Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus
Abstract
1. Introduction
2. Results
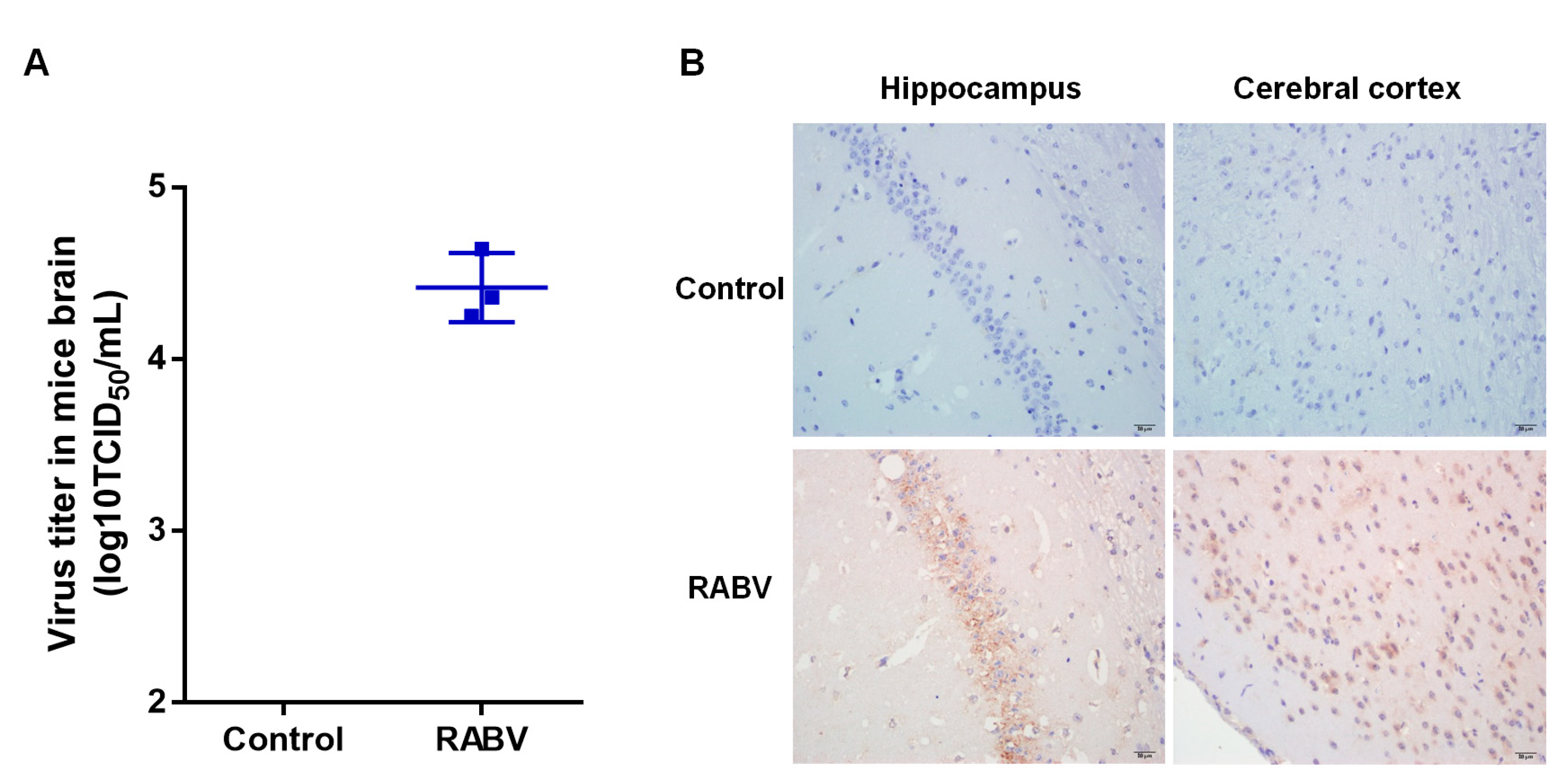
2.1. Validation of RABV Infection in Mice Brains
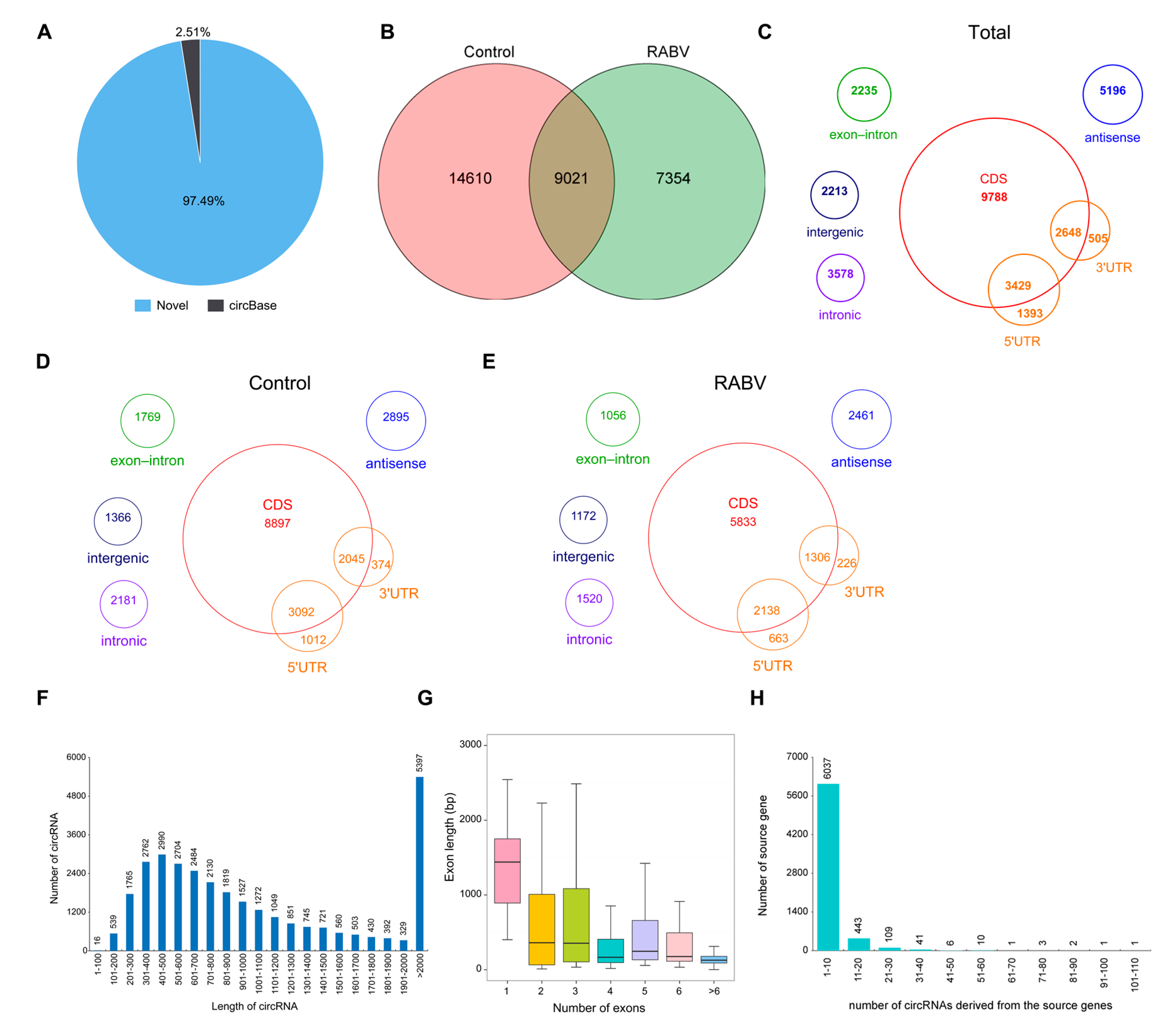
2.2. Characteristics of circRNAs Expressed in Mice Brain by RNA Sequencing
2.3. Identification of circRNAs Expression in RABV Infection
2.4. Expression Correlation of circRNAs and mRNAs
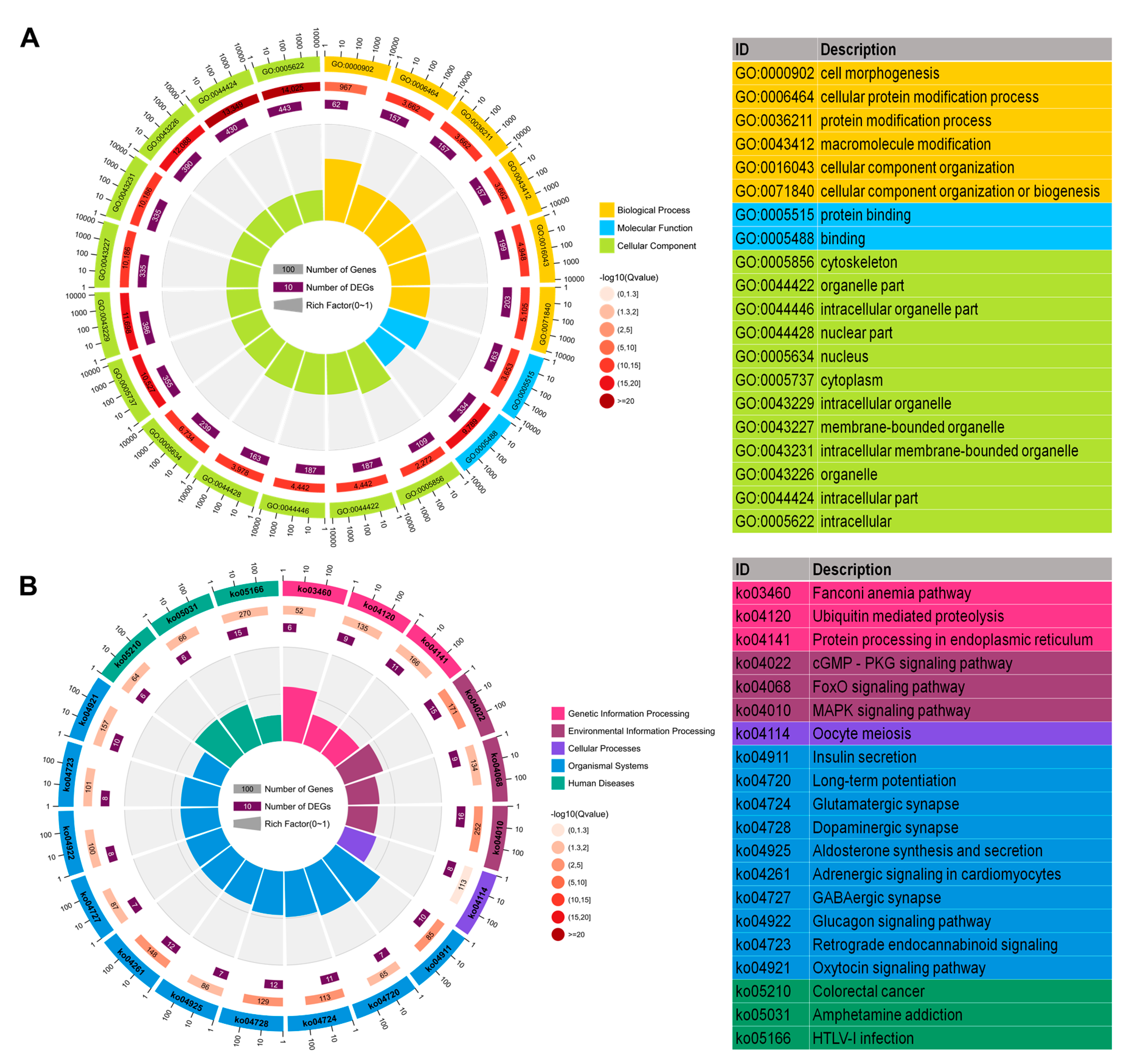
2.5. Functional Enrichment Analysis of Host Genes of Differentially Expressed circRNAs
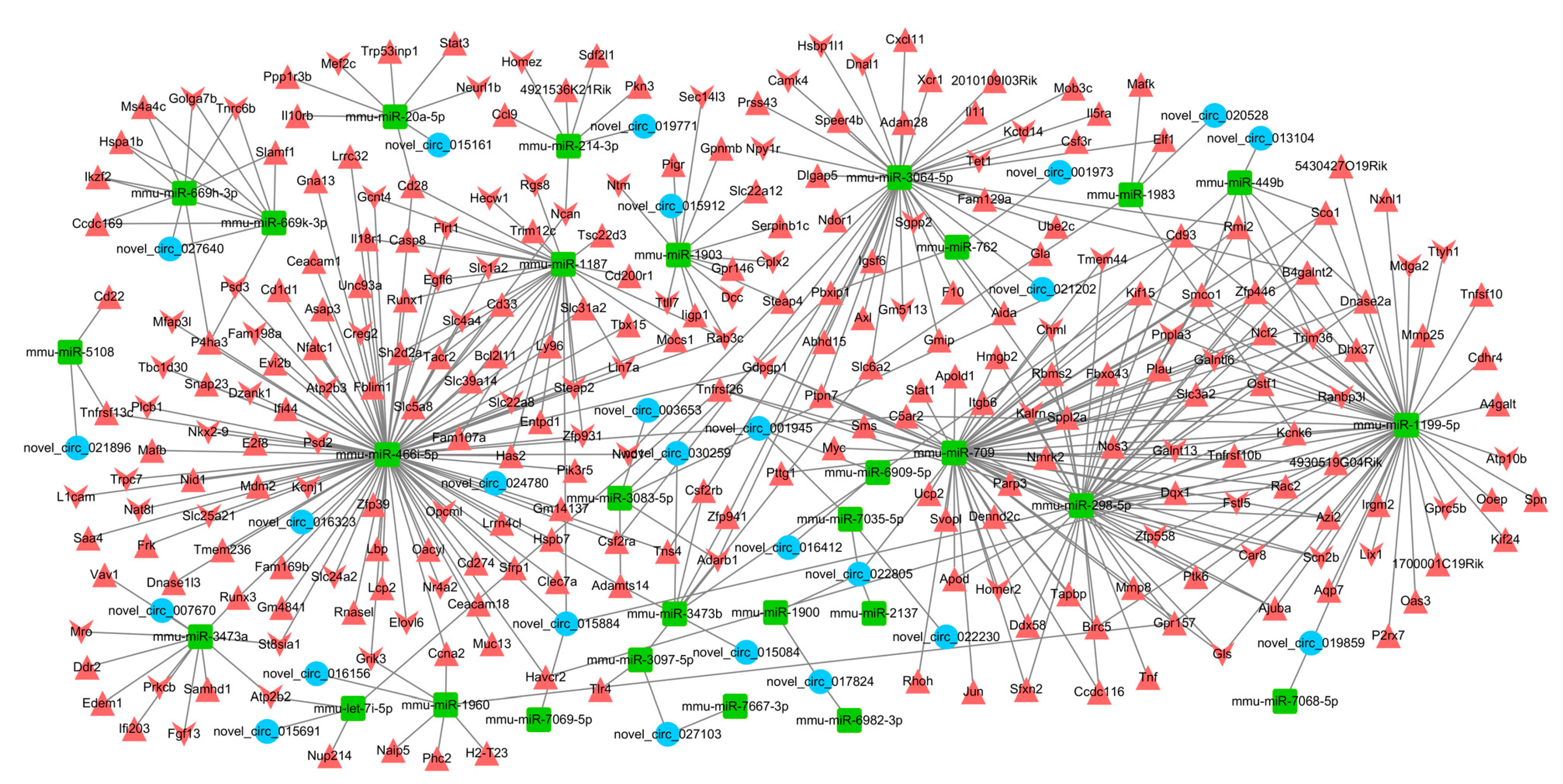
2.6. Interaction Network of circRNA–miRNA–mRNA
3. Discussion
4. Materials and Methods
4.1. RABV Infection
4.2. RNA Extraction and Quantitative Real-Time PCR
4.3. Immunohistochemical (IHC) Analysis
4.4. RNA Sequencing Analysis
4.5. Functional Enrichment Analysis
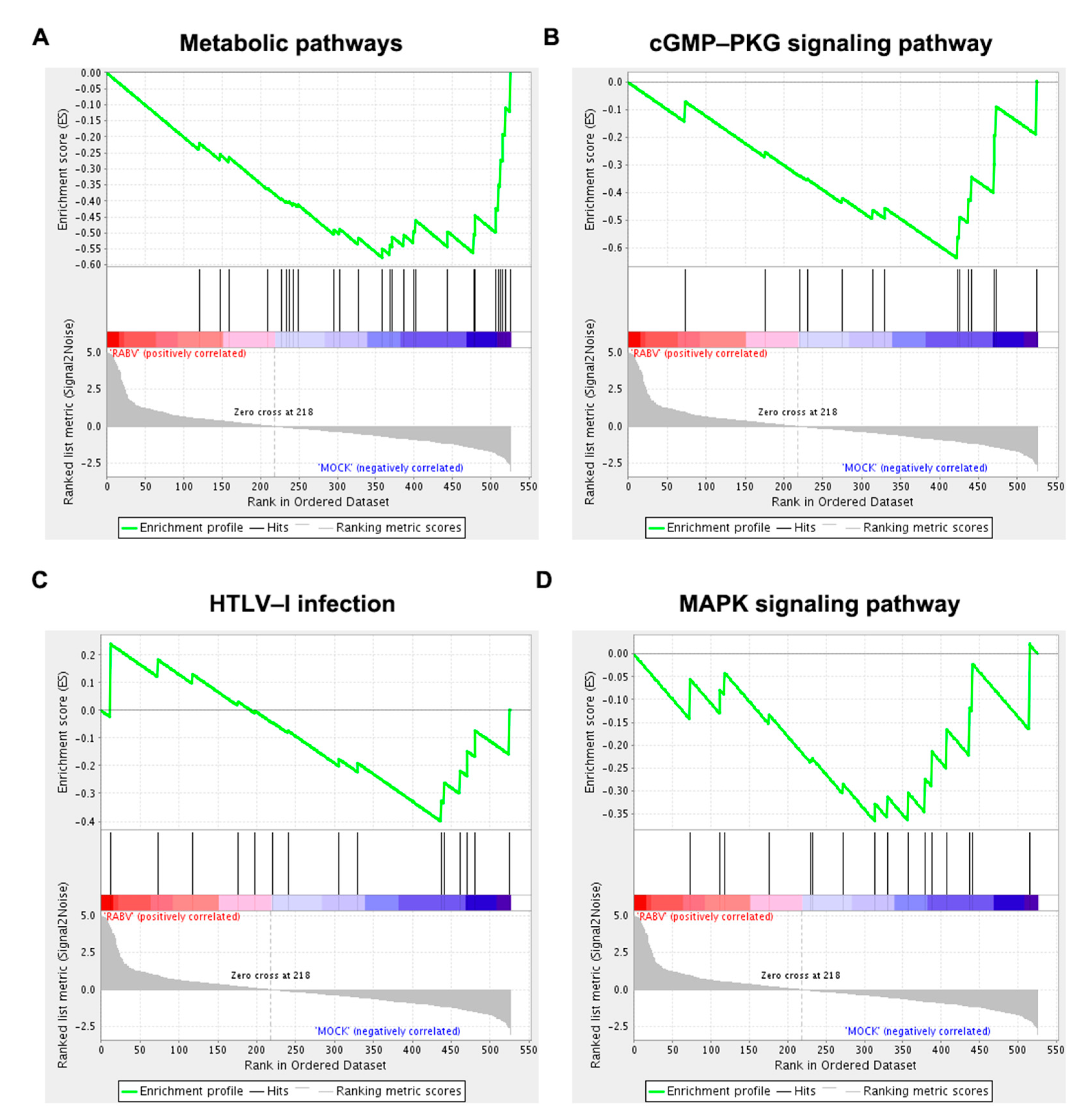
4.6. Gene Set Enrichment Analysis (GSEA)
4.7. Integrated Analysis of circRNAs-miRNAs-mRNAs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luco, S.; Delmas, O.; Vidalain, P.O.; Tangy, F.; Weil, R.; Bourhy, H. RelAp43, a member of the NF-kappaB family involved in innate immune response against Lyssavirus infection. PLoS Pathog. 2012, 8, e1003060. [Google Scholar] [CrossRef]
- Ben Khalifa, Y.; Luco, S.; Besson, B.; Sonthonnax, F.; Archambaud, M.; Grimes, J.M.; Larrous, F.; Bourhy, H. The matrix protein of rabies virus binds to RelAp43 to modulate NF-kappaB-dependent gene expression related to innate immunity. Sci. Rep. 2016, 6, 39420. [Google Scholar] [CrossRef]
- Masatani, T.; Ito, N.; Shimizu, K.; Ito, Y.; Nakagawa, K.; Sawaki, Y.; Koyama, H.; Sugiyama, M. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J. Virol. 2010, 84, 4002–4012. [Google Scholar] [CrossRef] [PubMed]
- Brzozka, K.; Finke, S.; Conzelmann, K.K. Identification of the rabies virus alpha/beta interferon antagonist: Phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 2005, 79, 7673–7681. [Google Scholar] [CrossRef]
- Rieder, M.; Brzozka, K.; Pfaller, C.K.; Cox, J.H.; Stitz, L.; Conzelmann, K.K. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: Inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J. Virol. 2011, 85, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Blondel, D.; Regad, T.; Poisson, N.; Pavie, B.; Harper, F.; Pandolfi, P.P.; De The, H.; Chelbi-Alix, M.K. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 2002, 21, 7957–7970. [Google Scholar] [CrossRef] [PubMed]
- Blondel, D.; Kheddache, S.; Lahaye, X.; Dianoux, L.; Chelbi-Alix, M.K. Resistance to rabies virus infection conferred by the PMLIV isoform. J. Virol. 2010, 84, 10719–10726. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Gu, J.; Deng, T.; Yuan, Z.; Hu, B.; Xu, Y.; Yan, Y.; Zan, J.; Liao, M.; et al. BECN1-dependent CASP2 incomplete autophagy induction by binding to rabies virus phosphoprotein. Autophagy 2017, 13, 739–753. [Google Scholar] [CrossRef]
- Zan, J.; Liu, J.; Zhou, J.W.; Wang, H.L.; Mo, K.K.; Yan, Y.; Xu, Y.B.; Liao, M.; Su, S.; Hu, R.L.; et al. Rabies virus matrix protein induces apoptosis by targeting mitochondria. Exp. Cell Res. 2016, 347, 83–94. [Google Scholar] [CrossRef]
- Baloul, L.; Camelo, S.; Lafon, M. Up-regulation of Fas ligand (FasL) in the central nervous system: A mechanism of immune evasion by rabies virus. J. Neurovirol. 2004, 10, 372–382. [Google Scholar] [CrossRef]
- Lafon, M.; Megret, F.; Meuth, S.G.; Simon, O.; Velandia Romero, M.L.; Lafage, M.; Chen, L.; Alexopoulou, L.; Flavell, R.A.; Prehaud, C.; et al. Detrimental contribution of the immuno-inhibitor B7-H1 to rabies virus encephalitis. J. Immunol. 2008, 180, 7506–7515. [Google Scholar] [CrossRef]
- Mégret, F.; Prehaud, C.; Lafage, M.; Moreau, P.; Rouas-Freiss, N.; Carosella, E.D.; Lafon, M. Modulation of HLA-G and HLA-E expression in human neuronal cells after rabies virus or herpes virus simplex type 1 infections. Hum. Immunol. 2007, 68, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.B.; Lu, Z.L.; Wei, X.K.; Zhong, T.Z.; Zhong, Y.Z.; Ouyang, L.X.; Luo, Y.; Xing, X.W.; Liao, F.; Peng, K.K.; et al. Viperin inhibits rabies virus replication via reduced cholesterol and sphingomyelin and is regulated upstream by TLR4. Sci. Rep. 2016, 6, 30529. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Diamond, M.S. Innate immune interactions within the central nervous system modulate pathogenesis of viral infections. Curr. Opin. Immunol. 2015, 36, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Lv, L.; Li, Y.; Sui, B.; Wu, Q.; Zhang, Y.; Pei, J.; Li, M.; Zhou, M.; Hooper, D.C.; et al. Dual Role of Toll-Like Receptor 7 in the Pathogenesis of Rabies Virus in a Mouse Model. J. Virol. 2020, 94, e00111-20. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Gibson, M.S.; Wash, R.S.; Ferrara, F.; Wright, E.; Temperton, N.; Kellam, P.; Fife, M. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. J. Virol. 2013, 87, 12957–12966. [Google Scholar] [CrossRef]
- Davis, B.M.; Fensterl, V.; Lawrence, T.M.; Hudacek, A.W.; Sen, G.C.; Schnell, M.J. Ifit2 Is a Restriction Factor in Rabies Virus Pathogenicity. J. Virol. 2017, 91, e00889-17. [Google Scholar] [CrossRef]
- Zhao, P.; Jiang, T.; Zhong, Z.; Zhao, L.; Yang, S.; Xia, X. Inhibition of rabies virus replication by interferon-stimulated gene 15 and its activating enzyme UBA7. Infect. Genet. Evol. 2017, 56, 44–53. [Google Scholar] [CrossRef]
- Tian, B.; Yuan, Y.; Yang, Y.; Luo, Z.; Sui, B.; Zhou, M.; Fu, Z.F.; Zhao, L. Interferon inducible GTPase 1 impedes the dimerization of rabies virus phosphoprotein and restricts viral replication. J. Virol. 2020, 94, e01203-20. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef]
- Garikipati, V.N.S.; Verma, S.K.; Cheng, Z.; Liang, D.; Truongcao, M.M.; Cimini, M.; Yue, Y.; Huang, G.; Wang, C.; Benedict, C.; et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019, 10, 4317. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, M.; Yang, X.; Zhang, M.; Luo, J.; Zhou, H.; Huang, N.; Xiao, F.; Lai, B.; Lv, W.; et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol. Cancer 2020, 19, 142. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, Y.; Han, B.; Bai, Y.; Zhou, R.; Gan, G.; Chao, J.; Hu, G.; Yao, H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 2017, 13, 1722–1741. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, S.; Das, S.; Sen, R.; Basak, P.; Chakrabarti, J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013, 4, 283. [Google Scholar] [CrossRef]
- Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes 2016, 7, 116. [Google Scholar] [CrossRef]
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yan, Y.; Lin, W.; Li, A.; Zhang, H.; Lei, X.; Dai, Z.; Li, X.; Li, H.; Chen, W.; et al. Circular RNA Vav3 sponges gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol. 2019, 16, 118–132. [Google Scholar] [CrossRef]
- Yu, T.; Ding, Y.; Zhang, Y.; Liu, Y.; Li, Y.; Lei, J.; Zhou, J.; Song, S.; Hu, B. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet. Microbiol. 2019, 231, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Chen, J.; Wang, H.; Jin, L.; Wang, L.; Huang, X.; Chen, W.; Yan, M.; Liu, G. Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells. Virology 2019, 527, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Nicolet, B.P.; Engels, S.; Aglialoro, F.; van den Akker, E.; von Lindern, M.; Wolkers, M.C. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018, 46, 8168–8180. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.K.; Wei, W.; Lee, J.A.; Lin, C.H.; Damianov, A.; de la Torre-Ubieta, L.; Halabi, R.; Otis, K.O.; Martin, K.C.; O’Dell, T.J.; et al. Rbfox1 Regulates Synaptic Transmission through the Inhibitory Neuron-Specific vSNARE Vamp1. Neuron 2018, 98, 127–141.e7. [Google Scholar] [CrossRef]
- Weissflog, L.; Scholz, C.J.; Jacob, C.P.; Nguyen, T.T.; Zamzow, K.; Gross-Lesch, S.; Renner, T.J.; Romanos, M.; Rujescu, D.; Walitza, S.; et al. KCNIP4 as a candidate gene for personality disorders and adult ADHD. Eur. Neuropsychopharmacol. 2013, 23, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Weyn-Vanhentenryck, S.M.; Mele, A.; Yan, Q.; Sun, S.; Farny, N.; Zhang, Z.; Xue, C.; Herre, M.; Silver, P.A.; Zhang, M.Q.; et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014, 6, 1139–1152. [Google Scholar] [CrossRef]
- Massone, S.; Vassallo, I.; Castelnuovo, M.; Fiorino, G.; Gatta, E.; Robello, M.; Borghi, R.; Tabaton, M.; Russo, C.; Dieci, G.; et al. RNA polymerase III drives alternative splicing of the potassium channel-interacting protein contributing to brain complexity and neurodegeneration. J. Cell Biol. 2011, 193, 851–866. [Google Scholar] [CrossRef]
- Bose, R.; Ain, R. Regulation of Transcription by Circular RNAs. Adv. Exp. Med. Biol. 2018, 1087, 81–94. [Google Scholar]
- Gokool, A.; Loy, C.T.; Halliday, G.M.; Voineagu, I. Circular RNAs: The Brain Transcriptome Comes Full Circle. Trends Neurosci. 2020, 43, 752–766. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, L.; Bai, Y.; Han, B.; He, C.; Gong, L.; Huang, R.; Shen, L.; Chao, J.; Liu, P.; et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry 2020, 25, 1175–1190. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.J.; Hafez, A.K.; Amoah, S.K.; Rodriguez, B.A.; Dell’Orco, M.; Lozano, E.; Hartley, B.J.; Alural, B.; Lalonde, J.; Chander, P.; et al. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol. Psychiatry 2020, 25, 2712–2727. [Google Scholar] [CrossRef]
- Han, M.G.; Park, J.S.; Lee, C.S.; Jeong, Y.E.; Ryou, J.S.; Cho, J.E.; Ju, Y.R.; Lee, K.K. Serum MicroRNA Expression Profiling in Mice Infected with Rabies Virus. Osong Public Health Res. Perspect. 2011, 2, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Zhu, M.; Zhang, J.; Cai, Y.; Zhai, X.; Wang, D.; Li, G.; Su, S.; Zhou, J. Microarray analysis of lncRNA expression in rabies virus infected human neuroblastoma cells. Infect. Genet. Evol. 2019, 67, 88–100. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, L.; Zhang, T.; Wang, H.; Qin, C.; Yang, S.; Xia, X. Changes in microRNA expression induced by rabies virus infection in mouse brains. Microb. Pathog. 2012, 52, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Liu, S.; Zhong, Z.; Jiang, T.; Weng, R.; Xie, M.; Yang, S.; Xia, X. Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci. Rep. 2018, 8, 11858. [Google Scholar] [CrossRef]
- Shi, N.; Zhang, X.Y.; Dong, C.Y.; Hou, J.L.; Zhang, M.L.; Guan, Z.H.; Li, Z.Y.; Duan, M. Alterations in microRNA expression profile in rabies virus-infected mouse neurons. Acta Virol. 2014, 58, 120–127. [Google Scholar] [CrossRef]
- Tu, Z.; Xu, M.; Zhang, J.; Feng, Y.; Hao, Z.; Tu, C.; Liu, Y. Pentagalloylglucose Inhibits the Replication of Rabies Virus via Mediation of the miR-455/SOCS3/STAT3/IL-6 Pathway. J. Virol. 2019, 93, e00539-19. [Google Scholar] [CrossRef]
- Sui, B.; Chen, D.; Liu, W.; Wu, Q.; Tian, B.; Li, Y.; Hou, J.; Liu, S.; Xie, J.; Jiang, H.; et al. A novel antiviral lncRNA, EDAL, shields a T309 O-GlcNAcylation site to promote EZH2 lysosomal degradation. Genome Biol. 2020, 21, 228. [Google Scholar] [CrossRef]
- Tuffereau, C.; Bénéjean, J.; Blondel, D.; Kieffer, B.; Flamand, A. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 1998, 17, 7250–7259. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, C.; Tang, H.; Gao, S.; Sun, F.; Yang, Y.; Zhou, W.; Hu, Y.; Ke, C.; Wu, Y.; et al. CD2-Associated Protein Contributes to Hepatitis C, Virus Propagation and Steatosis by Disrupting Insulin Signaling. Hepatology 2018, 68, 1710–1725. [Google Scholar] [CrossRef]
- Haqshenas, G.; Terradas, G.; Paradkar, P.N.; Duchemin, J.B.; McGraw, E.A.; Doerig, C. A Role for the Insulin Receptor in the Suppression of Dengue Virus and Zika Virus in Wolbachia-Infected Mosquito Cells. Cell Rep. 2019, 26, 529–535.e3. [Google Scholar] [CrossRef]
- Tsai, S.; Clemente-Casares, X.; Zhou, A.C.; Lei, H.; Ahn, J.J.; Chan, Y.T.; Choi, O.; Luck, H.; Woo, M.; Dunn, S.E.; et al. Insulin Receptor-Mediated Stimulation Boosts T Cell Immunity during Inflammation and Infection. Cell Metab. 2018, 28, 922–934.e4. [Google Scholar] [CrossRef]
- Sestan, M.; Marinovic, S.; Kavazovic, I.; Cekinovic, D.; Wueest, S.; Turk Wensveen, T.; Brizic, I.; Jonjic, S.; Konrad, D.; Wensveen, F.M.; et al. Virus-Induced Interferon-gamma Causes Insulin Resistance in Skeletal Muscle and Derails Glycemic Control in Obesity. Immunity 2018, 49, 164–177.e6. [Google Scholar] [CrossRef]
- Zelus, B.D.; Schickli, J.H.; Blau, D.M.; Weiss, S.R.; Holmes, K.V. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 degrees C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 2003, 77, 830–840. [Google Scholar] [CrossRef]
- Kim, W.M.; Huang, Y.H.; Gandhi, A.; Blumberg, R.S. CEACAM1 structure and function in immunity and its therapeutic implications. Semin. Immunol. 2019, 42, 101296. [Google Scholar] [CrossRef]
- Liu, T.M.; Wang, H.; Zhang, D.N.; Zhu, G.Z. Transcription Factor MafB Suppresses Type I Interferon, Production by CD14(+) Monocytes in Patients With Chronic Hepatitis, C. Front. Microbiol. 2019, 10, 1814. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Morishita, A.; Iwama, H.; Fujita, K.; Masaki, T.; Tani, J.; Tadokoro, T.; Nomura, T.; Sakamoto, T.; Oura, K.; et al. Identification of Micro RNA Associated with the Elimination of Hepatitis C Virus Genotype 1b by Direct-Acting Antivirals Therapies. J. Gastroenterol. Hepatol. 2020, 36, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H.A. A simple method of estimating fifty percent endpoints. Am. J. Trop. Med. Hyg. 1937, 27, 493–497. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]


| Oligonucleotide Sequences | 5′–3′ |
|---|---|
| novel_circ_015068 | TGCATAATCAGCTACGGTGGTG |
| TGGAGCCGTGATAGTAAACGC | |
| novel_circ_017745 | TTGGTTCTTCAAAATCGCTGCC |
| TCTGCCTTTGTATCAACCATCTTG | |
| novel_circ_015161 | GCTGTGCAATGGTGAAATTATGG |
| CACTGAGCATCACTCACTCCTTT | |
| novel_circ_011422 | GCTGTGTGACGTTCAGGAAGAA |
| TTCAAGGCCGCCTCTTCTTTG | |
| novel_circ_012539 | TTGGAATGTTGGACTCCTTGAGA |
| CCTTCATCCAACCAGGCACTT | |
| novel_circ_010735 | CCACAGCCTTGCTTGTGAAA |
| CTGTTTCCTCTACTGATGGTGGG | |
| novel_circ_008143 | ACCCGAAGAAAGATGGATCGG |
| CAGGCTCCCAAGATAAGCGA | |
| novel_circ_010627 | ACGGAGAGTATGTTGTCGAGGT |
| AACCACGGCTCCAAGTCAGT | |
| novel_circ_010564 | TGGACCCTGCTAAGAAGTCACC |
| TCTGCAATGTCCGGCCTCA | |
| novel_circ_026888 | TGCCTCTGTTTTATGCTGCTACC |
| TGGTTACGGTCAGTCAGTGGT | |
| novel_circ_006420 | AGTCAAAGTCCCACGGCCAC |
| TAGTGTTGAAGAGGGAGAGTGGT | |
| novel_circ_014267 | TTTCGACGTTTTCAGCGAGACA |
| GCCTTCTTCAGACATCATCAGTGG |
| circRNAs | Source Gene | Oligonucleotide Sequences (5′–3′) |
|---|---|---|
| novel_circ_015068 | PD-L1 | CCTGCTGTCACTTGCTACGG |
| TCCAGCTCCCGTTCTACAGG | ||
| novel_circ_015161 | Pcgf5 | TGCCCTTCTGCTACTGACCA |
| GGCAAGCGGAACACTGAGAA | ||
| novel_circ_011422 | Parp9 | ACAGGGAAGAGCAAAGGCGA |
| GTGGCCTGTTTCGGGTGATG | ||
| novel_circ_012539 | - | |
| novel_circ_017745 | Serinc3 | AGGAACATCAGCCTCGCTCT |
| GGACCGCTCAGGTTCATTGG | ||
| novel_circ_010735 | - | |
| novel_circ_008143 | Ocln | GGGAAAGCAGGAAAGGGCAA |
| CTGACCCAGTCCTCCTCCAG | ||
| novel_circ_010627 | Cntn1 | GGGCTGGGCATGACAAAGAA |
| TGGGTGTCGGGAAGAAGGTT | ||
| novel_circ_010564 | Shank3 | TTCCTCTCTGTGGGTGCCAT |
| CAGGGGAGGGGAGTAGCAAA | ||
| novel_circ_026888 | Slc20a2 | GGGTTTGGGGCAGAAGAGTG |
| TGTTGGAGGCAATCACCACG | ||
| novel_circ_006420 | Rtn1 | GGGTTTGGCACATCCCCTAA |
| TTCGCTACTCCCAAGCCTGT | ||
| novel_circ_014267 | Atp9b | ACAGTTCACGGGCTGGTTTC |
| ATGGGGGCATCTCGAAGTCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Su, J.; Wang, N.; Zhao, N.; Su, S. Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus. Int. J. Mol. Sci. 2021, 22, 6537. https://doi.org/10.3390/ijms22126537
Zhao W, Su J, Wang N, Zhao N, Su S. Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus. International Journal of Molecular Sciences. 2021; 22(12):6537. https://doi.org/10.3390/ijms22126537
Chicago/Turabian StyleZhao, Wen, Jingyin Su, Ningning Wang, Naiyu Zhao, and Shuo Su. 2021. "Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus" International Journal of Molecular Sciences 22, no. 12: 6537. https://doi.org/10.3390/ijms22126537
APA StyleZhao, W., Su, J., Wang, N., Zhao, N., & Su, S. (2021). Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus. International Journal of Molecular Sciences, 22(12), 6537. https://doi.org/10.3390/ijms22126537





