PubMed]
- Xiao, Y.; Wang, S.K.; Zhang, Y.; Rostami, A.; Kenkare, A.; Casella, G.; Yuan, Z.Q.; Li, X. Role of extracellular vesicles in neurodegenerative diseases. Prog. Neurobiol. 2021, 201, 102022. [Google Scholar] [CrossRef]
- Upadhya, R.; Zingg, W.; Shetty, S.; Shetty, A.K. Astrocyte-derived extracellular vesicles: Neuroreparative properties and role in the pathogenesis of neurodegenerative disorders. J. Control Release 2020, 323, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Sheng, S.; Wang, Y.; Ding, L.; Xu, X.; Xia, X.; Zheng, J.C. Astrocyte-derived extracellular vesicles: A double-edged sword in central nervous system disorders. Neurosci. Biobehav. Rev. 2021, 125, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Vassileff, N.; Cheng, L.; Hill, A.F. Extracellular vesicles—Propagators of neuropathology and sources of potential biomarkers and therapeutics for neurodegenerative diseases. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Huang, Y.; Zhang, H.; Lu, H.; Zheng, J.C. Exosomal miRNAs in central nervous system diseases: Biomarkers, pathological mediators, protective factors and therapeutic agents. Prog. Neurobiol. 2019, 183, 101694. [Google Scholar] [CrossRef]
- Gamez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Rene, R.; Alvarez, R.; Armengol, M.P.; Borras, F.E.; Beyer, K. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef]
- Aharon, A.; Spector, P.; Ahmad, R.S.; Horrany, N.; Sabbach, A.; Brenner, B.; Aharon-Peretz, J. Extracellular Vesicles of Alzheimer’s Disease Patients as a Biomarker for Disease Progression. Mol. Neurobiol. 2020, 57, 4156–4169. [Google Scholar] [CrossRef]
- Prada, I.; Gabrielli, M.; Turola, E.; Iorio, A.; D’Arrigo, G.; Parolisi, R.; De Luca, M.; Pacifici, M.; Bastoni, M.; Lombardi, M.; et al. Glia-to-neuron transfer of miRNAs via extracellular vesicles: A new mechanism underlying inflammation-induced synaptic alterations. Acta Neuropathol. 2018, 135, 529–550. [Google Scholar] [CrossRef]
- Ma, Y.; Li, C.; Huang, Y.; Wang, Y.; Xia, X.; Zheng, J.C. Exosomes released from neural progenitor cells and induced neural progenitor cells regulate neurogenesis through miR-21a. Cell Commun. Signal. 2019, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Giunti, D.; Marini, C.; Parodi, B.; Usai, C.; Milanese, M.; Bonanno, G.; Kerlero de Rosbo, N.; Uccelli, A. Role of miRNAs shuttled by mesenchymal stem cell-derived small extracellular vesicles in modulating neuroinflammation. Sci. Rep. 2021, 11, 1740. [Google Scholar] [CrossRef] [PubMed]
- Chivero, E.T.; Liao, K.; Niu, F.; Tripathi, A.; Tian, C.; Buch, S.; Hu, G. Engineered Extracellular Vesicles Loaded With miR-124 Attenuate Cocaine-Mediated Activation of Microglia. Front. Cell Dev. Biol. 2020, 8, 573. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, R.J.; Acharya, M.M.; Baulch, J.E.; Limoli, C.L. Extracellular Vesicle-Derived miR-124 Resolves Radiation-Induced Brain Injury. Cancer Res. 2020, 80, 4266–4277. [Google Scholar] [CrossRef]
| Cell Type/System | MicroRNA | Function in Brain Development | Brain Disease Relevance |
|---|---|---|---|
| Neuron | miR-124 | Promotes neuronal differentiation [50,51,52,53], neuron-glia communication | AD [173,174,175], HD [177], MDD [172], Frontotemporal dementia [176] |
| miR-137 | Neuronal differentiation, synaptic activity | SCZ [95], ASD [183], Intellectual disability [96] | |
| miR-9 | Promotes neuronal differentiation [27] | HD [26] | |
| miR-7 | Immediate early genes, synpatic transmission [147] | ||
| miR-125a | Neuronal translation, synaptic function | Fragile X [158] | |
| Microglia | miR-124 | Inhibits microglial activation | PD [178,179,180] |
| miR-146a-5p | Modulates synpase function [200] | Correlated with AD severity [199] | |
| Oligodendroglia | miR-219 | OL development, myelination and repair | SCZ [189], Multiple sclerosis [8] |
| miR-338 | OL development, myelination and repair | Multiple sclerosis [8] | |
| miR-146a | Myelination and repair | Multiple sclerosis [8] | |
| miR-212 | Prevents OL maturation [85] | Myelin disorders | |
| miR-125a | Prevents OL maturation [87] | Myelin disorders | |
| miR-335 | OL development [121] | Myelin disorders | |
| Astroglia | miR-451 | Glioma tumorigenesis [48,49] | |
| miR-125a | Extracellur vessicle | AD, PD, ALS, and stroke [194] | |
| miR-16-5p | Extracellur vessicle | AD, PD, ALS, and stroke [194] | |
| miR-302/367 | Reprogram astrocytes to neuronal or OL fate [88,89] | Potential repair of neurodegeneration or myelin lesion | |
| Brain exosomal miRNAs | miR-21a | Promotes neuronal differentiation [201] | |
| miR-467f | Pro-inflammatory phenotype of microglia | ALS [202] | |
| miR-466q | Pro-inflammatory phenotype of microglia | ALS [202] | |
| miR-23a-3p | Potential biomarker for AD | Reduced in AD [198] | |
| miR-126-3p | Potential biomarker for AD | Correlated with AD severity [199] | |
| miR-151a-3p | Potential biomarker for AD | Reduced in AD [198] | |
| miR-142-3p | Potential biomarker for AD | Correlated with AD severity [199] | |
| miR-223-3p | Potential biomarker for AD | Correlated with AD severity [199] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suster, I.; Feng, Y. Multifaceted Regulation of MicroRNA Biogenesis: Essential Roles and Functional Integration in Neuronal and Glial Development. Int. J. Mol. Sci. 2021, 22, 6765. https://doi.org/10.3390/ijms22136765
Suster I, Feng Y. Multifaceted Regulation of MicroRNA Biogenesis: Essential Roles and Functional Integration in Neuronal and Glial Development. International Journal of Molecular Sciences. 2021; 22(13):6765. https://doi.org/10.3390/ijms22136765
Chicago/Turabian StyleSuster, Izabela, and Yue Feng. 2021. "Multifaceted Regulation of MicroRNA Biogenesis: Essential Roles and Functional Integration in Neuronal and Glial Development" International Journal of Molecular Sciences 22, no. 13: 6765. https://doi.org/10.3390/ijms22136765
APA StyleSuster, I., & Feng, Y. (2021). Multifaceted Regulation of MicroRNA Biogenesis: Essential Roles and Functional Integration in Neuronal and Glial Development. International Journal of Molecular Sciences, 22(13), 6765. https://doi.org/10.3390/ijms22136765






