NK Cells in Chronic Lymphocytic Leukemia and Their Therapeutic Implications
Abstract
:1. Introduction
2. Overall View on NK Cells
2.1. Role of NK Cells in the Immune System
2.2. NK cell Receptors and Ligands and Their Role in Regulating NK Cell Activity
3. NK Cells in CLL
3.1. NK Cell Functions and Dysfunctions
3.2. Alterations in NK Cell Receptor–Ligand System
3.3. Escape of CLL Cells from NK Cell Antitumor Activity
4. Immunotherapeutic Approaches Involving NK Cells in CLL
4.1. Enhancement of NK Cell-Mediated ADCC
4.1.1. Monoclonal Antibodies
4.1.2. Bispecific and Trispecific Killer Cell Engagers
4.2. Restoring NK Cell Functions by Targeting The Immune Checkpoints
4.3. Allogeneic NK Cell Therapy
4.4. CAR-NK Cell Therapy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kipps, T.J.; Stevenson, F.K.; Wu, C.J.; Croce, C.M.; Packham, G.; Wierda, W.G.; O’Brien, S.; Gribben, J.; Rai, K. Chronic lymphocytic leukaemia. Nat. Rev. Dis. Primers 2017, 3, 16096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbri, G.; Dalla-Favera, R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat. Rev. Cancer 2016, 16, 145–162. [Google Scholar] [CrossRef]
- Puente, X.S.; Jares, P.; Campo, E. Chronic lymphocytic leukemia and mantle cell lymphoma: Crossroads of genetic and microenvironment interactions. Blood 2018, 131, 2283–2296. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef] [Green Version]
- Burger, J.A.; O’Brien, S. Evolution of CLL treatment—From chemoimmunotherapy to targeted and individualized therapy. Nat. Rev. Clin. Oncol. 2018, 15, 510–527. [Google Scholar] [CrossRef]
- Bond, D.A.; Woyach, J.A. Targeting BTK in CLL: Beyond Ibrutinib. Curr. Hematol. Malig. Rep. 2019, 14, 197–205. [Google Scholar] [CrossRef]
- Herling, C.D.; Abedpour, N.; Weiss, J.; Schmitt, A.; Jachimowicz, R.D.; Merkel, O.; Cartolano, M.; Oberbeck, S.; Mayer, P.; Berg, V.; et al. Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat. Commun. 2018, 9, 727. [Google Scholar] [CrossRef]
- Jain, N.; Keating, M.; Thompson, P.; Ferrajoli, A.; Burger, J.; Borthakur, G.; Takahashi, K.; Estrov, Z.; Fowler, N.; Kadia, T.; et al. Ibrutinib and Venetoclax for First-Line Treatment of CLL. N. Engl. J. Med. 2019, 380, 2095–2103. [Google Scholar] [CrossRef]
- Rogers, K.A.; Huang, Y.; Ruppert, A.S.; Abruzzo, L.V.; Andersen, B.L.; Awan, F.T.; Bhat, S.A.; Dean, A.; Lucas, M.; Banks, C.; et al. Phase II Study of Combination Obinutuzumab, Ibrutinib, and Venetoclax in Treatment-Naïve and Relapsed or Refractory Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2020, 38, 3626–3637. [Google Scholar] [CrossRef]
- Hillmen, P.; Rawstron, A.C.; Brock, K.; Muñoz-Vicente, S.; Yates, F.J.; Bishop, R.; Boucher, R.; MacDonald, D.; Fegan, C.; McCaig, A.; et al. Ibrutinib Plus Venetoclax in Relapsed/Refractory Chronic Lymphocytic Leukemia: The CLARITY Study. J. Clin. Oncol. 2019, 37, 2722–2729. [Google Scholar] [CrossRef]
- Arruga, F.; Gyau, B.B.; Iannello, A.; Vitale, N.; Vaisitti, T.; Deaglio, S. Immune Response Dysfunction in Chronic Lymphocytic Leukemia: Dissecting Molecular Mechanisms and Microenvironmental Conditions. Int. J. Mol. Sci. 2020, 21, 1825. [Google Scholar] [CrossRef] [Green Version]
- Forconi, F.; Moss, P. Perturbation of the normal immune system in patients with CLL. Blood 2015, 126, 573–581. [Google Scholar] [CrossRef] [Green Version]
- Vlachonikola, E.; Stamatopoulos, K.; Chatzidimitriou, A. T Cells in Chronic Lymphocytic Leukemia: A Two-Edged Sword. Front. Immunol. 2021, 11, 612244. [Google Scholar] [CrossRef]
- Roessner, P.M.; Seiffert, M. T-cells in chronic lymphocytic leukemia: Guardians or drivers of disease? Leukemia 2020, 34, 2012–2024. [Google Scholar] [CrossRef]
- Park, J.H.; Rivière, I.; Gonen, M.; Wang, X.; Sénéchal, B.; Curran, K.J.; Sauter, C.; Wang, Y.; Santomasso, B.; Mead, E.; et al. Long-Term Follow-up of CD19 CAR Therapy in Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2018, 378, 449–459. [Google Scholar] [CrossRef]
- Schuster, S.J.; Svoboda, J.; Chong, E.A.; Nasta, S.D.; Mato, A.R.; Anak, Ö.; Brogdon, J.L.; Pruteanu-Malinici, I.; Bhoj, V.; Landsburg, D.; et al. Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N. Engl. J. Med. 2017, 377, 2545–2554. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Zhou, J.; Young, K.H. PD-1 expression and clinical PD-1 blockade in B-cell lymphomas. Blood 2018, 131, 68–83. [Google Scholar] [CrossRef] [Green Version]
- Lorentzen, C.L.; Straten, P.T. CD19-Chimeric Antigen Receptor T Cells for Treatment of Chronic Lymphocytic Leukaemia and Acute Lymphoblastic Leukaemia. Scand. J. Immunol. 2015, 82, 307–319. [Google Scholar] [CrossRef]
- Fraietta, J.A.; Lacey, S.F.; Orlando, E.J.; Pruteanu-Malinici, I.; Gohil, M.; Lundh, S.; Boesteanu, A.C.; Wang, Y.; O’Connor, R.S.; Hwang, W.T.; et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 2018, 24, 563–571. [Google Scholar] [CrossRef]
- Ding, W.; LaPlant, B.R.; Call, T.G.; Parikh, S.A.; Leis, J.F.; He, R.; Shanafelt, T.D.; Sinha, S.; Le-Rademacher, J.; Feldman, A.L.; et al. Pembrolizumab in patients with CLL and Richter transformation or with relapsed CLL. Blood 2017, 129, 3419–3427. [Google Scholar] [CrossRef]
- Malmberg, K.J.; Carlsten, M.; Björklund, A.; Sohlberg, E.; Bryceson, Y.T.; Ljunggren, H.G. Natural killer cell-mediated immunosurveillance of human cancer. Semin. Immunol. 2017, 31, 20–29. [Google Scholar] [CrossRef]
- Perera Molligoda Arachchige, A.S. Human NK cells: From development to effector functions. Innate Immun. 2021, 27, 212–229. [Google Scholar] [CrossRef]
- Sivori, S.; Vacca, P.; Del Zotto, G.; Munari, E.; Mingari, M.C.; Moretta, L. Human NK cells: Surface receptors, inhibitory checkpoints, and translational applications. Cell. Mol. Immunol. 2019, 16, 430–441. [Google Scholar] [CrossRef]
- Ochoa, M.C.; Minute, L.; Rodriguez, I.; Garasa, S.; Perez-Ruiz, E.; Inogés, S.; Melero, I.; Berraondo, P. Antibody-dependent cell cytotoxicity: Immunotherapy strategies enhancing effector NK cells. Immunol. Cell Biol. 2017, 95, 347–355. [Google Scholar] [CrossRef]
- Shanehbandi, D.; Majidi, J.; Kazemi, T.; Baradaran, B.; Aghebati-Maleki, L. CD20-based Immunotherapy of B-cell Derived Hematologic Malignancies. Curr. Cancer Drug Targets 2017, 17, 423–444. [Google Scholar] [CrossRef]
- Marshall, M.; Stopforth, R.J.; Cragg, M.S. Therapeutic Antibodies: What Have We Learnt from Targeting CD20 and Where Are We Going? Front. Immunol. 2017, 8, 1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battella, S.; Cox, M.C.; Santoni, A.; Palmieri, G. Natural killer (NK) cells and anti-tumor therapeutic mAb: Unexplored interactions. J. Leukoc. Biol. 2016, 99, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Jaglowski, S.M.; Alinari, L.; Lapalombella, R.; Muthusamy, N.; Byrd, J.C. The clinical application of monoclonal antibodies in chronic lymphocytic leukemia. Blood 2010, 116, 3705–3714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggeri, L.; Capanni, M.; Urbani, E.; Perruccio, K.; Shlomchik, W.D.; Tosti, A.; Posati, S.; Rogaia, D.; Frassoni, F.; Aversa, F.; et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002, 295, 2097–2100. [Google Scholar] [CrossRef] [Green Version]
- Ruggeri, L.; Aversa, F.; Martelli, M.F.; Velardi, A. Allogeneic hematopoietic transplantation and natural killer cell recognition of missing self. Immunol. Rev. 2006, 214, 202–218. [Google Scholar] [CrossRef]
- Ziegler, H.W.; Kay, N.E.; Zarling, J.M. Deficiency of natural killer cell activity in patients with chronic lymphocytic leukemia. Int. J. Cancer 1981, 27, 321–327. [Google Scholar] [CrossRef]
- Kay, N.E.; Zarling, J.M. Impaired natural killer activity in patients with chronic lymphocytic leukemia is associated with a deficiency of azurophilic cytoplasmic granules in putative NK cells. Blood 1984, 63, 305–309. [Google Scholar] [CrossRef] [Green Version]
- Katrinakis, G.; Kyriakou, D.; Papadaki, H.; Kalokyri, I.; Markidou, F.; Eliopoulos, G.D. Defective natural killer cell activity in B-cell chronic lymphocytic leukaemia is associated with impaired release of natural killer cytotoxic factor(s) but not of tumour necrosis factor-alpha. Acta Haematol. 1996, 96, 16–23. [Google Scholar] [CrossRef]
- Le Garff-Tavernier, M.; Decocq, J.; de Romeuf, C.; Parizot, C.; Dutertre, C.A.; Chapiro, E.; Davi, F.; Debré, P.; Prost, J.F.; Teillaud, J.L.; et al. Analysis of CD16+CD56dim NK cells from CLL patients: Evidence supporting a therapeutic strategy with optimized anti-CD20 monoclonal antibodies. Leukemia 2011, 25, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Costello, R.T.; Knoblauch, B.; Sanchez, C.; Mercier, D.; Le Treut, T.; Sébahoun, G. Expression of natural killer cell activating receptors in patients with chronic lymphocytic leukaemia. Immunology 2012, 135, 151–157. [Google Scholar] [CrossRef]
- Kay, N.E.; Zarling, J. Restoration of impaired natural killer cell activity of B-chronic lymphocytic leukemia patients by recombinant interleukin-2. Am. J. Hematol. 1987, 24, 161–167. [Google Scholar] [CrossRef]
- Veuillen, C.; Aurran-Schleinitz, T.; Castellano, R.; Rey, J.; Mallet, F.; Orlanducci, F.; Pouyet, L.; Just-Landi, S.; Coso, D.; Ivanov, V.; et al. Primary B-CLL resistance to NK cell cytotoxicity can be overcome in vitro and in vivo by priming NK cells and monoclonal antibody therapy. J. Clin. Immunol. 2012, 32, 632–646. [Google Scholar] [CrossRef]
- Guven, H.; Gilljam, M.; Chambers, B.J.; Ljunggren, H.G.; Christensson, B.; Kimby, E.; Dilber, M.S. Expansion of natural killer (NK) and natural killer-like T (NKT)-cell populations derived from patients with B-chronic lymphocytic leukemia (B-CLL): A potential source for cellular immunotherapy. Leukemia 2003, 17, 1973–1980. [Google Scholar] [CrossRef]
- Hofland, T.; Endstra, S.; Gomes, C.; de Boer, R.; de Weerdt, I.; Bobkov, V.; Riedl, J.A.; Heukers, R.; Smit, M.J.; Eldering, E.; et al. Natural Killer Cell Hypo-responsiveness in Chronic Lymphocytic Leukemia can be Circumvented In Vitro by Adequate Activating Signaling. Hemasphere 2019, 3, e308. [Google Scholar] [CrossRef]
- Laprevotte, E.; Voisin, G.; Ysebaert, L.; Klein, C.; Daugrois, C.; Laurent, G.; Fournie, J.J.; Quillet-Mary, A. Recombinant human IL-15 trans-presentation by B leukemic cells from chronic lymphocytic leukemia induces autologous NK cell proliferation leading to improved anti-CD20 immunotherapy. J. Immunol. 2013, 191, 3634–3640. [Google Scholar] [CrossRef] [Green Version]
- Hemati, M.; Rasouli Nejad, Z.; Shokri, M.R.; Ghahremanfard, F.; Mir Mohammadkhani, M.; Kokhaei, P. IL-27 impact on NK cells activity: Implication for a robust anti-tumor response in chronic lymphocytic leukemia. Int. Immunopharmacol. 2020, 82, 106350. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Nassif Kerbauy, L.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N. Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef]
- Lam, V.C.; Lanier, L.L. NK cells in host responses to viral infections. Curr. Opin. Immunol. 2017, 44, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Artis, D.; Spits, H. The biology of innate lymphoid cells. Nature 2015, 517, 293–301. [Google Scholar] [CrossRef]
- Spits, H.; Bernink, J.H.; Lanier, L. NK cells and type 1 innate lymphoid cells: Partners in host disease. Nat. Immunol. 2016, 17, 758–764. [Google Scholar] [CrossRef]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef]
- Kumar, S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 2018, 154, 383–393. [Google Scholar] [CrossRef]
- Anfossi, N.; André, P.; Guia, S.; Falk, C.S.; Roetynck, S.; Stewart, C.A.; Breso, V.; Frassati, C.; Reviron, D.; Middleton, D.; et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 2006, 25, 331–342. [Google Scholar] [CrossRef]
- Kärre, K. Natural killer cell recognition of missing self. Nat. Immunol. 2008, 9, 477–480. [Google Scholar] [CrossRef]
- Parikh, B.A.; Bern, M.D.; Piersma, S.J.; Yang, L.; Beckman, D.L.; Poursine-Laurent, J.; Plougastel-Douglas, B.; Yokoyama, W.M. Control of Viral Infection by Natural Killer Cell Inhibitory Receptors. Cell Rep. 2020, 32, 107969. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; Ye, L.; He, Y.; Zhou, C. Killer immunoglobulin-like receptors/human leukocyte antigen class-I, a crucial immune pathway in cancer. Ann. Transl. Med. 2020, 8, 244. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, L.; Chiesa, M.D.; Sivori, S.; Mingari, M.C.; Pende, D.; Moretta, L. Human NK cells, their receptors and function. Eur. J. Immunol. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Smyth, M.J.; Trapani, J.A. Perforin-mediated target-cell death and immune homeostasis. Nat. Rev. Immunol. 2006, 6, 940–952. [Google Scholar] [CrossRef]
- Fauriat, C.; Long, E.O.; Ljunggren, H.G.; Bryceson, Y.T. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 2010, 115, 2167–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Huang, B.; Shi, J. Fas ligand and lytic granule differentially control cytotoxic dynamics of natural killer cell against cancer target. Oncotarget 2016, 7, 47163–47172. [Google Scholar] [CrossRef] [Green Version]
- Agaugué, S.; Marcenaro, E.; Ferranti, B.; Moretta, L.; Moretta, A. Human natural killer cells exposed to IL-2, IL-12, IL-18, or IL-4 differently modulate priming of naive T cells by monocyte-derived dendritic cells. Blood 2008, 112, 1776–1783. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Tian, Z.; Wei, H. Developmental and Functional Control of Natural Killer Cells by Cytokines. Front. Immunol. 2017, 8, 930. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Miller, M.; Stojanovic, A.; Garbi, N.; Cerwenka, A. Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J. Exp. Med. 2012, 209, 2351–2365. [Google Scholar] [CrossRef]
- Floros, T.; Tarhini, A.A. Anticancer Cytokines: Biology and Clinical Effects of Interferon-α2, Interleukin (IL)-2, IL-15, IL-21, and IL-12. Semin. Oncol. 2015, 42, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regis, S.; Dondero, A.; Caliendo, F.; Bottino, C.; Castriconi, R. NK Cell Function Regulation by TGF-β-Induced Epigenetic Mechanisms. Front. Immunol. 2020, 11, 311. [Google Scholar] [CrossRef] [Green Version]
- Scoville, S.D.; Freud, A.G.; Caligiuri, M.A. Cellular pathways in the development of human and murine innate lymphoid cells. Curr. Opin. Immunol. 2019, 56, 100–106. [Google Scholar] [CrossRef]
- Kennedy, M.K.; Glaccum, M.; Brown, S.N.; Butz, E.A.; Viney, J.L.; Embers, M.; Matsuki, N.; Charrier, K.; Sedger, L.; Willis, C.R.; et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 2000, 191, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Lodolce, J.P.; Boone, D.L.; Chai, S.; Swain, R.E.; Dassopoulos, T.; Trettin, S.; Ma, A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 1998, 9, 669–676. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, N.C.; Treiner, E.; Vance, R.E.; Jamieson, A.M.; Lemieux, S.; Raulet, D.H. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 2005, 105, 4416–4423. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Poursine-Laurent, J.; Truscott, S.M.; Lybarger, L.; Song, Y.J.; Yang, L.; French, A.R.; Sunwoo, J.B.; Lemieux, S.; Hansen, T.H.; et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 2005, 436, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Orr, M.T.; Lanier, L.L. Natural killer cell education and tolerance. Cell 2010, 142, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Geary, C.D.; Sun, J.C. Memory responses of natural killer cells. Semin. Immunol. 2017, 31, 11–19. [Google Scholar] [CrossRef]
- Freud, A.G.; Mundy-Bosse, B.L.; Yu, J.; Caligiuri, M.A. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity 2017, 47, 820–833. [Google Scholar] [CrossRef] [Green Version]
- Stabile, H.; Fionda, C.; Gismondi, A.; Santoni, A. Role of Distinct Natural Killer Cell Subsets in Anticancer Response. Front. Immunol. 2017, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Cichicki, F.; Schlums, H.; Theorell, J.; Tesi, B.; Miller, J.S.; Ljunggren, H.G.; Bryceson, Y.T. Diversification and Functional Specialization of Human NK Cell Subsets. Curr. Top Microbiol. Immunol. 2016, 395, 63–94. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M. Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity 2018, 48, 1104–1117. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, G.; Fan, X.; Dikiy, S.; Lee, S.Y.; Rudensky, A.Y. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 2015, 35, 981–985. [Google Scholar] [CrossRef] [Green Version]
- Daussy, C.; Faure, F.; Mayol, K.; Viel, S.; Gasteiger, G.; Charrier, E.; Bienvenu, J.; Henry, T.; Debien, E.; Hasan, U.A.; et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 2014, 211, 563–577. [Google Scholar] [CrossRef]
- Pikovskaya, O.; Chaix, J.; Rothman, N.J.; Collins, A.; Chen, Y.H.; Scipioni, A.M.; Vivier, E.; Reiner, S.L. Cutting edge: Eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 2016, 196, 1449–1454. [Google Scholar] [CrossRef]
- Mackay, L.K.; Minnich, M.; Kragten, N.A.; Liao, Y.; Nota, B.; Seillet, C.; Zaid, A.; Man, K.; Preston, S.; Freestone, D.; et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352, 459–463. [Google Scholar] [CrossRef] [Green Version]
- Filipovic, I.; Chiossone, L.; Vacca, P.; Hamilton, R.S.; Ingegnere, T.; Doisne, J.M.; Hawkes, D.A.; Mingari, M.C.; Sharkey, A.M.; Moretta, L.; et al. Molecular definition of group 1 innate lymphoid cells in the mouse uterus. Nat. Commun. 2018, 9, 4492. [Google Scholar] [CrossRef] [Green Version]
- Constantinides, M.G.; McDonald, B.D.; Verhoef, P.A.; Bendelac, A. A committed precursor to innate lymphoid cells. Nature 2014, 508, 397–401. [Google Scholar] [CrossRef] [Green Version]
- Klose, C.S.N.; Flach, M.; Mohle, L.; Rogell, L.; Hoyler, T.; Ebert, K.; Fabiunke, C.; Pfeifer, D.; Sexl, V.; Fonseca-Pereira, D.; et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 2014, 157, 340–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Cherrier, D.E.; Chea, S.; Vosshenrich, C.; Serafini, N.; Petit, M.; Liu, P.; Golub, R.; Di Santo, J.P. An Id2RFP-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity 2019, 50, 1054–1068.e3. [Google Scholar] [CrossRef] [Green Version]
- Bald, T.; Wagner, M.; Gao, Y.; Koyasu, S.; Smyth, M.J. Hide and seek: Plasticity of innate lymphoid cells in cancer. Semin. Immunol. 2019, 41, 101273. [Google Scholar] [CrossRef]
- Gao, Y.; Souza-Fonseca-Guimaraes, F.; Bald, T.; Foong, N.S.; Young, A.; Ngiow, S.F.; Rautela, J.; Straube, J.; Waddell, N.; Blake, S.J.; et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017, 18, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- McFarland, A.P.; Yalin, A.; Wang, S.Y.; Cortez, V.S.; Landsberger, T.; Sudan, R.; Peng, V.; Miller, H.L.; Ricci, B.; David, E.; et al. Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity 2021, 54, 1320–1337.e4. [Google Scholar] [CrossRef] [PubMed]
- Orange, J.S. Natural killer cell deficiency. J. Allergy Clin. Immunol. 2013, 132, 515–525. [Google Scholar] [CrossRef] [Green Version]
- Chijioke, O.; Müller, A.; Feederle, R.; Barros, M.H.; Krieg, C.; Emmel, V.; Marcenaro, E.; Leung, C.S.; Antsiferova, O.; Landtwing, V.; et al. Human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein-Barr virus infection. Cell Rep. 2013, 5, 1489–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, H.S.; Caligiuri, M.A. Molecular Basis for the Recognition of Herpes Simplex Virus Type 1 Infection by Human Natural Killer Cells. Front. Immunol. 2018, 9, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jost, S.; Altfeld, M. Control of human viral infections by natural killer cells. Annu. Rev. Immunol. 2013, 31, 163–194. [Google Scholar] [CrossRef] [PubMed]
- Foley, B.; Cooley, S.; Verneris, M.R.; Pitt, M.; Curtsinger, J.; Luo, X.; Lopez-Vergès, S.; Lanier, L.L.; Weisdorf, D.; Miller, J.S. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 2012, 119, 2665–2674. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, R.; Klein, E.; Pross, H.; Wigzell, H. “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur. J. Immunol. 1975, 5, 117–121. [Google Scholar] [CrossRef]
- Kärre, K.; Ljunggren, H.G.; Piontek, G.; Kiessling, R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986, 319, 675–678. [Google Scholar] [CrossRef]
- Wu, J.; Lanier, L.L. Natural killer cells and cancer. Adv. Cancer Res. 2003, 90, 127–156. [Google Scholar] [CrossRef]
- Guillerey, C.; Smyth, M.J. NK Cells and Cancer Immunoediting. Curr. Top. Microbiol. Immunol. 2016, 395, 115–145. [Google Scholar] [CrossRef] [PubMed]
- Marcus, A.; Gowen, B.G.; Thompson, T.W.; Iannello, A.; Ardolino, M.; Deng, W.; Wang, L.; Shifrin, N.; Raulet, D.H. Recognition of tumors by the innate immune system and natural killer cells. Adv. Immunol. 2014, 122, 91–128. [Google Scholar] [CrossRef] [Green Version]
- Guerra, N.; Tan, Y.X.; Joncker, N.T.; Choy, A.; Gallardo, F.; Xiong, N.; Knoblaugh, S.; Cado, D.; Greenberg, N.M.; Raulet, D.H. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 2008, 28, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Iguchi-Manaka, A.; Kai, H.; Yamashita, Y.; Shibata, K.; Tahara-Hanaoka, S.; Honda, S.; Yasui, T.; Kikutani, H.; Shibuya, K.; & Shibuya, A. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J. Exp. Med. 2008, 205, 2959–2964. [Google Scholar] [CrossRef] [Green Version]
- Imai, K.; Matsuyama, S.; Miyake, S.; Suga, K.; Nakachi, K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: An 11-year follow-up study of a general population. Lancet 2000, 356, 1795–1799. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, W.; Hu, B.; Wang, P.; Lv, X.; Chen, S.; Shao, Z. Prognostic Significance of Tumor-Infiltrating Natural Killer Cells in Solid Tumors: A Systematic Review and Meta-Analysis. Front. Immunol. 2020, 11, 1242. [Google Scholar] [CrossRef]
- Mastaglio, S.; Wong, E.; Perera, T.; Ripley, J.; Blombery, P.; Smyth, M.J.; Koldej, R.; Ritchie, D. Natural killer receptor ligand expression on acute myeloid leukemia impacts survival and relapse after chemotherapy. Blood Adv. 2018, 2, 335–346. [Google Scholar] [CrossRef] [Green Version]
- Luna, J.I.; Grossenbacher, S.K.; Murphy, W.J.; Canter, R.J. Targeting Cancer Stem Cells with Natural Killer Cell Immunotherapy. Expert Opin. Biol. Ther. 2017, 17, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Charap, A.J.; Enokida, T.; Brody, R.; Sfakianos, J.; Miles, B.; Bhardwaj, N.; Horowitz, A. Landscape of natural killer cell activity in head and neck squamous cell carcinoma. J. Immunother. Cancer. 2020, 8, e001523. [Google Scholar] [CrossRef]
- Melaiu, O.; Lucarini, V.; Cifaldi, L.; Fruci, D. Influence of the Tumor Microenvironment on NK Cell Function in Solid Tumors. Front. Immunol. 2020, 10, 3038. [Google Scholar] [CrossRef]
- Fauriat, C.; Just-Landi, S.; Mallet, F.; Arnoulet, C.; Sainty, D.; Olive, D.; Costello, R.T. Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood 2007, 109, 323–330. [Google Scholar] [CrossRef]
- Carlsten, M.; Baumann, B.C.; Simonsson, M.; Jädersten, M.; Forsblom, A.M.; Hammarstedt, C.; Bryceson, Y.T.; Ljunggren, H.G.; Hellström-Lindberg, E.; Malmberg, K.J. Reduced DNAM-1 expression on bone marrow NK cells associated with impaired killing of CD34+ blasts in myelodysplastic syndrome. Leukemia 2010, 24, 1607–1616. [Google Scholar] [CrossRef]
- Liu, S.; Galat, V.; Galat, Y.; Lee, Y.; Wainwright, D.; Wu, J. NK cell-based cancer immunotherapy: From basic biology to clinical development. J. Hematol. Oncol. 2021, 14, 7. [Google Scholar] [CrossRef]
- Barrow, A.D.; Martin, C.J.; Colonna, M. The Natural Cytotoxicity Receptors in Health and Disease. Front. Immunol. 2019, 10, 909. [Google Scholar] [CrossRef] [Green Version]
- Hecht, M.L.; Rosental, B.; Horlacher, T.; Hershkovitz, O.; De Paz, J.L.; Noti, C.; Schauer, S.; Porgador, A.; Seeberger, P.H. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J. Proteome Res. 2009, 8, 712–720. [Google Scholar] [CrossRef]
- Luczo, J.M.; Ronzulli, S.L.; Tompkins, S.M. Influenza A Virus Hemagglutinin and Other Pathogen Glycoprotein Interactions with NK Cell Natural Cytotoxicity Receptors NKp46, NKp44, and NKp30. Viruses 2021, 13, 156. [Google Scholar] [CrossRef]
- Brandt, C.S.; Baratin, M.; Yi, E.C.; Kennedy, J.; Gao, Z.; Fox, B.; Haldeman, B.; Ostrander, C.D.; Kaifu, T.; Chabannon, C.; et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 2009, 206, 1495–1503. [Google Scholar] [CrossRef] [Green Version]
- Daßler-Plenker, J.; Reiners, K.S.; van den Boorn, J.G.; Hansen, H.P.; Putschli, B.; Barnert, S.; Schuberth-Wagner, C.; Schubert, R.; Tüting, T.; Hallek, M.; et al. RIG-I activation induces the release of extracellular vesicles with antitumor activity. Oncoimmunology 2016, 5, e1219827. [Google Scholar] [CrossRef]
- Binici, J.; Hartmann, J.; Herrmann, J.; Schreiber, C.; Beyer, S.; Güler, G.; Vogel, V.; Tumulka, F.; Abele, R.; Mäntele, W.; et al. A soluble fragment of the tumor antigen BCL2-associated athanogene 6 (BAG-6) is essential and sufficient for inhibition of NKp30 receptor-dependent cytotoxicity of natural killer cells. J. Biol. Chem. 2013, 288, 34295–34303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baychelier, F.; Sennepin, A.; Ermonval, M.; Dorgham, K.; Debré, P.; Vieillard, V. Identification of a cellular ligand for the natural cytotoxicity receptor NKp44. Blood 2013, 122, 2935–2942. [Google Scholar] [CrossRef] [Green Version]
- Rosental, B.; Brusilovsky, M.; Hadad, U.; Oz, D.; Appel, M.Y.; Afergan, F.; Yossef, R.; Rosenberg, L.A.; Aharoni, A.; Cerwenka, A.; et al. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J. Immunol. 2011, 187, 5693–5702. [Google Scholar] [CrossRef]
- Barrow, A.D.; Edeling, M.A.; Trifonov, V.; Luo, J.; Goyal, P.; Bohl, B.; Bando, J.K.; Kim, A.H.; Walker, J.; Andahazy, M.; et al. Natural Killer Cells Control Tumor Growth by Sensing a Growth Factor. Cell 2018, 172, 534–548. [Google Scholar] [CrossRef] [Green Version]
- Gaggero, S.; Bruschi, M.; Petretto, A.; Parodi, M.; Del Zotto, G.; Lavarello, C.; Prato, C.; Santucci, L.; Barbuto, A.; Bottino, C.; et al. Nidogen-1 is a novel extracellular ligand for the NKp44 activating receptor. Oncoimmunology 2018, 7, e1470730. [Google Scholar] [CrossRef]
- Narni-Mancinelli, E.; Gauthier, L.; Baratin, M.; Guia, S.; Fenis, A.; Deghmane, A.E.; Rossi, B.; Fourquet, P.; Escalière, B.; Kerdiles, Y.M.; et al. Complement factor P is a ligand for the natural killer cell-activating receptor NKp46. Sci. Immunol. 2017, 2, eaam9628. [Google Scholar] [CrossRef] [Green Version]
- Nevins, A.M.; Subramanian, A.; Tapia, J.L.; Delgado, D.P.; Tyler, R.C.; Jensen, D.R.; Ouellette, A.J.; Volkman, B.F. A requirement for metamorphic interconversion in the antimicrobial activity of chemokine XCL1. Biochemistry 2016, 55, 3784–3793. [Google Scholar] [CrossRef]
- Alexandre, Y.O.; Ghilas, S.; Sanchez, C.; Le Bon, A.; Crozat, K.; Dalod, M. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med. 2016, 213, 75–92. [Google Scholar] [CrossRef] [Green Version]
- Dhar, P.; Wu, J.D. NKG2D and its ligands in cancer. Curr. Opin. Immunol. 2018, 51, 55–61. [Google Scholar] [CrossRef]
- Xing, S.; Ferrari de Andrade, L. NKG2D and MICA/B shedding: A ‘tag game’ between NK cells and malignant cells. Clin. Transl. Immunol. 2020, 9, e1230. [Google Scholar] [CrossRef]
- Hilpert, J.; Grosse-Hovest, L.; Grünebach, F.; Buechele, C.; Nuebling, T.; Raum, T.; Steinle, A.; Salih, H.R. Comprehensive analysis of NKG2D ligand expression and release in leukemia: Implications for NKG2D-mediated NK cell responses. J. Immunol. 2012, 189, 1360–1371. [Google Scholar] [CrossRef] [Green Version]
- Zingoni, A.; Vulpis, E.; Loconte, L.; Santoni, A. NKG2D Ligand Shedding in Response to Stress: Role of ADAM10. Front. Immunol. 2020, 11, 447. [Google Scholar] [CrossRef]
- Berhani, O.; Nachmani, D.; Yamin, R.; Schmiedel, D.; Bar-On, Y.; Mandelboim, O. Vigilin Regulates the Expression of the Stress-Induced Ligand MICB by Interacting with Its 5′ Untranslated Region. J. Immunol. 2017, 198, 3662–3670. [Google Scholar] [CrossRef] [Green Version]
- Lazarova, M.; Steinle, A. Impairment of NKG2D-Mediated Tumor Immunity by TGF-β. Front. Immunol. 2019, 10, 2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pende, D.; Spaggiari, G.M.; Marcenaro, S.; Martini, S.; Rivera, P.; Capobianco, A.; Falco, M.; Lanino, E.; Pierri, I.; Zambello, R.; et al. Analysis of the receptor-ligand interactions in the natural killer-mediated lysis of freshly isolated myeloid or lymphoblastic leukemias: Evidence for the involvement of the Poliovirus receptor (CD155) and Nectin-2 (CD112). Blood 2005, 105, 2066–2073. [Google Scholar] [CrossRef]
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in regulating immunity and tolerance. Clin. Immunol. 2016, 164, 10–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcenaro, E.; Augugliaro, R.; Falco, M.; Castriconi, R.; Parolini, S.; Sivori, S.; Romeo, E.; Millo, R.; Moretta, L.; Bottino, C.; et al. CD59 is physically and functionally associated with natural cytotoxicity receptors and activates human NK cell-mediated cytotoxicity. Eur. J. Immunol. 2003, 33, 3367–3376. [Google Scholar] [CrossRef]
- Bryceson, Y.T.; March, M.E.; Barber, D.F.; Ljunggren, H.G.; Long, E.O. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J. Exp. Med. 2005, 202, 1001–1012. [Google Scholar] [CrossRef]
- Stewart, C.A.; Laugier-Anfossi, F.; Vély, F.; Saulquin, X.; Riedmuller, J.; Tisserant, A.; Gauthier, L.; Romagné, F.; Ferracci, G.; Arosa, F.A.; et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 13224–13229. [Google Scholar] [CrossRef] [Green Version]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Söderström, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Blunt, M.D.; Khakoo, S.I. Activating killer cell immunoglobulin-like receptors: Detection, function and therapeutic use. Int. J. Immunogenet. 2020, 47, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef] [Green Version]
- Gauthier, M.; Laroye, C.; Bensoussan, D.; Boura, C.; Decot, V. Natural Killer cells and monoclonal antibodies: Two partners for successful antibody dependent cytotoxicity against tumor cells. Crit. Rev. Oncol. Hematol. 2021, 160, 103261. [Google Scholar] [CrossRef]
- Felices, M.; Lenvik, T.R.; Davis, Z.B.; Miller, J.S.; Vallera, D.A. Generation of BiKEs and TriKEs to Improve NK Cell-Mediated Targeting of Tumor Cells. Methods Mol. Biol. 2016, 1441, 333–346. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.M.; Kulski, J.K.; Gaudieri, S.; Witt, C.S.; Freitas, E.M.; Trowsdale, J.; Christiansen, F.T. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene 2004, 335, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Parham, P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005, 5, 201–214. [Google Scholar] [CrossRef]
- Horowitz, A.; Djaoud, Z.; Nemat-Gorgani, N.; Blokhuis, J.; Hilton, H.G.; Béziat, V.; Malmberg, K.J.; Norman, P.J.; Guethlein, L.A.; Parham, P. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci. Immunol. 2016, 1, eaag1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manser, A.R.; Weinhold, S.; Uhrberg, M. Human KIR repertoires: Shaped by genetic diversity and evolution. Immunol. Rev. 2015, 267, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, S.; Long, E.O. KIR2DL4 (CD158d): An activation receptor for HLA-G. Front. Immunol. 2012, 3, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- André, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Bléry, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Samaridis, J.; Cella, M.; Angman, L.; Allen, R.L.; O’Callaghan, C.A.; Dunbar, R.; Ogg, G.S.; Cerundolo, V.; Rolink, A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J. Immunol. 1998, 160, 3096–3100. [Google Scholar]
- Chiossone, L.; Vienne, M.; Kerdiles, Y.M.; Vivier, E. Natural killer cell immunotherapies against cancer: Checkpoint inhibitors and more. Semin. Immunol. 2017, 31, 55–63. [Google Scholar] [CrossRef]
- Pesce, S.; Greppi, M.; Tabellini, G.; Rampinelli, F.; Parolini, S.; Olive, D.; Moretta, L.; Moretta, A.; Marcenaro, E. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J. Allergy Clin. Immunol. 2017, 139, 335–346. [Google Scholar] [CrossRef] [Green Version]
- Pesce, S.; Greppi, M.; Grossi, F.; Del Zotto, G.; Moretta, L.; Sivori, S.; Genova, C.; Marcenaro, E. PD/1-PD-Ls Checkpoint: Insight on the Potential Role of NK Cells. Front. Immunol. 2019, 10, 1242. [Google Scholar] [CrossRef]
- Guan, J.; Wang, R.; Hasan, S.; Tao, L.; Wazir, M.; Jain, A.G.; Zhu, X.; Perkins, S.; Mohamed, S.; Chang, C.C.; et al. Prognostic Significance of the Dynamic Change of Programmed Death-ligand 1 Expression in Patients with Multiple Myeloma. Cureus 2019, 11, e4401. [Google Scholar] [CrossRef] [Green Version]
- Dougall, W.C.; Kurtulus, S.; Smyth, M.J.; Anderson, A.C. TIGIT and CD96: New checkpoint receptor targets for cancer immunotherapy. Immunol. Rev. 2017, 276, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.M.; Li, W.Q.; Wu, Y.H.; Han, L.; Cao, X.G.; Yang, X.M.; Wang, H.F.; Zhao, W.S.; Zhai, W.J.; Qi, Y.M.; et al. Intrinsic Expression of Immune Checkpoint Molecule TIGIT Could Help Tumor Growth in vivo by Suppressing the Function of NK and CD8(+) T Cells. Front. Immunol. 2018, 9, 2821. [Google Scholar] [CrossRef] [Green Version]
- Kandel, S.; Adhikary, P.; Li, G.; Cheng, K. The TIM3/Gal9 signaling pathway: An emerging target for cancer immunotherapy. Cancer Lett. 2021, 510, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Ruffo, E.; Wu, R.C.; Bruno, T.C.; Workman, C.J.; Vignali, D.A.A. Lymphocyte-activation gene 3 (LAG3): The next immune checkpoint receptor. Semin. Immunol. 2019, 42, 101305. [Google Scholar] [CrossRef]
- Reiners, K.S.; Topolar, D.; Henke, A.; Simhadri, V.R.; Kessler, J.; Sauer, M.; Bessler, M.; Hansen, H.P.; Tawadros, S.; Herling, M.; et al. Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 2013, 121, 3658–3665. [Google Scholar] [CrossRef] [PubMed]
- Maki, G.; Hayes, G.M.; Naji, A.; Tyler, T.; Carosella, E.D.; Rouas-Freiss, N.; Gregory, S.A. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: Implication of HLA-G. Leukemia 2008, 22, 998–1006. [Google Scholar] [CrossRef] [Green Version]
- Jewell, A.P.; Worman, C.P.; Giles, F.J.; Goldstone, A.H.; Lydyard, P.M. Resistance of chronic lymphocytic leukaemia cells to interferon-alpha generated lymphokine activated killer cells. Leuk. Lymphoma 1992, 7, 473–480. [Google Scholar] [CrossRef]
- Spitz, D.L.; Zucker-Franklin, D.; Nabi, Z.F. Unmasking of cryptic natural killer (NK) cell recognition sites on chronic lymphocytic leukemia lymphocytes. Am. J. Hematol. 1988, 28, 155–161. [Google Scholar] [CrossRef]
- Huergo-Zapico, L.; Gonzalez-Rodriguez, A.P.; Contesti, J.; Gonzalez, E.; López-Soto, A.; Fernandez-Guizan, A.; Acebes-Huerta, A.; de Los Toyos, J.R.; Lopez-Larrea, C.; Groh, V.; et al. Expression of ERp5 and GRP78 on the membrane of chronic lymphocytic leukemia cells: Association with soluble MICA shedding. Cancer Immunol. Immunother. 2012, 61, 1201–1210. [Google Scholar] [CrossRef]
- Eskelund, C.W.; Nederby, L.; Thysen, A.H.; Skovbo, A.; Roug, A.S.; Hokland, M.E. Interleukin-21 and rituximab enhance NK cell functionality in patients with B-cell chronic lymphocytic leukaemia. Leuk. Res. 2011, 35, 914–920. [Google Scholar] [CrossRef]
- Le Garff-Tavernier, M.; Herbi, L.; de Romeuf, C.; Nguyen-Khac, F.; Davi, F.; Grelier, A.; Boudjoghra, M.; Maloum, K.; Choquet, S.; Urbain, R.; et al. Antibody-dependent cellular cytotoxicity of the optimized anti-CD20 monoclonal antibody ublituximab on chronic lymphocytic leukemia cells with the 17p deletion. Leukemia 2014, 28, 230–233. [Google Scholar] [CrossRef]
- Hofland, T.; Eldering, E.; Kater, A.P.; Tonino, S.H. Engaging Cytotoxic T and NK Cells for Immunotherapy in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2019, 20, 4315. [Google Scholar] [CrossRef] [Green Version]
- Demanet, C.; Mulder, A.; Deneys, V.; Worsham, M.J.; Maes, P.; Claas, F.H.; Ferrone, S. Down-regulation of HLA-A and HLA-Bw6, but not HLA-Bw4, allospecificities in leukemic cells: An escape mechanism from CTL and NK attack? Blood 2004, 103, 3122–3130. [Google Scholar] [CrossRef] [Green Version]
- Verheyden, S.; Ferrone, S.; Mulder, A.; Claas, F.H.; Schots, R.; De Moerloose, B.; Benoit, Y.; Demanet, C. Role of the inhibitory KIR ligand HLA-Bw4 and HLA-C expression levels in the recognition of leukemic cells by Natural Killer cells. Cancer Immunol. Immunother. 2009, 58, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.T.; Zhu, H.Y.; Wu, Y.J.; Xia, Y.; Wu, J.Z.; Wu, W.; Liang, J.H.; Wang, L.; Fan, L.; Li, J.Y.; et al. Elevated absolute NK cell counts in peripheral blood predict good prognosis in chronic lymphocytic leukemia. J. Cancer Res. Clin. Oncol. 2018, 144, 449–457. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, A.P.; Contesti, J.; Huergo-Zapico, L.; Lopez-Soto, A.; Fernández-Guizán, A.; Acebes-Huerta, A.; Gonzalez-Huerta, A.J.; Gonzalez, E.; Fernandez-Alvarez, C.; Gonzalez, S. Prognostic significance of CD8 and CD4 T cells in chronic lymphocytic leukemia. Leuk. Lymphoma 2010, 51, 1829–1836. [Google Scholar] [CrossRef]
- Palmer, S.; Hanson, C.A.; Zent, C.S.; Porrata, L.F.; Laplant, B.; Geyer, S.M.; Markovic, S.N.; Call, T.G.; Bowen, D.A.; Jelinek, D.F.; et al. Prognostic importance of T and NK-cells in a consecutive series of newly diagnosed patients with chronic lymphocytic leukaemia. Br. J. Haematol. 2008, 141, 607–614. [Google Scholar] [CrossRef]
- Hadadi, L.; Hafezi, M.; Amirzargar, A.A.; Sharifian, R.A.; Abediankenari, S.; Asgarian-Omran, H. Dysregulated Expression of Tim-3 and NKp30 Receptors on NK Cells of Patients with Chronic Lymphocytic Leukemia. Oncol. Res. Treat. 2019, 42, 202–208. [Google Scholar] [CrossRef]
- Huergo-Zapico, L.; Acebes-Huerta, A.; Gonzalez-Rodriguez, A.P.; Contesti, J.; Gonzalez-García, E.; Payer, A.R.; Villa-Alvarez, M.; Fernández-Guizán, A.; López-Soto, A.; Gonzalez, S. Expansion of NK cells and reduction of NKG2D expression in chronic lymphocytic leukemia. Correlation with progressive disease. PLoS ONE 2014, 9, e108326. [Google Scholar] [CrossRef]
- MacFarlane, A.W., 4th; Jillab, M.; Smith, M.R.; Alpaugh, R.K.; Cole, M.E.; Litwin, S.; Millenson, M.M.; Al-Saleem, T.; Cohen, A.D.; Campbell, K.S. NK cell dysfunction in chronic lymphocytic leukemia is associated with loss of the mature cells expressing inhibitory killer cell Ig-like receptors. Oncoimmunology 2017, 6, e1330235. [Google Scholar] [CrossRef]
- Gumá, M.; Angulo, A.; Vilches, C.; Gómez-Lozano, N.; Malats, N.; López-Botet, M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004, 104, 3664–3671. [Google Scholar] [CrossRef] [Green Version]
- López-Botet, M.; Muntasell, A.; Vilches, C. The CD94/NKG2C+ NK-cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin. Immunol. 2014, 26, 145–151. [Google Scholar] [CrossRef]
- Parry, H.M.; Stevens, T.; Oldreive, C.; Zadran, B.; McSkeane, T.; Rudzki, Z.; Paneesha, S.; Chadwick, C.; Stankovic, T.; Pratt, G.; et al. NK cell function is markedly impaired in patients with chronic lymphocytic leukaemia but is preserved in patients with small lymphocytic lymphoma. Oncotarget 2016, 7, 68513–68526. [Google Scholar] [CrossRef] [Green Version]
- Lotz, M.; Ranheim, E.; Kipps, T.J. Transforming growth factor beta as endogenous growth inhibitor of chronic lymphocytic leukemia B cells. J. Exp. Med. 1994, 179, 999–1004. [Google Scholar] [CrossRef]
- Lad, D.; Hoeppli, R.; Huang, Q.; Garcia, R.; Xu, L.; Toze, C.; Broady, R.; Levings, M. Regulatory T-cells drive immune dysfunction in CLL. Leuk. Lymphoma 2018, 59, 486–489. [Google Scholar] [CrossRef]
- Kowalska, W.; Bojarska-Junak, A. Monocytic MDSC as a source of immunosuppressive cytokines in chronic lymphocytic leukemia (CLL) microenvironment. Folia Histochem. Cytobiol. 2020, 58, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Nückel, H.; Switala, M.; Sellmann, L.; Horn, P.A.; Dürig, J.; Dührsen, U.; Küppers, R.; Grosse-Wilde, H.; Rebmann, V. The prognostic significance of soluble NKG2D ligands in B-cell chronic lymphocytic leukemia. Leukemia 2010, 24, 1152–1159. [Google Scholar] [CrossRef]
- Sánchez-Martínez, D.; Lanuza, P.M.; Gómez, N.; Muntasell, A.; Cisneros, E.; Moraru, M.; Azaceta, G.; Anel, A.; Martínez-Lostao, L.; Villalba, M.; et al. Activated Allogeneic NK Cells Preferentially Kill Poor Prognosis B-Cell Chronic Lymphocytic Leukemia Cells. Front. Immunol. 2016, 7, 454. [Google Scholar] [CrossRef]
- Puiggros, A.; Blanco, G.; Muntasell, A.; Rodríguez-Rivera, M.; Nonell, L.; Altadill, M.; Puigdecanet, E.; Arnal, M.; Calvo, X.; Gimeno, E.; et al. Reduced expansion of CD94/NKG2C+ NK cells in chronic lymphocytic leukemia and CLL-like monoclonal B-cell lymphocytosis is not related to increased human cytomegalovirus seronegativity or NKG2C deletions. Int. J. Lab. Hematol. 2021, in press. [Google Scholar] [CrossRef]
- Petersen, L.; Roug, A.S.; Skovbo, A.; Thysen, A.H.; Eskelund, C.W.; Hokland, M.E. The CD94/NKG2C-expressing NK cell subset is augmented in chronic lymphocytic leukemia patients with positive human cytomegalovirus serostatus. Viral Immunol. 2009, 22, 333–337. [Google Scholar] [CrossRef]
- McWilliams, E.M.; Mele, J.M.; Cheney, C.; Timmerman, E.A.; Fiazuddin, F.; Strattan, E.J.; Mo, X.; Byrd, J.C.; Muthusamy, N.; Awan, F.T. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology 2016, 5, e1226720. [Google Scholar] [CrossRef] [Green Version]
- Wagner, B.; da Silva Nardi, F.; Schramm, S.; Kraemer, T.; Celik, A.A.; Dürig, J.; Horn, P.A.; Dührsen, U.; Nückel, H.; Rebmann, V. HLA-E allelic genotype correlates with HLA-E plasma levels and predicts early progression in chronic lymphocytic leukemia. Cancer 2017, 123, 814–823. [Google Scholar] [CrossRef]
- Linet, M.S.; Bias, W.B.; Dorgan, J.F.; McCaffrey, L.D.; Humphrey, R.L. HLA antigens in chronic lymphocytic leukemia. Tissue Antigens 1988, 31, 71–78. [Google Scholar] [CrossRef]
- Mueller, L.P.; Machulla, H.K. Increased frequency of homozygosity for HLA class II loci in female patients with chronic lymphocytic leukemia. Leuk. Lymphoma 2002, 43, 1013–1019. [Google Scholar] [CrossRef]
- Villa-Álvarez, M.; Lorenzo-Herrero, S.; Gonzalez-Rodriguez, A.P.; López-Soto, A.; Payer, A.R.; Gonzalez-Garcia, E.; Huergo-Zapico, L.; Gonzalez, S. Ig-like transcript 2 (ILT2) suppresses T cell function in chronic lymphocytic leukemia. Oncoimmunology 2017, 6, e1353856. [Google Scholar] [CrossRef]
- Własiuk, P.; Tomczak, W.; Zając, M.; Dmoszyńska, A.; Giannopoulos, K. Total expression of HLA-G and TLR-9 in chronic lymphocytic leukemia patients. Hum. Immunol. 2013, 74, 1592–1597. [Google Scholar] [CrossRef]
- Nückel, H.; Rebmann, V.; Dürig, J.; Dührsen, U.; Grosse-Wilde, H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood 2005, 105, 1694–1698. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, R.; Audrito, V.; Vacca, P.; Rossi, D.; Brusa, D.; Stignani, M.; Bortolotti, D.; D’Arena, G.; Coscia, M.; Laurenti, L.; et al. HLA-G is a component of the chronic lymphocytic leukemia escape repertoire to generate immune suppression: Impact of the HLA-G 14 base pair (rs66554220) polymorphism. Haematologica 2014, 99, 888–896. [Google Scholar] [CrossRef]
- Taghiloo, S.; Allahmoradi, E.; Ebadi, R.; Tehrani, M.; Hosseini-Khah, Z.; Janbabaei, G.; Shekarriz, R.; Asgarian-Omran, H. Upregulation of Galectin-9 and PD-L1 Immune Checkpoints Molecules in Patients with Chronic Lymphocytic Leukemia. Asian Pac. J. Cancer Prev. 2017, 18, 2269–2274. [Google Scholar] [CrossRef]
- Wdowiak, K.; Gallego-Colon, E.; Francuz, T.; Czajka-Francuz, P.; Ruiz-Agamez, N.; Kubeczko, M.; Grochoła, I.; Wybraniec, M.T.; Chudek, J.; Wojnar, J. Increased serum levels of Galectin-9 in patients with chronic lymphocytic leukemia. Oncol. Lett. 2019, 17, 1019–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallois, A.; Silva, I.; Osman, I.; Bhardwaj, N. Reversal of natural killer cell exhaustion by TIM-3 blockade. Oncoimmunology 2015, 3, e946365. [Google Scholar] [CrossRef] [Green Version]
- Brusa, D.; Serra, S.; Coscia, M.; Rossi, D.; D’Arena, G.; Laurenti, L.; Jaksic, O.; Fedele, G.; Inghirami, G.; Gaidano, G.; et al. The PD-1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia. Haematologica 2013, 98, 953–963. [Google Scholar] [CrossRef]
- Palma, M.; Gentilcore, G.; Heimersson, K.; Mozaffari, F.; Näsman-Glaser, B.; Young, E.; Rosenquist, R.; Hansson, L.; Österborg, A.; Mellstedt, H. T cells in chronic lymphocytic leukemia display dysregulated expression of immune checkpoints and activation markers. Haematologica 2017, 102, 562–572. [Google Scholar] [CrossRef] [Green Version]
- Beldi-Ferchiou, A.; Lambert, M.; Dogniaux, S.; Vély, F.; Vivier, E.; Olive, D.; Dupuy, S.; Levasseur, F.; Zucman, D.; Lebbé, C.; et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget 2016, 7, 72961–72977. [Google Scholar] [CrossRef] [Green Version]
- Grzywnowicz, M.; Karczmarczyk, A.; Skorka, K.; Zajac, M.; Zaleska, J.; Chocholska, S.; Tomczak, W.; Giannopoulos, K. Expression of Programmed Death 1 Ligand in Different Compartments of Chronic Lymphocytic Leukemia. Acta Haematol. 2015, 134, 255–262. [Google Scholar] [CrossRef]
- Holdenrieder, S.; Stieber, P.; Peterfi, A.; Nagel, D.; Steinle, A.; Salih, H.R. Soluble MICA in malignant diseases. Int. J. Cancer 2006, 118, 684–687. [Google Scholar] [CrossRef]
- Zhang, J.; Basher, F.; Wu, J.D. NKG2D Ligands in Tumor Immunity: Two Sides of a Coin. Front. Immunol. 2015, 6, 97. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Lu, S.; Wang, X.; Page, S.T.; Higano, C.S.; Plymate, S.R.; Greenberg, N.M.; Sun, S.; Li, Z.; Wu, J.D. Perturbation of NK cell peripheral homeostasis accelerates prostate carcinoma metastasis. J. Clin. Investig. 2013, 123, 4410–4422. [Google Scholar] [CrossRef]
- Reinart, N.; Nguyen, P.H.; Boucas, J.; Rosen, N.; Kvasnicka, H.M.; Heukamp, L.; Rudolph, C.; Ristovska, V.; Velmans, T.; Mueller, C.; et al. Delayed development of chronic lymphocytic leukemia in the absence of macrophage migration inhibitory factor. Blood 2013, 121, 812–821. [Google Scholar] [CrossRef] [Green Version]
- Carosella, E.D. The tolerogenic molecule HLA-G. Immunol. Lett. 2011, 138, 22–24. [Google Scholar] [CrossRef]
- Liu, L.; Wang, L.; Zhao, L.; He, C.; Wang, G. The Role of HLA-G in Tumor Escape: Manipulating the Phenotype and Function of Immune Cells. Front. Oncol. 2020, 10, 597468. [Google Scholar] [CrossRef]
- LeMaoult, J.; Krawice-Radanne, I.; Dausset, J.; Carosella, E.D. HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc. Natl. Acad. Sci. USA 2004, 101, 7064–7069. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Yan, W.H. Human Leukocyte Antigen-G (HLA-G) Expression in Cancers: Roles in Immune Evasion, Metastasis and Target for Therapy. Mol. Med. 2015, 21, 782–791. [Google Scholar] [CrossRef]
- Erikci, A.A.; Karagoz, B.; Ozyurt, M.; Ozturk, A.; Kilic, S.; Bilgi, O. HLA-G expression in B chronic lymphocytic leukemia: A new prognostic marker? Hematology 2009, 14, 101–105. [Google Scholar] [CrossRef]
- Perez-Chacon, G.; Rosado, S.; Rebolleda, N.; Losada-Fernandez, I.; Vargas, J.A.; Morado, M.; Jorda, J.; Perez-Aciego, P. Prognostic irrelevance of HLA-G in B-cell chronic lymphocytic leukemia. Int. J. Lab. Hematol. 2009, 31, 327–337. [Google Scholar] [CrossRef]
- Sebti, Y.; Le Friec, G.; Pangault, C.; Gros, F.; Drénou, B.; Guilloux, V.; Bernard, M.; Lamy, T.; Fauchet, R.; Amiot, L. Soluble HLA-G molecules are increased in lymphoproliferative disorders. Hum. Immunol. 2003, 64, 1093–1101. [Google Scholar] [CrossRef]
- Giannopoulos, K.; Schmitt, M.; Kowal, M.; Własiuk, P.; Bojarska-Junak, A.; Roliński, J.; Dmoszyńska, A. The significance of soluble HLA-G plasma levels as well as messenger HLA-G for B-cell chronic lymphocytic leukemia (B-CLL). Leuk. Res. 2008, 32, 1815–1819. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; Mendes-Junior, C.T.; Veiga-Castelli, L.C.; Roger, M.; Moreau, P.; Donadi, E.A. A comprehensive study of polymorphic sites along the HLA-G gene: Implication for gene regulation and evolution. Mol. Biol. Evol. 2011, 28, 3069–3086. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yu, S.; Han, Y.; Wang, Y.; Sun, Y. Human leukocyte antigen-G expression and polymorphisms promote cancer development and guide cancer diagnosis/treatment. Oncol. Lett. 2018, 15, 699–709. [Google Scholar] [CrossRef]
- Rousseau, P.; Le Discorde, M.; Mouillot, G.; Marcou, C.; Carosella, E.D.; Moreau, P. The 14 bp deletion-insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum. Immunol. 2003, 64, 1005–1010. [Google Scholar] [CrossRef]
- Weiss, L.; Melchardt, T.; Egle, A.; Grabmer, C.; Greil, R.; Tinhofer, I. Regulatory T cells predict the time to initial treatment in early stage chronic lymphocytic leukemia. Cancer 2011, 117, 2163–2169. [Google Scholar] [CrossRef] [PubMed]
- Buechele, C.; Baessler, T.; Wirths, S.; Schmohl, J.U.; Schmiedel, B.J.; Salih, H.R. Glucocorticoid-induced TNFR-related protein (GITR) ligand modulates cytokine release and NK cell reactivity in chronic lymphocytic leukemia (CLL). Leukemia 2012, 26, 991–1000. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Keating, M.J.; Manshouri, T.; Giles, F.J.; Dey, A.; Estrov, Z.; Koller, C.A.; Kurzrock, R.; Thomas, D.A.; Faderl, S.; et al. The clinical significance of tumor necrosis factor-alpha plasma level in patients having chronic lymphocytic leukemia. Blood 2002, 100, 1215–1219. [Google Scholar] [CrossRef]
- Lai, R.; O’Brien, S.; Maushouri, T.; Rogers, A.; Kantarjian, H.; Keating, M.; Albitar, M. Prognostic value of plasma interleukin-6 levels in patients with chronic lymphocytic leukemia. Cancer 2002, 95, 1071–1075. [Google Scholar] [CrossRef]
- Binsky, I.; Haran, M.; Starlets, D.; Gore, Y.; Lantner, F.; Harpaz, N.; Leng, L.; Goldenberg, D.M.; Shvidel, L.; Berrebi, A.; et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc. Natl. Acad. Sci. USA 2007, 104, 13408–13413. [Google Scholar] [CrossRef] [Green Version]
- Buechele, C.; Baessler, T.; Schmiedel, B.J.; Schumacher, C.E.; Grosse-Hovest, L.; Rittig, K.; Salih, H.R. 4-1BB ligand modulates direct and Rituximab-induced NK-cell reactivity in chronic lymphocytic leukemia. Eur. J. Immunol. 2012, 42, 737–748. [Google Scholar] [CrossRef]
- Wild, J.; Schmiedel, B.J.; Maurer, A.; Raab, S.; Prokop, L.; Stevanović, S.; Dörfel, D.; Schneider, P.; Salih, H.R. Neutralization of (NK-cell-derived) B-cell activating factor by Belimumab restores sensitivity of chronic lymphoid leukemia cells to direct and Rituximab-induced NK lysis. Leukemia 2015, 29, 1676–1683. [Google Scholar] [CrossRef] [Green Version]
- Boross, P.; Leusen, J.H.; Boross, P.; Leusen, J.H. Mechanisms of action of CD20 antibodies. Am. J. Cancer Res. 2012, 2, 676–690. [Google Scholar]
- Jaglowski, S.M.; Byrd, J.C. Rituximab in chronic lymphocytic leukemia. Semin. Hematol. 2010, 47, 156–169. [Google Scholar] [CrossRef]
- Fischer, K.; Bahlo, J.; Fink, A.M.; Goede, V.; Herling, C.D.; Cramer, P.; Langerbeins, P.; von Tresckow, J.; Engelke, A.; Maurer, C.; et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: Updated results of the CLL8 trial. Blood 2016, 127, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Eichhorst, B.; Fink, A.M.; Bahlo, J.; Busch, R.; Kovacs, G.; Maurer, C.; Lange, E.; Köppler, H.; Kiehl, M.; Sökler, M.; et al. First-line chemoimmunotherapy with bendamustine and rituximab versus fludarabine, cyclophosphamide, and rituximab in patients with advanced chronic lymphocytic leukaemia (CLL10): An international, open-label, randomised, phase 3, non-inferiority trial. Lancet Oncol. 2016, 17, 928–942. [Google Scholar] [CrossRef]
- Stilgenbauer, S.; Schnaiter, A.; Paschka, P.; Zenz, T.; Rossi, M.; Döhner, K.; Bühler, A.; Böttcher, S.; Ritgen, M.; Kneba, M.; et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 2014, 123, 3247–3254. [Google Scholar] [CrossRef] [Green Version]
- Sportoletti, P.; Baldoni, S.; Del Papa, B.; Aureli, P.; Dorillo, E.; Ruggeri, L.; Plebani, S.; Amico, V.; Di Tommaso, A.; Rosati, E.; et al. A revised NOTCH1 mutation frequency still impacts survival while the allele burden predicts early progression in chronic lymphocytic leukemia. Leukemia 2014, 28, 436–439. [Google Scholar] [CrossRef]
- De Falco, F.; Sabatini, R.; Del Papa, B.; Falzetti, F.; Di Ianni, M.; Sportoletti, P.; Baldoni, S.; Screpanti, I.; Marconi, P.; Rosati, E. Notch signaling sustains the expression of Mcl-1 and the activity of eIF4E to promote cell survival in CLL. Oncotarget 2015, 6, 16559–16572. [Google Scholar] [CrossRef] [Green Version]
- Vyas, M.; Müller, R.; Pogge von Strandmann, E. Antigen Loss Variants: Catching Hold of Escaping Foes. Front. Immunol. 2017, 8, 175. [Google Scholar] [CrossRef] [Green Version]
- Cartron, G.; Dacheux, L.; Salles, G.; Solal-Celigny, P.; Bardos, P.; Colombat, P.; Watier, H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 2002, 99, 754–758. [Google Scholar] [CrossRef] [Green Version]
- Werlenius, O.; Aurelius, J.; Hallner, A.; Akhiani, A.A.; Simpanen, M.; Martner, A.; Andersson, P.O.; Hellstrand, K.; Thorén, F.B. Reactive oxygen species induced by therapeutic CD20 antibodies inhibit natural killer cell-mediated antibody-dependent cellular cytotoxicity against primary CLL cells. Oncotarget 2016, 7, 32046–32053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Martinez, D.; Allende-Vega, N.; Orecchioni, S.; Talarico, G.; Cornillon, A.; Vo, D.N.; Rene, C.; Lu, Z.Y.; Krzywinska, E.; Anel, A.; et al. Expansion of allogeneic NK cells with efficient antibody-dependent cell cytotoxicity against multiple tumors. Theranostics 2018, 8, 3856–3869. [Google Scholar] [CrossRef] [PubMed]
- van der Horst, H.J.; Nijhof, I.S.; Mutis, T.; Chamuleau, M. Fc-Engineered Antibodies with Enhanced Fc-Effector Function for the Treatment of B-Cell Malignancies. Cancers 2020, 12, 3041. [Google Scholar] [CrossRef] [PubMed]
- Offner, F.; Robak, T.; Janssens, A.; Govind Babu, K.; Kloczko, J.; Grosicki, S.; Mayer, J.; Panagiotidis, P.; Schuh, A.; Pettitt, A.; et al. A five-year follow-up of untreated patients with chronic lymphocytic leukaemia treated with ofatumumab and chlorambucil: Final analysis of the Complement 1 phase 3 trial. Br. J. Haematol. 2020, 190, 736–740. [Google Scholar] [CrossRef]
- Jones, J.A.; Robak, T.; Brown, J.R.; Awan, F.T.; Badoux, X.; Coutre, S.; Loscertales, J.; Taylor, K.; Vandenberghe, E.; Wach, M.; et al. Efficacy and safety of idelalisib in combination with ofatumumab for previously treated chronic lymphocytic leukaemia: An open-label, randomised phase 3 trial. Lancet Haematol. 2017, 4, e114–e126. [Google Scholar] [CrossRef]
- Moreno, C.; Greil, R.; Demirkan, F.; Tedeschi, A.; Anz, B.; Larratt, L.; Simkovic, M.; Samoilova, O.; Novak, J.; Ben-Yehuda, D.; et al. Ibrutinib plus obinutuzumab versus chlorambucil plus obinutuzumab in first-line treatment of chronic lymphocytic leukaemia (iLLUMINATE): A multicentre, randomised, open-label, phase 3 trial [published correction appears in Lancet Oncol. 2019 Jan;20(1):e10]. Lancet Oncol. 2019, 20, 43–56. [Google Scholar] [CrossRef]
- Sharman, J.P.; Egyed, M.; Jurczak, W.; Skarbnik, A.; Pagel, J.M.; Flinn, I.W.; Kamdar, M.; Munir, T.; Walewska, R.; Corbett, G.; et al. Acalabrutinib with or without obinutuzumab versus chlorambucil and obinutuzumab for treatment-naive chronic lymphocytic leukaemia (ELEVATE TN): A randomised, controlled, phase 3 trial. Lancet 2020, 395, 1278–1291. [Google Scholar] [CrossRef]
- Fischer, K.; Al-Sawaf, O.; Bahlo, J.; Fink, A.M.; Tandon, M.; Dixon, M.; Robrecht, S.; Warburton, S.; Humphrey, K.; Samoylova, O.; et al. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2019, 380, 2225–2236. [Google Scholar] [CrossRef]
- Sawas, A.; Farber, C.M.; Schreeder, M.T.; Khalil, M.Y.; Mahadevan, D.; Deng, C.; Amengual, J.E.; Nikolinakos, P.G.; Kolesar, J.M.; Kuhn, J.G.; et al. A phase 1/2 trial of ublituximab, a novel anti-CD20 monoclonal antibody, in patients with B-cell non-Hodgkin lymphoma or chronic lymphocytic leukaemia previously exposed to rituximab. Br. J. Haematol. 2017, 177, 243–253. [Google Scholar] [CrossRef]
- Sharman, J.P.; Farber, C.M.; Mahadevan, D.; Schreeder, M.T.; Brooks, H.D.; Kolibaba, K.S.; Fanning, S.; Klein, L.; Greenwald, D.R.; Sportelli, P.; et al. Ublituximab (TG-1101), a novel glycoengineered anti-CD20 antibody, in combination with ibrutinib is safe and highly active in patients with relapsed and/or refractory chronic lymphocytic leukaemia: Results of a phase 2 trial. Br. J. Haematol. 2017, 176, 412–420. [Google Scholar] [CrossRef]
- Sharman, J.P.; Brander, D.M.; Mato, A.R.; Ghosh, N.; Schuster, S.J.; Kambhampati, S.; Burke, J.M.; Lansigan, F.; Schreeder, M.T.; Lunin, S.D.; et al. Ublituximab plus ibrutinib versus ibrutinib alone for patients with relapsed or refractory high-risk chronic lymphocytic leukaemia (GENUINE): A phase 3, multicentre, open-label, randomised trial. Lancet Haematol. 2021, 8, e254–e266. [Google Scholar] [CrossRef]
- Cardarelli, P.M.; Rao-Naik, C.; Chen, S.; Huang, H.; Pham, A.; Moldovan-Loomis, M.C.; Pan, C.; Preston, B.; Passmore, D.; Liu, J.; et al. A nonfucosylated human antibody to CD19 with potent B-cell depletive activity for therapy of B-cell malignancies. Cancer Immunol. Immunother. 2010, 59, 257–265. [Google Scholar] [CrossRef]
- Horton, H.M.; Bernett, M.J.; Pong, E.; Peipp, M.; Karki, S.; Chu, S.Y.; Richards, J.O.; Vostiar, I.; Joyce, P.F.; Repp, R.; et al. Potent in vitro and in vivo activity of an Fc-engineered anti-CD19 monoclonal antibody against lymphoma and leukemia. Cancer Res. 2008, 68, 8049–8057. [Google Scholar] [CrossRef] [Green Version]
- Ohmachi, K.; Ogura, M.; Suehiro, Y.; Ando, K.; Uchida, T.; Choi, I.; Ogawa, Y.; Kobayashi, M.; Fukino, K.; Yokoi, Y.; et al. A multicenter phase I study of inebilizumab, a humanized anti-CD19 monoclonal antibody, in Japanese patients with relapsed or refractory B-cell lymphoma and multiple myeloma. Int. J. Hematol. 2019, 109, 657–664. [Google Scholar] [CrossRef]
- Jurczak, W.; Zinzani, P.L.; Gaidano, G.; Goy, A.; Provencio, M.; Nagy, Z.; Robak, T.; Maddocks, K.; Buske, C.; Ambarkhane, S.; et al. Phase IIa study of the CD19 antibody MOR208 in patients with relapsed or refractory B-cell non-Hodgkin’s lymphoma. Ann. Oncol. 2018, 29, 1266–1272. [Google Scholar] [CrossRef]
- Payandeh, Z.; Noori, E.; Khalesi, B.; Mard-Soltani, M.; Abdolalizadeh, J.; Khalili, S. Anti-CD37 targeted immunotherapy of B-Cell malignancies. Biotechnol. Lett. 2018, 40, 1459–1466. [Google Scholar] [CrossRef]
- Witkowska, M.; Smolewski, P.; Robak, T. Investigational therapies targeting CD37 for the treatment of B-cell lymphoid malignancies. Expert. Opin. Investig. Drugs 2018, 27, 171–177. [Google Scholar] [CrossRef]
- Betrian, S.; Ysebaert, L.; Heider, K.H.; Delord, J.P.; Fournié, J.J.; Quillet-Mary, A. Idelalisib improves CD37 antibody BI 836826 cytotoxicity against chemo-resistant /relapse-initiating CLL cells: A rationale for combination treatment. Blood Cancer J. 2016, 6, e496. [Google Scholar] [CrossRef]
- Stilgenbauer, S.; Aurran Schleinitz, T.; Eichhorst, B.; Lang, F.; Offner, F.; Rossi, J.F.; Schroyens, W.; Van Den Neste, E.; Ysebaert, L.; von Wangenheim, U.; et al. Phase 1 first-in-human trial of the anti-CD37 antibody BI 836826 in relapsed/refractory chronic lymphocytic leukemia. Leukemia 2019, 33, 2531–2535. [Google Scholar] [CrossRef]
- Robak, T.; Robak, P.; Smolewski, P. TRU-016, a humanized anti-CD37 IgG fusion protein for the potential treatment of B-cell malignancies. Curr. Opin. Investig. Drugs 2009, 10, 1383–1390. [Google Scholar]
- Byrd, J.C.; Pagel, J.M.; Awan, F.T.; Forero, A.; Flinn, I.W.; Deauna-Limayo, D.P.; Spurgeon, S.E.; Andritsos, L.A.; Gopal, A.K.; Leonard, J.P.; et al. A phase 1 study evaluating the safety and tolerability of otlertuzumab, an anti-CD37 mono-specific ADAPTIR therapeutic protein in chronic lymphocytic leukemia. Blood 2014, 123, 1302–1308. [Google Scholar] [CrossRef] [Green Version]
- Robak, T.; Hellmann, A.; Kloczko, J.; Loscertales, J.; Lech-Maranda, E.; Pagel, J.M.; Mato, A.; Byrd, J.C.; Awan, F.T.; Hebart, H.; et al. Randomized phase 2 study of otlertuzumab and bendamustine versus bendamustine in patients with relapsed chronic lymphocytic leukaemia. Br. J. Haematol. 2017, 176, 618–628. [Google Scholar] [CrossRef]
- Labrijn, A.F.; Janmaat, M.L.; Reichert, J.M.; Parren, P. Bispecific antibodies: A mechanistic review of the pipeline. Nat. Rev. Drug Discov. 2019, 18, 585–608. [Google Scholar] [CrossRef]
- Goebeler, M.E.; Bargou, R.C. T cell-engaging therapies—BiTEs and beyond. Nat. Rev. Clin. Oncol. 2020, 17, 418–434. [Google Scholar] [CrossRef]
- Bruenke, J.; Fischer, B.; Barbin, K.; Schreiter, K.; Wachter, Y.; Mahr, K.; Titgemeyer, F.; Niederweis, M.; Peipp, M.; Zunino, S.J.; et al. A recombinant bispecific single-chain Fv antibody against HLA class II and FcgammaRIII (CD16) triggers effective lysis of lymphoma cells. Br. J. Haematol. 2004, 125, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Kellner, C.; Bruenke, J.; Horner, H.; Schubert, J.; Schwenkert, M.; Mentz, K.; Barbin, K.; Stein, C.; Peipp, M.; Stockmeyer, B.; et al. Heterodimeric bispecific antibody-derivatives against CD19 and CD16 induce effective antibody-dependent cellular cytotoxicity against B-lymphoid tumor cells. Cancer Lett. 2011, 303, 128–139. [Google Scholar] [CrossRef]
- Gleason, M.K.; Verneris, M.R.; Todhunter, D.A.; Zhang, B.; McCullar, V.; Zhou, S.X.; Panoskaltsis-Mortari, A.; Weiner, L.M.; Vallera, D.A.; Miller, J.S. Bispecific and trispecific killer cell engagers directly activate human NK cells through CD16 signaling and induce cytotoxicity and cytokine production. Mol. Cancer Ther. 2012, 11, 2674–2684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felices, M.; Kodal, B.; Hinderlie, P.; Kaminski, M.F.; Cooley, S.; Weisdorf, D.J.; Vallera, D.A.; Miller, J.S.; Bachanova, V. Novel CD19-targeted TriKE restores NK cell function and proliferative capacity in CLL. Blood Adv. 2019, 3, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Vyas, M.; Schneider, A.C.; Shatnyeva, O.; Reiners, K.S.; Tawadros, S.; Kloess, S.; Köhl, U.; Hallek, M.; Hansen, H.P.; Pogge von Strandmann, E. Mono- and dual-targeting triplebodies activate natural killer cells and have anti-tumor activity in vitro and in vivo against chronic lymphocytic leukemia. Oncoimmunology 2016, 5, e1211220. [Google Scholar] [CrossRef] [Green Version]
- Muntasell, A.; Ochoa, M.C.; Cordeiro, L.; Berraondo, P.; López-Díaz de Cerio, A.; Cabo, M.; López-Botet, M.; Melero, I. Targeting NK-cell checkpoints for cancer immunotherapy. Curr. Opin. Immunol. 2017, 45, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Riches, J.C.; Davies, J.K.; McClanahan, F.; Fatah, R.; Iqbal, S.; Agrawal, S.; Ramsay, A.G.; Gribben, J.G. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood 2013, 121, 1612–1621. [Google Scholar] [CrossRef]
- Ramsay, A.G.; Clear, A.J.; Fatah, R.; Gribben, J.G. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: Establishing a reversible immune evasion mechanism in human cancer. Blood 2012, 120, 1412–1421. [Google Scholar] [CrossRef] [PubMed]
- McClanahan, F.; Hanna, B.; Miller, S.; Clear, A.J.; Lichter, P.; Gribben, J.G.; Seiffert, M. PD-L1 checkpoint blockade prevents immune dysfunction and leukemia development in a mouse model of chronic lymphocytic leukemia. Blood 2015, 126, 203–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younes, A.; Brody, J.; Carpio, C.; Lopez-Guillermo, A.; Ben-Yehuda, D.; Ferhanoglu, B.; Nagler, A.; Ozcan, M.; Avivi, I.; Bosch, F.; et al. Safety and activity of ibrutinib in combination with nivolumab in patients with relapsed non-Hodgkin lymphoma or chronic lymphocytic leukaemia: A phase 1/2a study. Lancet Haematol. 2019, 6, e67–e78. [Google Scholar] [CrossRef]
- Condoluci, A.; Rossi, D. Richter Syndrome. Curr. Oncol. Rep. 2021, 23, 26. [Google Scholar] [CrossRef]
- Vey, N.; Karlin, L.; Sadot-Lebouvier, S.; Broussais, F.; Berton-Rigaud, D.; Rey, J.; Charbonnier, A.; Marie, D.; André, P.; Paturel, C.; et al. A phase 1 study of lirilumab (antibody against killer immunoglobulin-like receptor antibody KIR2D.; IPH2102) in patients with solid tumors and hematologic malignancies. Oncotarget 2018, 9, 17675–17688. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, M.; Herishanu, Y.; Katz, B.Z.; Dezorella, N.; Sun, C.; Kay, S.; Polliack, A.; Avivi, I.; Wiestner, A.; Perry, C. Lymphocyte activation gene 3: A novel therapeutic target in chronic lymphocytic leukemia. Haematologica 2017, 102, 874–882. [Google Scholar] [CrossRef] [Green Version]
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; González-Rodríguez, A.P.; Payer, Á.R.; González-García, E.; López-Soto, A.; Gonzalez, S. LAG-3 Blockade with Relatlimab (BMS-986016) Restores Anti-Leukemic Responses in Chronic Lymphocytic Leukemia. Cancers 2021, 13, 2112. [Google Scholar] [CrossRef]
- Dreger, P.; Corradini, P.; Kimby, E.; Michallet, M.; Milligan, D.; Schetelig, J.; Wiktor-Jedrzejczak, W.; Niederwieser, D.; Hallek, M.; Montserrat, E. Indications for allogeneic stem cell transplantation in chronic lymphocytic leukemia. Chronic Leukemia Working Party of the EBMT. Leukemia 2007, 21, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Burger, J.A.; Barr, P.M.; Robak, T.; Owen, C.; Ghia, P.; Tedeschi, A.; Bairey, O.; Hillmen, P.; Coutre, S.E.; Devereux, S.; et al. Long-term efficacy and safety of first-line ibrutinib treatment for patients with CLL/SLL: 5 years of follow-up from the phase 3 RESONATE-2 study. Leukemia 2020, 34, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Flinn, I.W.; Hillmen, P.; Montillo, M.; Nagy, Z.; Illés, Á.; Etienne, G.; Delgado, J.; Kuss, B.J.; Tam, C.S.; Gasztonyi, Z.; et al. The phase 3 DUO trial: Duvelisib vs ofatumumab in relapsed and refractory CLL/SLL. Blood 2018, 132, 2446–2455. [Google Scholar] [CrossRef] [Green Version]
- Stilgenbauer, S.; Eichhorst, B.; Schetelig, J.; Hillmen, P.; Seymour, J.F.; Coutre, S.; Jurczak, W.; Mulligan, S.P.; Schuh, A.; Assouline, S.; et al. Venetoclax for patients with chronic lymphocytic leukemia with 17p deletion: Results from the full population of a phase II pivotal trial. J. Clin. Oncol. 2018, 36, 1973–1980. [Google Scholar] [CrossRef]
- Gribben, J.G. How and when I do allogeneic transplant in CLL. Blood 2018, 132, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Dreger, P.; Schetelig, J.; Andersen, N.; Corradini, P.; van Gelder, M.; Gribben, J.; Kimby, E.; Michallet, M.; Moreno, C.; Stilgenbauer, S.; et al. Managing high-risk CLL during transition to a new treatment era: Stem cell transplantation or novel agents? European Research Initiative on CLL (ERIC) and the European Society for Blood and Marrow Transplantation (EBMT). Blood 2014, 124, 3841–3849. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, A.; Fretham, C.; Lee, S.J.; Arora, M.; Brunner, J.; Chhabra, S.; Devine, S.; Eapen, M.; Hamadani, M.; Hari, P.; et al. Current Use of and Trends in Hematopoietic Cell Transplantation in the United States. Biol. Blood Marrow Transplant. 2020, 26, E177–E182. [Google Scholar] [CrossRef]
- Locatelli, F.; Pende, D.; Falco, M.; Della Chiesa, M.; Moretta, A.; Moretta, L. NK Cells Mediate a Crucial Graft-versus-Leukemia Effect in Haploidentical-HSCT to Cure High-Risk Acute Leukemia. Trends Immunol. 2018, 39, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, B.K.; Copelan, E.A. Concise review: The role of hematopoietic stem cell transplantation in the treatment of acute myeloid leukemia. Stem Cells 2012, 30, 1581–1586. [Google Scholar] [CrossRef]
- Gribben, J.G.; Zahrieh, D.; Stephans, K.; Bartlett-Pandite, L.; Alyea, E.P.; Fisher, D.C.; Freedman, A.S.; Mauch, P.; Schlossman, R.; Sequist, L.V.; et al. Autologous and allogeneic stem cell transplantations for poor-risk chronic lymphocytic leukemia. Blood 2005, 106, 389–396. [Google Scholar] [CrossRef] [Green Version]
- Bachanova, V.; Weisdorf, D.J.; Wang, T.; Marsh, S.G.E.; Cereb, N.; Haagenson, M.D.; Spellman, S.R.; Lee, S.J.; Guethlein, L.A.; Parham, P.; et al. Donor Killer Cell Immunoglobulin-Like Receptor Genotype Does Not Improve Graft-versus-Leukemia Responses in Chronic Lymphocytic Leukemia after Unrelated Donor Transplant: A Center for International Blood and Marrow Transplant Research Analysis. Biol. Blood Marrow Transplant. 2019, 25, 949–954. [Google Scholar] [CrossRef]
- van Gorkom, G.; van Gelder, M.; Eikema, D.J.; Blok, H.J.; van Lint, M.T.; Koc, Y.; Ciceri, F.; Beelen, D.; Chevallier, P.; Selleslag, D.; et al. CLL subcommittee; Chronic Malignancies Working Party of the EBMT. Outcomes of haploidentical stem cell transplantation for chronic lymphocytic leukemia: A retrospective study on behalf of the chronic malignancies working party of the EBMT. Bone Marrow Transplant. 2018, 53, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Calvo, T.; Reina-Ortiz, C.; Giraldos, D.; Gascón, M.; Woods, D.; Asenjo, J.; Marco-Brualla, J.; Azaceta, G.; Izquierdo, I.; Palomera, L.; et al. Expanded and activated allogeneic NK cells are cytotoxic against B-chronic lymphocytic leukemia (B-CLL) cells with sporadic cases of resistance. Sci. Rep. 2020, 10, 19398. [Google Scholar] [CrossRef]
- Di Ianni, M.; Del Papa, B.; Zei, T.; Iacucci Ostini, R.; Cecchini, D.; Cantelmi, M.G.; Baldoni, S.; Sportoletti, P.; Cavalli, L.; Carotti, A.; et al. T regulatory cell separation for clinical application. Transfus. Apher. Sci. 2012, 47, 213–216. [Google Scholar] [CrossRef]
- Olson, J.A.; Leveson-Gower, D.B.; Gill, S.; Baker, J.; Beilhack, A.; Negrin, R.S. NK cells mediate reduction of GVHD by inhibiting activated, alloreactive T cells while retaining GVT effects. Blood 2010, 115, 4293–4301. [Google Scholar] [CrossRef] [Green Version]
- Fesnak, A.D.; June, C.H.; Levine, B.L. Engineered T cells: The promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 2016, 16, 566–581. [Google Scholar] [CrossRef]
- Rafei, H.; Daher, M.; Rezvani, K. Chimeric antigen receptor (CAR) natural killer (NK)-cell therapy: Leveraging the power of innate immunity. Br. J. Haematol. 2021, 193, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.L.; Levine, B.L.; Kalos, M.; Bagg, A.; June, C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011, 365, 725–733. [Google Scholar] [CrossRef] [Green Version]
- Brentjens, R.J.; Rivière, I.; Park, J.H.; Davila, M.L.; Wang, X.; Stefanski, J.; Taylor, C.; Yeh, R.; Bartido, S.; Borquez-Ojeda, O.; et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 2011, 118, 4817–4828. [Google Scholar] [CrossRef] [PubMed]
- Kalos, M.; Levine, B.L.; Porter, D.L.; Katz, S.; Grupp, S.A.; Bagg, A.; June, C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011, 95, 95ra73. [Google Scholar] [CrossRef] [Green Version]
- Porter, D.L.; Hwang, W.T.; Frey, N.V.; Lacey, S.F.; Shaw, P.A.; Loren, A.W.; Bagg, A.; Marcucci, K.T.; Shen, A.; Gonzalez, V.; et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 2015, 7, 303ra139. [Google Scholar] [CrossRef] [Green Version]
- Turtle, C.J.; Hay, K.A.; Hanafi, L.A.; Li, D.; Cherian, S.; Chen, X.; Wood, B.; Lozanski, A.; Byrd, J.C.; Heimfeld, S.; et al. Durable molecular remissions in chronic lymphocytic leukemia treated with CD19-specific chimeric antigen receptor-modified T cells after failure of ibrutinib. J. Clin. Oncol. 2017, 35, 3010–3020. [Google Scholar] [CrossRef]
- Geyer, M.B.; Rivière, I.; Sénéchal, B.; Wang, X.; Wang, Y.; Purdon, T.J.; Hsu, M.; Devlin, S.M.; Halton, E.; Lamanna, N.; et al. Autologous CD19-targeted CAR T cells in patients with residual CLL following initial purine analog-based therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 1896–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skorka, K.; Ostapinska, K.; Malesa, A.; Giannopoulos, K. The Application of CAR-T Cells in Haematological Malignancies. Arch. Immunol. Ther. Exp. 2020, 68, 34. [Google Scholar] [CrossRef] [PubMed]
- Lemal, R.; Tournilhac, O. State-of-the-art for CAR T-cell therapy for chronic lymphocytic leukemia in 2019. J. Immunother. Cancer 2019, 7, 202. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, J.M.; Schubert, M.L.; Wang, L.; Huckelhoven, A.; Sellner, L.; Stock, S.; Schmitt, A.; Kleist, C.; Gern, U.; Loskog, A.; et al. Differences in expansion potential of naive chimeric antigen receptor T cells from healthy donors and untreated chronic lymphocytic leukemia patients. Front. Immunol. 2018, 8, 1956. [Google Scholar] [CrossRef] [Green Version]
- Magalhaes, I.; Kalland, I.; Kochenderfer, J.N.; Osterborg, A.; Uhlin, M.; Mattsson, J. CD19 Chimeric Antigen Receptor T Cells From Patients With Chronic Lymphocytic Leukemia Display an Elevated IFN-gamma Production Profile. J. Immunother. 2018, 41, 73–83. [Google Scholar] [CrossRef]
- Goulmy, E. Human minor histocompatibility antigens: New concepts for marrow transplantation and adoptive immunotherapy. Immunol. Rev. 1997, 157, 125–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauthier, J.; Hirayama, A.V.; Purushe, J.; Hay, K.A.; Lymp, J.; Li, D.H.; Yeung, C.C.S.; Sheih, A.; Pender, B.S.; Hawkins, R.M.; et al. Feasibility and efficacy of CD19-targeted CAR T cells with concurrent ibrutinib for CLL after ibrutinib failure. Blood 2020, 135, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.S.; Johnstone, T.G.; Baturevych, A.; Hause, R.J.; Ragan, S.P.; Clouser, C.R.; Jones, J.C.; Ponce, R.; Krejsa, C.M.; Salmon, R.A.; et al. Antitumor Potency of an Anti-CD19 Chimeric Antigen Receptor T-Cell Therapy, Lisocabtagene Maraleucel in Combination with Ibrutinib or Acalabrutinib. J. Immunother. 2020, 43, 107–120. [Google Scholar] [CrossRef]
- Long, M.; Beckwith, K.; Do, P.; Mundy, B.L.; Gordon, A.; Lehman, A.M.; Maddocks, K.J.; Cheney, C.; Jones, J.A.; Flynn, J.M.; et al. Ibrutinib treatment improves T cell number and function in CLL patients. J. Clin. Investig. 2017, 127, 3052–3064. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dou, M.; Ma, Q.; Yao, R.; Liu, J. Chimeric antigen receptor (CAR)-modified NK cells against cancer: Opportunities and challenges. Int. Immunopharmacol. 2019, 74, 105695. [Google Scholar] [CrossRef]
- Rotolo, R.; Leuci, V.; Donini, C.; Cykowska, A.; Gammaitoni, L.; Medico, G.; Valabrega, G.; Aglietta, M.; Sangiolo, D. CAR-Based Strategies beyond T Lymphocytes: Integrative Opportunities for Cancer Adoptive Immunotherapy. Int. J. Mol. Sci. 2019, 20, 2839. [Google Scholar] [CrossRef] [Green Version]
- Rezvani, K. Adoptive cell therapy using engineered natural killer cells. Bone Marrow Transplant. 2019, 54, 785–788. [Google Scholar] [CrossRef]
- Sotillo, E.; Barrett, D.M.; Black, K.L.; Bagashev, A.; Oldridge, D.; Wu, G.; Sussman, R.; Lanauze, C.; Ruella, M.; Gazzara, M.R.; et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015, 5, 1282–1295. [Google Scholar] [CrossRef] [Green Version]
- Herrera, L.; Santos, S.; Vesga, M.A.; Anguita, J.; Martin-Ruiz, I.; Carrascosa, T.; Juan, M.; Eguizabal, C. Adult peripheral blood and umbilical cord blood NK cells are good sources for effective CAR therapy against CD19 positive leukemic cells. Sci. Rep. 2019, 9, 18729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boissel, L.; Betancur, M.; Wels, W.S.; Tuncer, H.; Klingemann, H. Transfection with mRNA for CD19 specific chimeric antigen receptor restores NK cell mediated killing of CLL cells. Leuk. Res. 2009, 33, 1255–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, T.; Uherek, C.; Maki, G.; Chow, K.U.; Schimpf, A.; Klingemann, H.G.; Tonn, T.; Wels, W.S. Expression of a CD20-specific chimeric antigen receptor enhances cytotoxic activity of NK cells and overcomes NK-resistance of lymphoma and leukemia cells. Cancer Immunol. Immunother. 2008, 57, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Grote, S.; Mittelstaet, J.; Baden, C.; Chun-Ho Chan, K.; Seitz, C.; Schlegel, P.; Kaiser, A.; Handgretinger, R.; Schleicher, S. Adapter chimeric antigen receptor (AdCAR)-engineered NK-92 cells: An off-the-shelf cellular therapeutic for universal tumor targeting. Oncoimmunology 2020, 9, 1825177. [Google Scholar] [CrossRef]
- Fujisaki, H.; Kakuda, H.; Shimasaki, N.; Imai, C.; Ma, J.; Lockey, T.; Eldridge, P.; Leung, W.H.; Campana, D. Expansion of highly cytotoxic human natural killer cells for cancer cell therapy. Cancer Res. 2009, 69, 4010–4017. [Google Scholar] [CrossRef] [Green Version]
- Clausen, J.; Petzer, A.L.; Vergeiner, B.; Enk, M.; Stauder, R.; Gastl, G.; Gunsilius, E. Optimal timing for the collection and in vitro expansion of cytotoxic CD56(+) lymphocytes from patients undergoing autologous peripheral blood stem cell transplantation. J. Hematother. Stem Cell Res. 2001, 10, 513–521. [Google Scholar] [CrossRef]
- Imamura, M.; Shook, D.; Kamiya, T.; Shimasaki, N.; Chai, S.M.; Coustan-Smith, E.; Imai, C.; Campana, D. Autonomous growth and increased cytotoxicity of natural killer cells expressing membrane-bound interleukin-15. Blood 2014, 124, 1081–1088. [Google Scholar] [CrossRef] [Green Version]
- Nagashima, S.; Mailliard, R.; Kashii, Y.; Reichert, T.E.; Herberman, R.B.; Robbins, P.; Whiteside, T.L. Stable transduction of the interleukin-2 gene into human natural killer cell lines and their phenotypic and functional characterization in vitro and in vivo. Blood 1998, 91, 3850–3861. [Google Scholar] [CrossRef] [Green Version]
- Liu, E.; Tong, Y.; Dotti, G.; Shaim, H.; Savoldo, B.; Mukherjee, M.; Orange, J.; Wan, X.; Lu, X.; Reynolds, A. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 2018, 32, 520–531. [Google Scholar] [CrossRef] [PubMed]
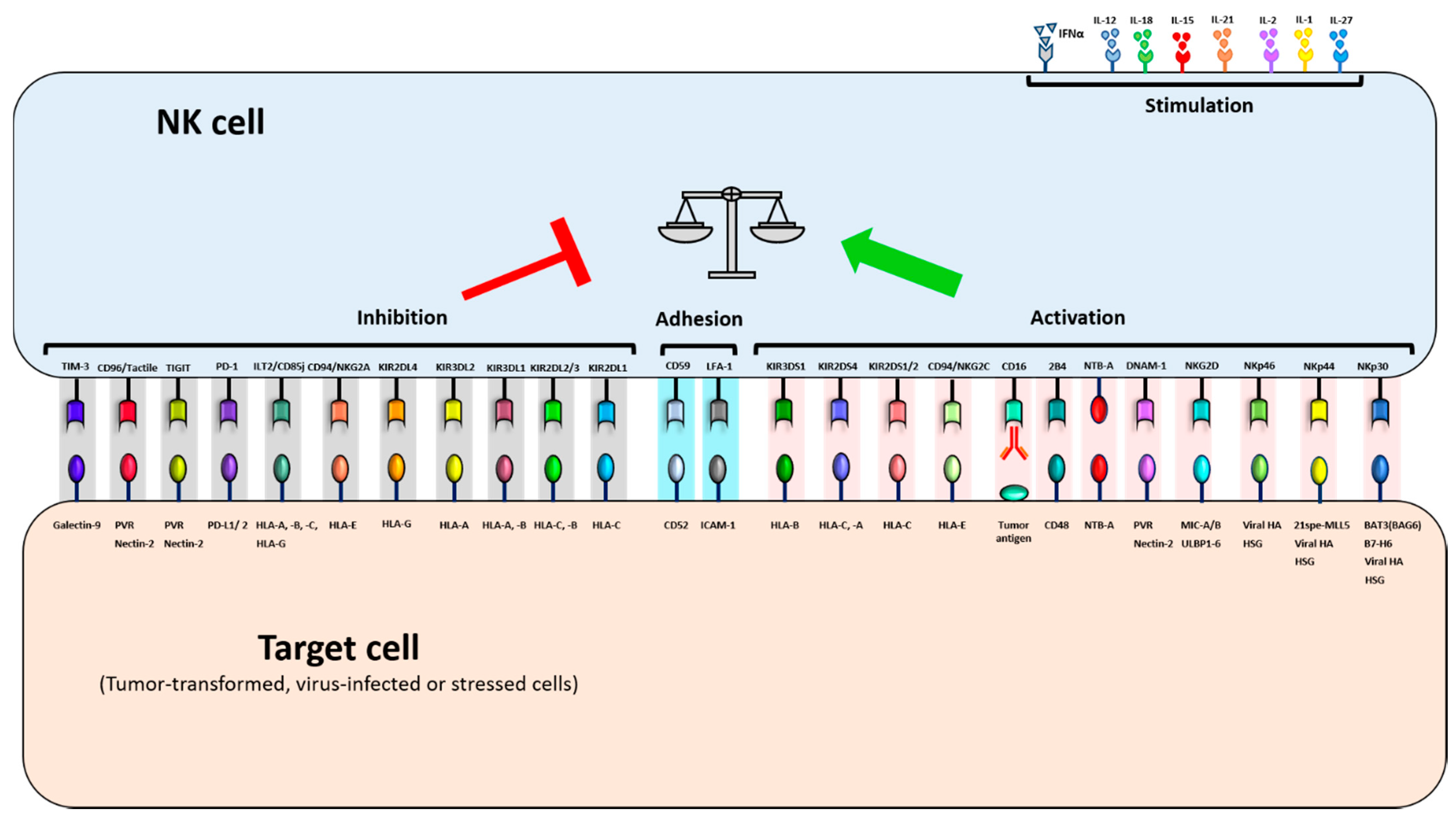
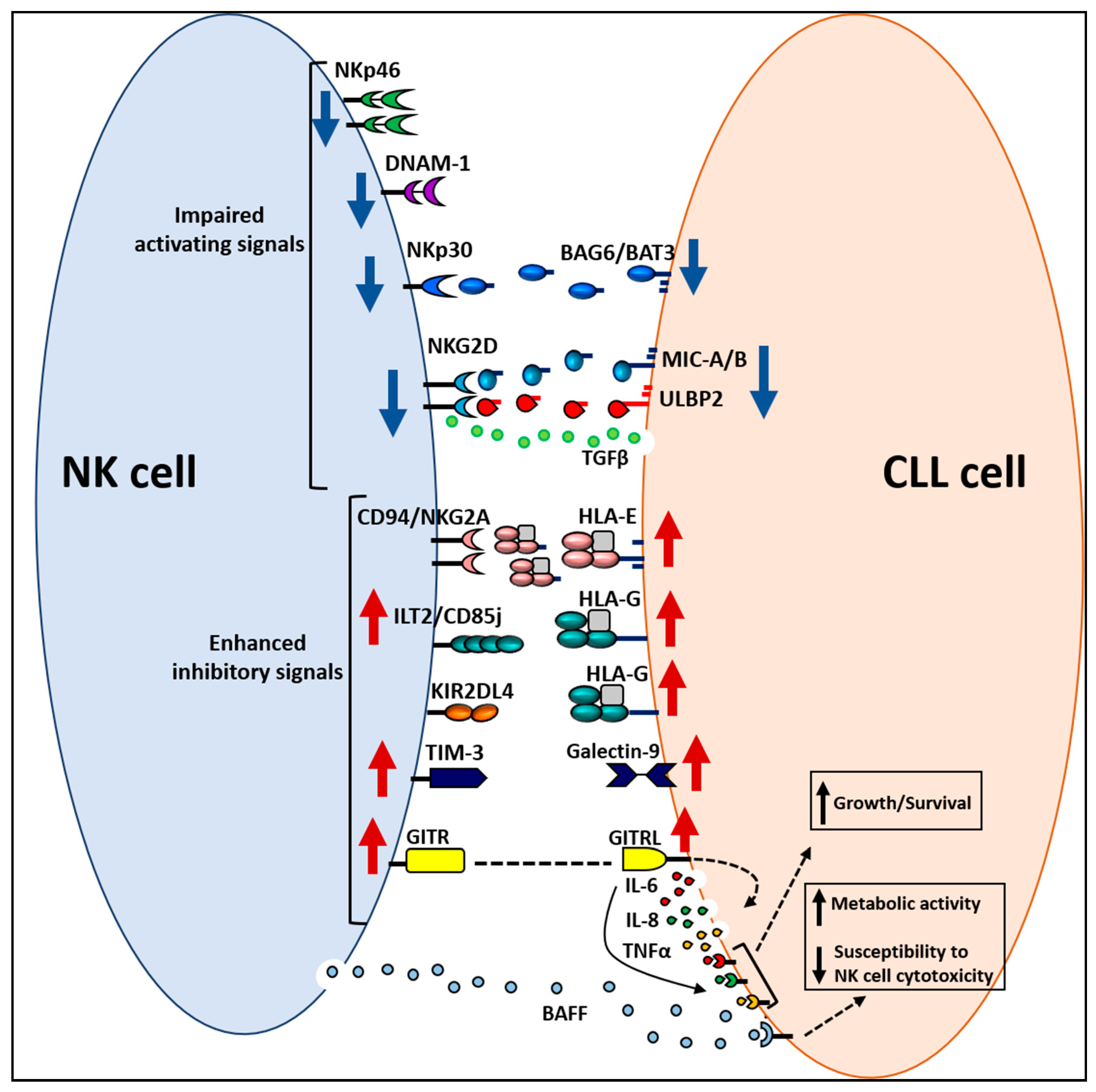
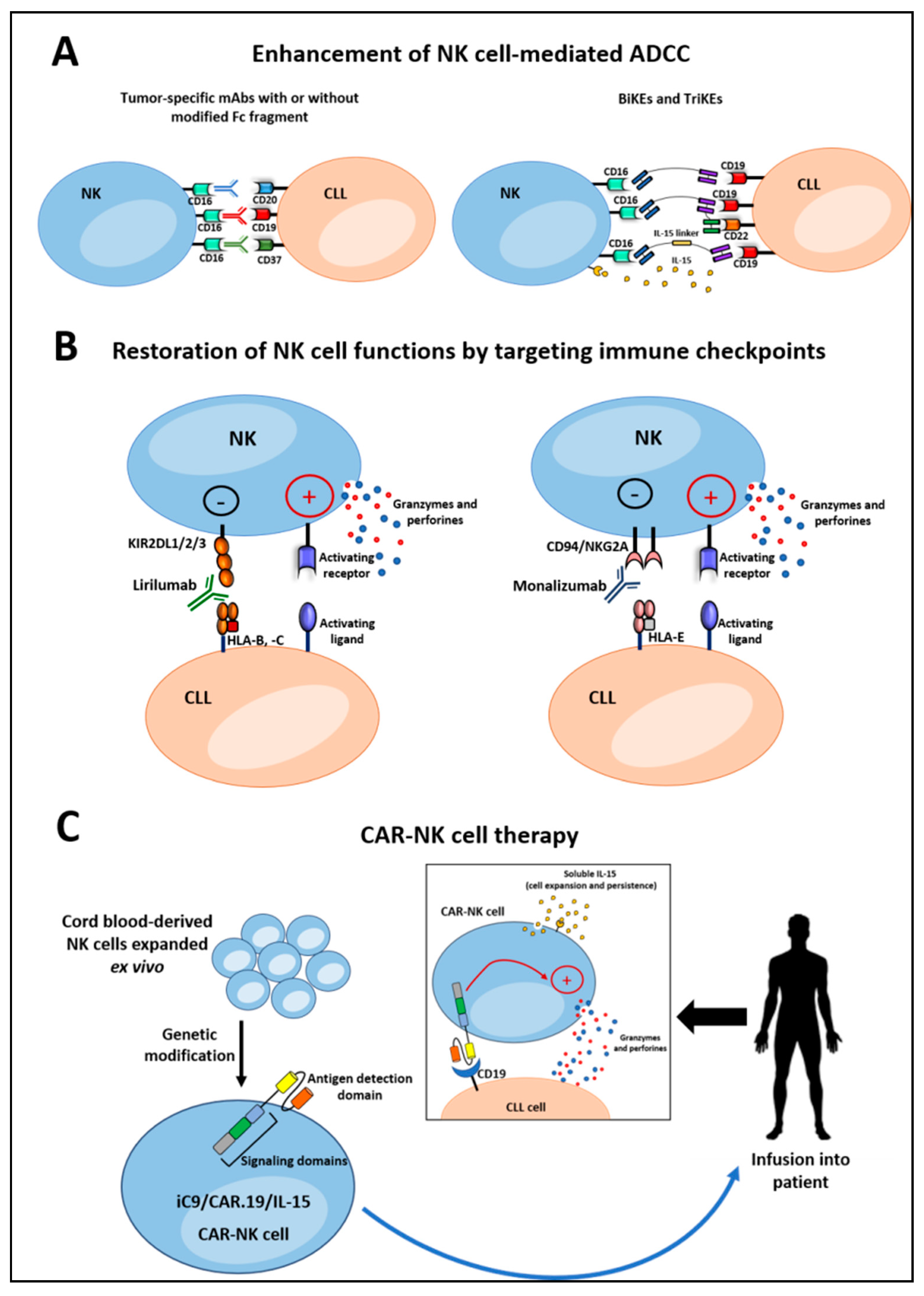
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sportoletti, P.; De Falco, F.; Del Papa, B.; Baldoni, S.; Guarente, V.; Marra, A.; Dorillo, E.; Rompietti, C.; Adamo, F.M.; Ruggeri, L.; et al. NK Cells in Chronic Lymphocytic Leukemia and Their Therapeutic Implications. Int. J. Mol. Sci. 2021, 22, 6665. https://doi.org/10.3390/ijms22136665
Sportoletti P, De Falco F, Del Papa B, Baldoni S, Guarente V, Marra A, Dorillo E, Rompietti C, Adamo FM, Ruggeri L, et al. NK Cells in Chronic Lymphocytic Leukemia and Their Therapeutic Implications. International Journal of Molecular Sciences. 2021; 22(13):6665. https://doi.org/10.3390/ijms22136665
Chicago/Turabian StyleSportoletti, Paolo, Filomena De Falco, Beatrice Del Papa, Stefano Baldoni, Valerio Guarente, Andrea Marra, Erica Dorillo, Chiara Rompietti, Francesco Maria Adamo, Loredana Ruggeri, and et al. 2021. "NK Cells in Chronic Lymphocytic Leukemia and Their Therapeutic Implications" International Journal of Molecular Sciences 22, no. 13: 6665. https://doi.org/10.3390/ijms22136665
APA StyleSportoletti, P., De Falco, F., Del Papa, B., Baldoni, S., Guarente, V., Marra, A., Dorillo, E., Rompietti, C., Adamo, F. M., Ruggeri, L., Di Ianni, M., & Rosati, E. (2021). NK Cells in Chronic Lymphocytic Leukemia and Their Therapeutic Implications. International Journal of Molecular Sciences, 22(13), 6665. https://doi.org/10.3390/ijms22136665





