Abstract
Osteoarthritis (OA) is the most common musculoskeletal disorder causing a great disability and a reduction in the quality of life. In OA, articular chondrocytes (AC) and synovial fibroblasts (SF) release innate-derived immune mediators that initiate and perpetuate inflammation, inducing cartilage extracellular matrix (ECM) degradation. Given the lack of therapies for the treatment of OA, in this study, we explore biomarkers that enable the development of new therapeutical approaches. We analyze the set of secreted proteins in AC and SF co-cultures by stable isotope labeling with amino acids (SILAC). We describe, for the first time, 115 proteins detected in SF-AC co-cultures stimulated by fibronectin fragments (Fn-fs). We also study the role of the vasoactive intestinal peptide (VIP) in this secretome, providing new proteins involved in the main events of OA, confirmed by ELISA and multiplex analyses. VIP decreases proteins involved in the inflammatory process (CHI3L1, PTX3), complement activation (C1r, C3), and cartilage ECM degradation (DCN, CTSB and MMP2), key events in the initiation and progression of OA. Our results support the anti-inflammatory and anti-catabolic properties of VIP in rheumatic diseases and provide potential new targets for OA treatment.
Keywords:
osteoarthritis; synovial fibroblasts; chondrocytes; VIP; CHI3L1; PTX3; complement system; decorin; cathepsin B; MMP2 1. Introduction
Osteoarthritis (OA) is the most common musculoskeletal disorder that affects nearly 300 million people in the world population that are over 60 years of age. It causes great disability and a substantial reduction in the quality of life, representing a global public health problem with no current treatment. It is classically established that chronic joint overload alters its mechanical function, triggering joint inflammation and cartilage degeneration [1]. Current information proposes that OA is a more complex pathology that includes other causal factors, such as genetic and metabolic factors. The confluence of all these factors generates the disruption of the homeostasis of the whole joint, including the cartilage, subchondral bone, synovium, meniscus and the infrapatellar fat pad, by the release of catabolic and inflammatory factors [2,3].
Although the final consequence is the loss of articular cartilage, all joint tissues, including bone and synovium, participate in the production of these factors concerning both different cellular types and the extracellular matrix (ECM) [4,5,6]. In OA, articular chondrocytes (AC) and synovial fibroblasts (SF) mainly release innate-derived immune mediators that initiate and perpetuate inflammation, inducing cartilage ECM degradation [5,7,8,9]. The fact that SF are active drivers of joint destruction in rheumatoid arthritis (RA) is well established [10], but their behavior in OA patients is less understood.
The ECM is a complex mixture of macromolecules secreted by cells into the extracellular space that provides the supportive architecture on which cells adhere, migrate, and regulate tissue development [11], interacting with cell surface receptors on joint resident cells. Therefore, the ECM not only functions as a structural support for a group of cells in a tissue, but also actively communicates with the cells to ensure homeostasis. Thus, variations in the composition and physical properties of the ECM lead to the development of many diseases, including cancer and rheumatic diseases, such as OA and RA, among others [12,13,14]. In this sense, the integrity of ECM composition is crucial to the regular function of load-bearing tissues, such as cartilage in OA [15].
The glycoprotein fibronectin (Fn) is a component of the pericellular matrix that plays an important role in the maintenance of the mechanical properties of the cartilage [16]. During OA progression, the injury induces tissue proteolysis and ECM damage, generating ECM-degradation products, including Fn fragments (Fn-fs), that induce cytokine and proteinase expressions, chronifying the inflammation [12,17,18].
Joint tissues are sources of bioactive neuropeptides, such as neuropeptide Y, pituitary adenylate cyclase-activating polypeptide and vasoactive intestinal peptide (VIP) that induces changes in the cell metabolism in degenerative conditions such as OA [19]. VIP and its G protein-coupled receptors (GPCRs), VPAC1, and VPAC2, form a signaling axis that modulates both the innate and acquired immunity in several inflammatory/autoimmune diseases, including OA [20,21]. VIP sources in the joint comprise both nerve fibers of the sympathetic nervous system and a cellular origin, including lymphocytes and SF from OA and RA patients [20,21,22]. Functionally, a decrease in the number of these nerve endings is described in OA and RA [19]. Moreover, VIP levels are reduced in cartilage and synovial fluid of OA patients, compared to healthy controls, which could participate in the pathology development [23]. Clinically, VIP levels in synovial fluid and cartilage of OA patients are negatively related with joint impairment, being a prospective indicator of disease severity [24].
Given the absence of specific therapies for the treatment of OA, the exploration of biomarkers that enable the development of new therapeutical approaches is crucial. In this context, we proposed to analyze the set of secreted proteins in chondrocyte and synoviocyte co-cultures, joint cell types involved in crucial physiological processes in OA. Moreover, we also studied the modulating role of the endogenous peptide VIP in this secretome, providing novel VIP-modulated proteins involved in the main events that take place during the OA pathology: the inflammatory process, the activation of the complement system, and the cartilage ECM-degradation.
2. Results
2.1. SF-AC Co-Cultures Secretome Profiling
SF and AC grown in a stable isotope labeling with amino acids (SILAC) medium were put together in co-culture and treated with 45 kDa Fn-fs as a stimulus of inflammation and ECM destruction in the presence and absence of VIP to elucidate which proteins are modulated by this neuropeptide. A total of 115 proteins were detected in the secretome of the co-cultures (Table 1, Figure 1).

Table 1.
Proteins identified by SILAC analysis in the secretome of OA SF-AC co-cultures.
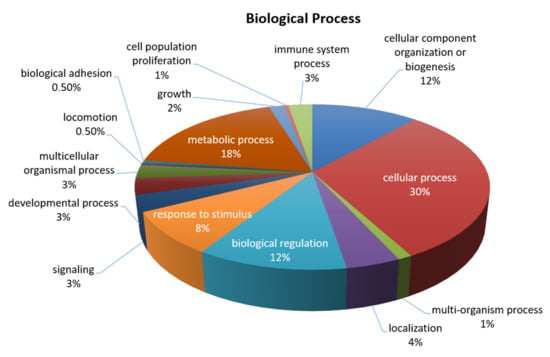
Figure 1.
General biological processes involving the proteins detected in the OA SF-AC co-cultures secretome identified by SILAC. Protein classification was performed, using the PANTHER (protein analysis through evolutionary relationships) database.
2.2. Modulation of SF-AC Co-Cultures Secretome by VIP
Among the proteins detected by SILAC, 28 proteins were significantly modulated by VIP in one of the replicates (forward or reverse) for the four patients (Supplementary Table S1). Nine of these proteins were consistently modulated in both replicates (forward and reverse) decreasing 8 of them, as shown in the ratio Fn-fs+VIP:Fn-fs (Figure 2a, Table 2, Supplementary Table S1). Proteins downregulated by VIP included those involved in the immune response: chitinase-3-like protein 1 (CHI3L1), pentraxin-related protein 3 (PTX3), complement C1r subcomponent (C1r), complement C3 (C3), sulfhydryl oxidase 1 (QSOX1), and cathepsin B (CTSB). Among them, CHI3L1, PTX3 and C3 were also implicated in inflammation, and C1r, C3 took part in complement activation. In addition, VIP also decreased proteins involved in ECM degradation, including DCN, CTSB, and MMP2 (Figure 2b).
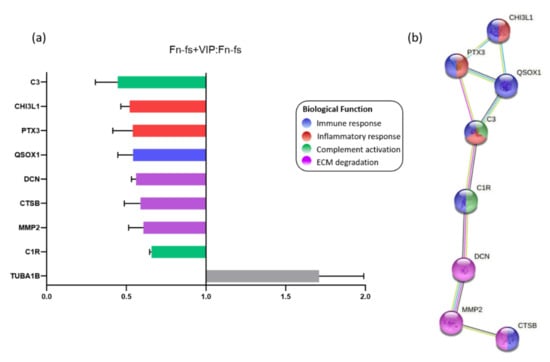
Figure 2.
VIP-modulated proteins in the SF-AC co-cultures secretome. (a) Average SILAC ratios (n = 4) that represent the relative protein abundance in Fn-fs+VIP versus Fn-fs treated SF-AC co-cultures at the same time point (48 h). Ratios are presented as the media of the replicates forward and reverse for each patient. (b) Functional protein association networks and biological processes where the proteins downregulated by VIP are involved, according to the STRING database.

Table 2.
VIP-modulated proteins in the in the Fn-fs-stimulated secretome of OA SF-AC co-cultures identified by SILAC.
2.3. VIP-Modulated Inflammatory Proteins
Among the proteins modulated by VIP in the SILAC analysis, we decided to validate those involved in the inflammatory response, CHI3L1 and PTX3, which is a common process that takes place in OA, worsening the disease progression. Firstly, we evaluated the constitutive production of these proteins by SF and AC cultured alone, as well as in co-culture for 48 h, by ELISA and Multiplex for CHI3L1 and PTX3, respectively. The results showed that AC produced more CHI3L1 than SF. This increase was also observed in the co-culture (Figure 3a). No differences were observed in PTX3 (Figure 3b). Next, the SF-AC co-cultures were treated with 45 kDa Fn-fs, as a pro-inflammatory stimulus, in the presence and absence of VIP. No effects were observed with the Fn-fs treatment alone, while in presence of VIP, the production of both CHI3L1 and PTX3 was decreased in the secretome of the co-cultures, corroborating the anti-inflammatory effect of VIP (Figure 3c,d). No significant effects were observed when the SF and AC were cultured alone (Supplementary Figure S1).
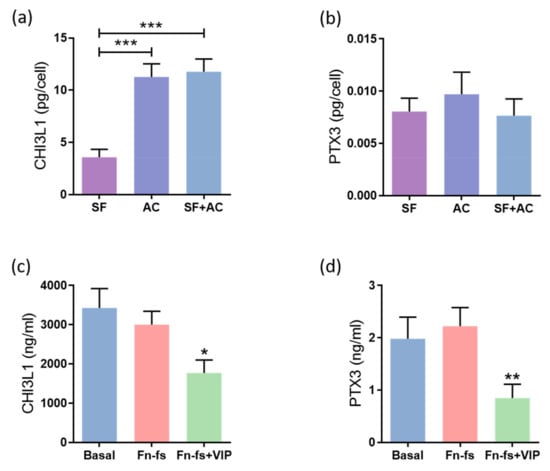
Figure 3.
VIP-modulated inflammatory proteins. Secreted levels of (a,c) CHI3L1 and (b,d) PTX3 were determined by ELISA and Multiplex, respectively, in the secretomes from SF and AC alone and in co-culture (n = 6). Data are presented as mean ± SEM of triplicate determinations. (a,b) Constitutive protein expression of (a) CHI3L1 and (b) PTX3 in the secretomes from SF and AC alone and in co-culture at 48 h. Results are presented as pg corrected by the number of cells for each condition. *** p < 0.001. (c,d) Protein expression of (c) CHI3L1 and (d) PTX3 in the SF-AC co-cultures secretomes at 48 h of treatment with and without 10 nM 45 kDa Fn-fs in the presence and absence of 10 nM VIP. * p < 0.05, ** p < 0.01 Fn-fs+VIP vs. Fn-fs.
2.4. VIP-Modulated Complement System Proteins
In addition to the inflammatory proteins mentioned above, results derived from the SILAC analysis showed that VIP tends to decrease the complement cascade proteins CFB, complement C1r, C1s subcomponents, C3, as well as the complement-related protein clusterin. Among them, VIP significantly decreased C1r, C1s, C3 and clusterin in any of the replicates for the 4 patients, as observed in the ratio Fn-fs+VIP:Fn-fs, and the complement system proteins C1r and C3 in both replicates (Supplementary Table S1). Regarding the constitutive production, no significant differences were observed between the different cultures in the secreted levels of C1r nor in C3 (Figure 4a,b). On the other hand, the results obtained from ELISA showed a decrease in the production of C1r in the SF-AC co-cultures treated with Fn-fs+VIP in comparison to the inflammatory stimulus alone, while no effects were observed with the Fn-fs (Figure 4c). However, despite VIP tended to decrease the levels of C3, no significant differences were observed by Multiplex analysis (Figure 4d). No significant effects were observed in the isolated cultures either (Supplementary Figure S2).
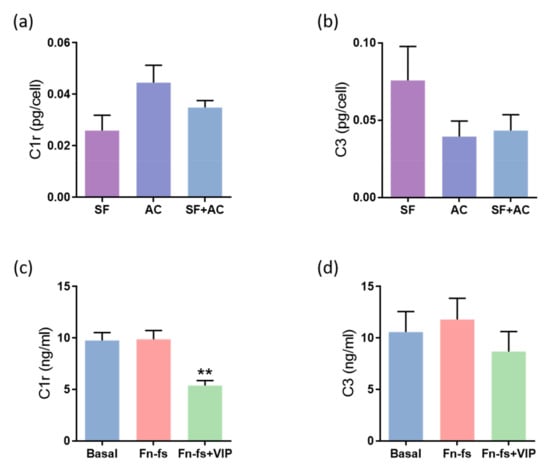
Figure 4.
VIP-modulated complement system proteins. Secreted levels of (a,c) C1r and (b,d) C3 were determined by ELISA and Multiplex, respectively, in the secretomes from SF and AC alone and in co-culture (n = 6). Data are presented as mean ± SEM of triplicate determinations. (a,b) Constitutive protein expression of (a) C1r and (b) C3 PTX3 in the secretomes from SF and AC alone and in co-culture at 48 h. Results are presented as pg corrected by the number of cells for each condition. (c,d) Protein expression of (c) C1r and (d) C3 in the SF-AC co-cultures secretomes at 48 h of treatment with and without 10 nM 45 kDa Fn-fs in the presence and absence of 10 nM VIP. ** p < 0.01 Fn-fs+VIP vs. Fn-fs.
2.5. VIP-Modulated ECM Degradation Proteins
Degradation of cartilage ECM is one of the key events that take place in the OA pathology. SILAC analysis showed that VIP modulated some of these proteins, significantly decreasing DCN, MMP2 and CTSB in the secretome from SF-AC co-cultures. Therefore, these molecules were validated by ELISA for DCN and CSTB, and Multiplex for MMP2. Firstly, we evaluated the constitutive production of these proteins by the SF and AC cultured alone as well as in co-culture for 48 h. We observed that SF and AC had an additive effect in the production of DCN (Figure 5a), while the SF produced more CTSB and MMP2 than the AC, whose production was also reduced in the co-culture (Figure 5b,c). Next, the SF-AC co-cultures were treated with 45 kDa Fn-fs as a stimulus of ECM destruction in the presence or absence of VIP. While no effects were observed with the Fn-fs treatment alone, the presence of VIP reduced the production of DCN, CTSB and MMP2 in the secretome of the co-cultures, corroborating the protective effect of VIP in the ECM degradation (Figure 5d–f). A decrease in the CTSB and MMP2 production was also observed in the secretome of SF cultured alone in the presence of VIP when stimulated with Fn-fs (Supplementary Figure S3b,c), while this decrease was not significant in the AC isolated cultures (Supplementary Figure S3e,f). No significant effects were observed in the levels of DCN in the case of the isolated cultures (Supplementary Figure S3a,d).
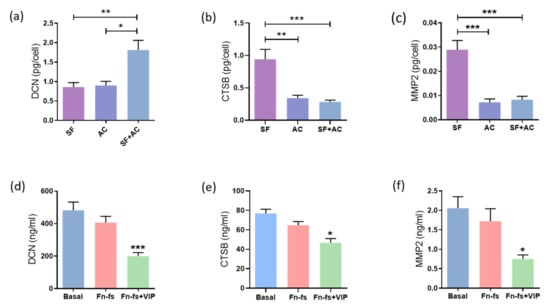
Figure 5.
VIP-modulated ECM degradation proteins. Secreted levels of (a,d) DCN, (b,e) CTSB, and (c,f) MMP2 were determined by ELISA and Multiplex, respectively, in the secretomes from SF and AC alone and in co-culture (n = 6). Data are presented as mean ± SEM of triplicate determinations. (a–c) Constitutive protein expression of (a) DCN, (b) CTSB, and (c) MMP2 in the secretomes from SF and AC alone and in co-culture at 48 h. * p < 0.05, ** p < 0.01, *** p < 0.001. Results are presented as pg corrected by the number of cells for each condition. (d–f) Protein expression of (d) DCN, (e) DCN, and (f) MMP2 in the SF-AC co-cultures secretomes at 48 h of treatment with and without 10 nM 45 kDa Fn-fs in the presence and absence of 10 nM VIP. * p < 0.05, *** p < 0.001 Fn-fs+VIP vs. Fn-fs.
3. Discussion
As far as we know, this is the first study that analyzed the proteins detected in the secretome of SF-AC co-cultures in OA patients. We described a total of 115 proteins, 74 of which had been previously reported in the secretome of IL-1β-stimulated AC from healthy donors by SILAC [25]. When comparing with our results, 39 were commonly identified in both secretomes, while 76 proteins were exclusively detected in the present Fn-fs-stimulated SF-AC co-cultures, and 35 in the previous IL-1β-stimulated AC cultures (Supplementary Table S2).
Discrepancies regarding the number of proteins identified could be explained by the different experimental designs used in the studies. The main differences include the stimuli (IL1β in the previous analysis vs. Fn-fs in the present one), the cell type (AC vs. SF-AC co-cultures), as well as the samples origin (healthy donors vs. OA patients). In another proteomic study, 30 proteins were over-expressed in the lysate of SF from RA patients compared with controls [26].
In the present study, we also reported, for the first time, a proteomic analysis by SILAC of the effect of a neuropeptide present in the joint in OA patients. Among the proteins detected in the SF-AC co-cultures secretome, VIP consistently decreased 8 of them, including proteins involved in the inflammatory process and complement activation, as well as in the cartilage ECM degradation, key events in the initiation and progression of OA.
Regarding pro-inflammatory proteins, we described the modulation by VIP of two immune-related proteins: CHI3L1 (also known as human cartilage glycoprotein-39 or YKL-40) and PTX3.
CHI3L1 is produced by several cell types, including synoviocytes, chondrocytes, osteoblasts, macrophages, monocytes, neutrophils, and vascular smooth muscle cells [27,28,29,30]. Increased levels of CHI3L1 were reported in several pathologies, including rheumatic diseases and cancer, being involved in inflammation, tissue remodeling and tissue injury in health, as well as during disease [29,31,32,33]. CHI3L1 is also related to bone resorption activity [32]. Its levels in serum and synovial fluid correlate with OA and RA severity [34,35,36,37,38,39,40,41]. Moreover, higher levels of CHI3L1 were addressed in OA synovial fluid and sclerotic osteoblasts compared to healthy donors [27,40]. In addition, CHI3L1 is positively associated with inflammation and cartilage destruction mediators in OA [42]. All in all, CHI3L1 represents a potential biomarker in several pathologies, including rheumatic diseases, such as psoriasis, psoriatic arthritis, RA, and OA [43,44].
Conversely, PTX3 belongs to the group of acute-phase proteins pentraxins, which play a key role in inflammation, also being involved in cell proliferation, migration, and ECM remodeling [45,46]. Production of PTX3 is induced in different cell types in response to pro-inflammatory mediators, including macrophages, endothelial cells and synoviocytes. Increased levels of PTX3 were reported in patients with systemic inflammation, including septic shock, myocardial infarction, and systemic vasculitis [47]. Regarding rheumatic diseases, PTX3 is involved in osteoblast proliferation in osteoporosis [46] and its expression is induced by TNF-α in OA SF [48]. In RA, increased levels of PTX3 are associated with disease severity [45,47,48].
In the present study, we corroborated the production of CHI3L1 and PTX3 by both OA-SF and -AC, where AC produces more CHI3L1 than SF, consistent with previous studies, suggesting that cartilage is the main source of this protein [42]. In addition, its levels in AC are related to tissue degeneration in OA [49]. We validated the SILAC results by ELISA or Multiplex, confirming that VIP decreases the pro-inflammatory mediators CHI3L1 and PTX3 in the secretome from SF-AC co-cultures, validating the anti-inflammatory properties of VIP [20,21,50,51,52].
Complement system proteins are involved in the pathogenesis of OA by means of ECM degradation, AC and SF inflammatory responses and synovitis, cartilage angiogenesis, bone remodeling and osteophyte formation, cell lysis, and stem cell recruitment. Several studies have focused on the complement pathway as a target for the treatment of rheumatic diseases [53]. During complement activation, its inflammatory cleavage products bind to their receptors and mediate several inflammatory effects. The complement can be activated by cartilage ECM components and their cleavage products [54,55,56,57,58]. Specifically, fibronectin binds to the C1q complement subcomponent [59,60]. Cartilage, synovium and bone are key sources of the classical and alternative complement pathway components [54,61], produced by articular cells, including AC and SF in healthy donors, RA, and OA patients [62,63,64,65]. Increased levels of complement factors have been reported in the synovial fluid from OA and RA joints and acute knee injuries [54,66]. In addition, complement activation is also associated with inflammation and osteochondral fractures in OA [67], and antiangiogenic, anti-inflammatory, and anti-catabolic effects of chondroitin sulphate have been related to the reduction of complement components, including CFB, C1r, C1s and C3 [25].
C1 is the first component of the classical pathway of the complement system, a multimolecular protease consisting of two catalytic subunits, C1r and C1s, and recognition protein, C1q [68]. In a previous proteomic study complement C1r was the most significantly upregulated protein in OA synovial fluid, whose levels correlated with severity [69]. Increased levels of C1 complex were also previously described in serum and synovial fluid from RA patients [70,71].
On the other hand, cleavage of C3 is the common point of the three complement activation pathways (classical, alternative, and lectin pathway) [54]. C3 is one the most expressed complement components in OA osteochondral biopsies, cartilage, AC and SF [61], and may be involved in bone remodeling in OA [72]. Moreover, in OA and other arthritic diseases, C3 cleavage fragments in serum and synovial fluid are related to disease severity, pain, and clinical symptoms [73,74,75,76,77,78,79].
Consistent with previous findings, our results derived from SILAC analysis in the SF-AC co-cultures corroborated the production of the complement cascade proteins CFB and CFH, complement C1r and C1s subcomponents, C3, and clusterin. However, no presence of C2, C4, and C1q was detected, as reported by other authors in human AC [25,64]. Regarding VIP, it tends to reduce these complement cascade proteins, consistently decreasing complement C1r, as well as C3, results corroborated by ELISA for C1r. C3 Multiplex analysis also showed a decrease, although it was not significant, which was due to the limited number of samples. These data demonstrate, for the first time, the regulatory character of VIP in the complement system, an important component of innate immunity.
Finally, we reported the VIP-mediated reduction of three proteins involved in ECM homeostasis: DCN, CTSB and MMP2.
ECM-degradation is a key feature in OA and an important focus of investigation due to the irreversible nature of the damaged cartilage. DCN is a structural protein highly present in the articular cartilage interterritorial and pericellular ECM [80,81,82,83,84,85,86,87]. This small, leucine-rich proteoglycan is involved in the regulation of different biological functions, including ECM organization, cell adhesion, migration and proliferation [87]. In rheumatic diseases, it was detected in the synovial fluid from OA and RA patients [88]. Increased DCN expression was described in OA cartilage [89,90,91], as well as in fibroblast-like chondrocytes, being upregulated in late stages of the disease [87]. In addition, DCN possess pro-inflammatory properties through activation of TLR4 and the NF-κB pathway [88]. Previous studies have reported a VIP-mediated downregulation of TLR4 expression inhibiting NF-κB signaling [92]. Increased levels of DCN-specific antibodies have been identified in the synovial fluid from patients with inflammatory rheumatic diseases [93]. Soluble isoforms of DCN released by ADAMTSs and MMPs in damaged cartilage can act as endogenous danger signals in OA and RA [88,89,90,93,94]. Binding of DCN to C1q has also been reported, but without induction of complement activation [58]. Conversely, addition of DCN to synovial fluid inhibits SF expansion in OA patients [95].
Cysteine cathepsins and MMPs are highly involved in the development of OA [18,96,97,98,99,100,101]. CTSB is the main cysteine peptidase in OA cartilage [102], involved in cartilage-ECM degradation including collagen and aggrecan [103,104]. In turn, fragments derived from collagen type II induce expression of cathepsins, including CTSB, in OA AC [99]. In addition, CTSB degrades the MMPs inhibitors TIMP-1 and TIMP-2, promoting angiogenesis, mineralization and osteophyte formation [105]. However, it is considered a perpetuator rather than an initiator of cartilage destruction [102,106]. CTSB is reported to be a marker of chondrocyte differentiation since its levels increase by subculturing cells in monolayers, suggesting dedifferentiation toward a SF phenotype [106,107]. In addition, CTSB action is associated with joint inflammation [108], and elevated levels of cysteine cathepsins are present in synovial fluid and SF from RA patients [100,109,110,111,112] as well as in the synovial membrane from RA and OA patients compared to controls [113,114]. Moreover, increased CTSB activity and expression are shown in AC and cartilage from OA patients [102,115,116,117,118,119,120], and its levels in serum and synovial fluid correlate with severity and joint inflammation [119]. Furthermore, it is hypothesized that anomalous trafficking of cathepsin B found in pathologies, such as OA, depend on alternative splicing of its pre-mRNA [121].
MMPs are the main proteases involved in the degradation of collagen type II, produced by chondrocytes as well as by synovial cells, whose contribution in rheumatic diseases has been widely studied [122,123]. MMP2, also known as gelatinase A, degrades denatured collagens and gelatins, as wells as other ECM components [122,124]. Several studies have noted increased MMP2 levels in cartilage [98,125,126,127,128], serum [129], synovial fluid [130,131,132,133], as well as in the synovium and pannus-like tissue in OA [128,134]. This increased expression was also detected in SF from RA and OA patients [135,136], in synoviocytes co-cultured with fibroblasts [137] and platelets [130], as well as in AC co-cultured with subchondral bone osteoblasts in OA [138]. Consequently, MMP2 is defined as a potential biomarker for OA [139,140,141], and is positively correlated to histopathology severity in OA cartilage [142]. Conversely, Thorson et al. detected lower MMP2 plasma levels in OA patients compared to controls, suggesting a secondary role of this MMP [143].
All in all, regarding proteins involved in ECM homeostasis, we showed that SF and AC have an additive effect in the production of DCN in co-culture. We also demonstrated that SF produce more MMP2, which is inhibited by the presence of AC in the co-culture, compared to SF cultured alone. Regarding VIP, it also reduces the levels of DCN, CTSB and MMP2 in the SF-AC co-cultures. Decrease in MMP2 expression mediated by VIP was also reported in renal-cell carcinoma [144]. The present study supports previous data from our laboratory reporting the beneficial effects of VIP in the protection of several components of the ECM, modulating uPA, MMP9 and MMP13 as well as ADAMTS-4, -5, -7, and -12 production. [8,145]. Moreover, since DCN, CTSB and MMP2 also promote the inflammatory process in OA, reduction mediated by VIP again corroborates its well-known anti-inflammatory role in rheumatic diseases.
We used 45 KDa Fn-fs as a pro-inflammatory and degradative stimulus as previously described [18]. However, despite the fact that no significant effects were detected in the proteins analyzed in this study with the Fn-fs treatment alone, this stimulus is necessary for VIP to exert its effects (data not shown).
Since a pioneering study demonstrated the beneficial role of VIP in a murine arthritis model, downregulating both the inflammatory and the autoimmune components of the disease [146], different studies valued its therapeutic potential. Gradually these results were corroborated in different inflammatory/autoimmune pathologies. In rheumatic diseases, its protective effect was also reported in bone erosion through NF-κB and AP-1 modulation [147]. In early arthritis (EA) patients, disease activity inversely correlated with VIP serum levels and after a two-year follow-up, those patients with low baseline levels of VIP showed higher disease activity and received more intensive treatment [148]. Confirming these data, an association between serum VIP levels and variants in the VIP gene was reported [149].
Although most of the effects point to a beneficial role of the VIP axis specifically in OA, it was described in murine models that VIP might promote mechanosensitivity and pain [150]. In patients, it was also reported a pro-inflammatory role of VIP [151]. However, VIP is downregulated in the synovial fluid of patients, which entails a rise in the expression of pro-inflammatory cytokines contributing to OA development [24]. Thus, it can be hypothesized that a defeat of VIP contributes to the pathogenesis and that the reestablishment of VIP levels could pause or suppress disease evolution [23,24]. In addition, since no significant effects of the VIP treatment were observed in the isolated SF and AC cultures, with the exception of CTSB and MMP2 in Fn-fs+VIP-treated SF, our study highlights the significance of both cell types in the microenvironment of the OA joint, further supporting the worth of our study in co-cultures.
Some limitations of the study should be noted, including the limited number of samples, the lack of controls, and the low number of identified proteins. Undoubtedly, further studies employing high resolution equipment would allow the identification of a higher number of proteins in the secretomes of the co-cultures not detected in the present study. In addition, future studies with more samples and comparisons between OA patients and healthy donors would be also of interest. Nevertheless, we honestly believe that the limitations due to a rapidly obsoleting technology do not reduce the interest and impact of our findings.
In summary, the novelty of the current study consists in the identification of the proteins released by two cell populations present in the OA joint and key in the pathology of the disease. Our results support the anti-inflammatory and anti-catabolic properties of VIP by reducing several crucial mediators involved in inflammatory/immune response and ECM degradation. Finally, we demonstrate, for the first time, the modulation of the complement cascade proteins mediated VIP.
4. Materials and Methods
4.1. Subjects and Samples Procurement
Osteoarthritic cartilage and synovial tissue were obtained from the knee of ten patients (five women and five men) aged between 50 and 92, undergoing leg amputations caused by trauma or individuals who died and were tissue and organ donors. All tissue samples were provided by the Autopsy Service at Hospital Universitario de A Coruña. Informed consent was obtained from the patients before surgery. OA patients were diagnosed following the American College of Rheumatology (ACR) criteria for OA classification [152]. The study was approved by the local Ethics Committee (Galicia, Spain).
4.2. Cell Cultures
AC were isolated as previously described [153]. Briefly, cartilage surfaces were rinsed with a saline buffer; scalpels were used to cut parallel vertical sections 5 mm apart from the cartilage surface to the subchondral bone. These cartilage strips were dissected from the bone, and the tissue was incubated with trypsin at 37 °C for 10 min and then digested with type IV clostridial collagenase. The release of AC from cartilage was achieved after 16 h of digestion in an incubator at 37 °C on a microplate shaker. SF cultures were established by explant growth of synovial biopsies, cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 25 mM HEPES and 4.5 g/L glucose, completed with 10% heat-inactivated fetal bovine serum (FBS) (Lonza Ibérica SAU, Barcelona, Spain), 1% L-glutamine and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA), at 37 °C and 5% CO2. After three passages, residual contamination by macrophages was avoided, previously assessed by flow cytometry analysis of SF with a purity of 95% [92].
4.3. Chondrocyte and Synovial Fibroblasts Stable Isotope Labeling by Amino Acids in Cell Cultures (SILAC)
AC and SF were labeled as previously described [154]. Briefly, isolated cells were recovered and plated at low density in SILAC DMEM-Flex (Invitrogen), lacking arginine (R) and lysine (K) and supplemented with 10% dialyzed fetal bovine serum (dFBS) (Invitrogen), 4.5 g/L glucose, 2mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin (Merck, Darmstadt, Germany). In the case of light media, standard L-lysine and L-arginine were used, while isotope-labeled D-lysine (D4) and C-arginine (13C6), and isotope-labeled L-lysine (13C6,15N2) and L-arginine (13C6,15N4) were used for medium and heavy conditions, respectively. For the initial cell expansion, 5 × 104 AC and SF from each patient were seeded in three T-25 and T-75 cell culture flasks, respectively (one flask per condition: light, medium and heavy, for each cell type). At confluence, cells were recovered from each culture flask by trypsinization and plated into 6-well transwells (Corning Inc., Corning, NY, USA): AC were seeded into 6-well plates (5 × 104 AC per well) and SF were seeded in 6-well inserts (1.5 × 104 SF per insert), separately.
4.4. Co-Cultures and Treatments
When 80% of confluence was reached, corresponding to 100% of the labeling as previously described [154], the FBS-containing medium was removed and cells were washed thoroughly to remove abundant serum proteins. The inserts with adherent SF were placed onto the corresponding 6-well plates with AC and incubated in serum-free medium with and without 10 nM 45 kDa Fn-fs (Merck) in the presence and absence of 10 nM VIP (Bachem, Bubendorf, CH, Switzerland) for 48 h. Experiments were performed in replicate: one replicate of forward (Fn-fs medium/Fn-fs+VIP heavy) and one replicate of reverse labeling (Fn-fs heavy/Fn-fs+VIP medium) (Figure 6).
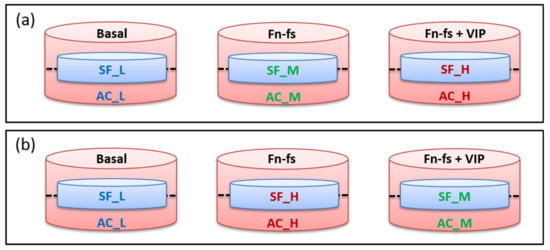
Figure 6.
Experimental design of SF-AC co-cultures for SILAC analysis. (a) Forward replicate (Fn-fs medium/Fn-fs+VIP heavy). (b) Reverse replicate (Fn-fs heavy/Fn-fs+VIP medium). SF, synovial fibroblasts; AC, articular chondrocytes; L, light; M, medium; H, heavy; Fn-fs, fibronectin fragments; VIP, vasoactive intestinal peptide.
4.5. Collection and Preparation of Conditioned Media, One-Dimensional Gel Electrophoresis and In-Gel Digestion of Proteins
Secretomes from each condition (light, medium and heavy) were mixed 1:1 and concentrated, using Amicon® Ultra 10K centrifugal filter devices (Merck). About 20 µg were separated on a 10% SDS-PAGE gel. The gel was stained with Coomassie blue and the resulting lanes were cut into 7 slices (Figure 7), and subjected to in-gel digestion as previously described [154]. Briefly, in-gel reduction was done for 45 min at 56 °C using 10 mM DTT in 25 mM ammonium bicarbonate followed by in-gel alkylation, using 55 mM iodoacetamide in 25 mM ammonium bicarbonate for 30 min in the dark. Digestion was performed overnight with 12.5 ng/L Sequencing grade Modified Trypsin (Promega, Madison, WI, USA) at 37 °C. The extracted peptide mixtures were desalted and concentrated via NuTips (Glygen Corp., Columbia, MD, USA) as previously described [154].
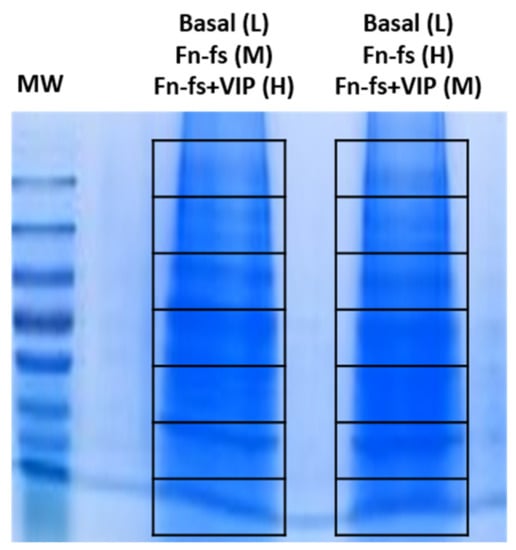
Figure 7.
One-dimensional gel electrophoresis separation of SF-AC co-cultures secretome proteins. In each lane, 20 μg of mixed samples (L + M + H) were resolved. Then, the whole gel column was sliced into 7 sections and each lane was subjected to nanoLC-MS/MS analysis. MW, molecular weight markers; L, light; M, medium; H, heavy; Fn-fs, fibronectin fragments; VIP, vasoactive intestinal peptide.
4.6. NanoLC-MALDI-TOF/TOF Analysis
Peptide fractions were resolved, using RP-nLC in a Tempo nanoLC system (Eksigent, Dublin, CA, USA). The peptide mixture (5 µL) was injected on a C18 precolumn (Michrom, 0.5 × 2 mm) coupled to a reversed-phase column (Integrafit C18, Proteopep II, 75 µm id, 10.2 cm, 5 µm, 300 Å pore size; New Objective, Woburn, MA, USA). Peptides were then eluted in a linear gradient of 5–50% ACN (45 min gradient, flow rate = 350 nL/min), mixed with α-cyano-4-hydroxycinnamic acid matrix (4 mg/mL, flow rate = 1.2 µL/min) and deposited onto a MALDI plate (Sun Collect; Sunchrom, Friedrichsdorf, Germany). Chromatograms corresponding to each gel section were composed of 180 spots. Data acquisition was carried out using a 4800 MALDI-TOF/TOF instrument (Sciex, Foster City, CA, USA), employing the 4000 Series ExplorerTM software version 3.7 (Sciex). MS full-scan spectra from 800 to 4000 m/z were acquired for each peptide-containing LC spots, using 1500 laser shots and a laser intensity of 3800 kV. After screening of all the spots in MS-positive reflector mode, the fragmentation of automatically selected precursors was performed at a collision energy of 1 kV with a collision-induced dissociation (CID) gas (air). Up to 12 of the most intense ion signals per spot position with signal/noise ratios (S/N) above 80 were selected as precursors for tandem mass spectrometry (MS/MS) analysis, excluding common trypsin autolysis peaks and matrix peaks. The number of shots was 1800 for MS/MS, and the laser intensity was set to 4700 kV. A second MS/MS was performed, excluding the precursors selected in the previous MS/MS run. Precursors with S/N > 50 were selected to identify proteins that were not identified in the first MS/MS analysis.
4.7. Proteomics Data Analysis
Protein identification was carried out by ProteinPilot 4.5 (Sciex) by searching in the UniProtKB/Swiss-Prot database 2017_02 (http://www.expasy.ch/sprot; 553,655 sequences). Alternatively, hits were contrasted with UniProt_all database for the estimation of contaminating events. Search parameters included SILAC labeling, IAA alkylation, in-gel trypsin digestion and urea denaturalization as specific factors.
Proteomic data analysis was performed on four different OA biological replicates and two technical replicates by isotope switching. Protein identification and quantification were carried out using ProteinPilot™ software v.4.5 (Sciex). Each MS/MS spectrum was searched in the Uniprot/Swissprot database (UniProt 2017_02 release version containing 553,655, with taxonomy restriction_Homo sapiens), using the Paragon Algorithm. The following ProteinPilot search parameters were used: sample type set as SILAC (Lys +6, Arg +10), oxidation of methionine residue as variable modification, iodoacetamide alkylation of cysteine residue as a fixed modification, in-gel trypsin digestion and urea denaturalization as specific factors and a maximum of one missed cleavage allowed for trypsin. Proteins identified with ≥2 distinct peptides with 95% confidence and a ProtScore ≥1.3 were considered for relative quantification. Proteins identified with one single peptide were manually inspected. Quantified proteins were considered significant when protein ratios ≥ 1.2 or ≤0.83, and p-value < 0.05. Data were also normalized for loading error by bias correction. Searches against a concatenated database containing both forward and reversed sequences allowed the false discovery rate to be kept at 1%. Common contaminants, such as albumin, were excluded from the analysis. The results obtained from ProteinPilot were exported to Microsoft Excel for further analyses. Gene ontology enrichment analysis on the set of detected and modulated proteins was performed, using PANTHER and STRING databases, respectively.
4.8. ELISA and Multiplex Analysis
Isolated AC and SF were plated into 12-well transwells (Corning): AC were seeded into 12-well plates (2 × 105 AC per well) and SF into 12-well inserts (4 × 104 SF per insert). When 80% confluence was reached, the inserts with adherent SF were placed onto the corresponding 12-well plates with AC and incubated in a serum-free medium with or without 10 nM 45 kDa Fn-fs (Merck) in the presence or absence of 10 nM VIP (Bachem) for 48 h. Secretomes were collected from the cultures and selected proteins were measured, using commercial ELISA kits for CHI3L1, C1r, DCN (Merck), and CTSB (Vitro S.A., Madrid, Spain), and a 3-Plex Multiplex for human MMP-2, PTX-3 and C3 (Merck), following manufacturer’s instructions (Figure 8b). AC and SF were also plated separately into 12-well plates and inserts, respectively, for comparisons between the secretome of each cell type and the co-culture under basal conditions (Figure 8a). In parallel, isolated cultures of SF and AC were also performed in 12-well plates (2 × 105 cells per well) with the same treatments described above, for analysis by ELISA and Multiplex (Figure 8c).
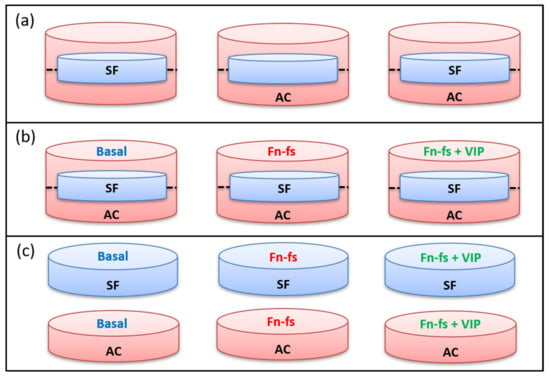
Figure 8.
Experimental design of SF and AC co-cultures and isolated cultures for ELISA and Multiplex analysis. (a) Basal conditions in the SF-AC transwells. (b) Treatments of SF-AC co-cultures. (c) Treatments of SF and AC isolated cultures. SF, synovial fibroblasts; AC, articular chondrocytes; Fn-fs, fibronectin fragments; VIP, vasoactive intestinal peptide.
A schematic representation of the whole experimental design is shown in Figure 9.
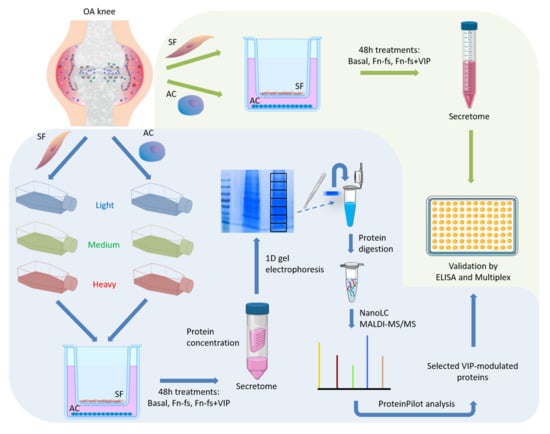
Figure 9.
Schematic representation of the experimental design.
4.9. Statistical Analysis
In the proteomic analysis, normalization tools and the statistical package from Protein Pilot software were employed. In the ELISAS and Multiplex, statistical analysis was performed, using GraphPad Prism software version 8 (GraphPad Inc, San Diego, CA, USA). Data were subjected to a normality test (Kolmogórov–Smirnov test) and equal variance test (F-test). Differences were assessed using Student’s two-tailed t test or Mann–Whitney test for two group comparisons. For comparisons between more than two groups, One-way analysis of variance (ANOVA), Brown–Forsythe or Kruskal–Wallis tests were performed, with Turkey, Games–Howell or Dunn’s multiple comparisons post hoc tests, respectively. Results are presented as mean ± SEM. The p-values < 0.05 were considered statistically significant.
5. Conclusions
The present study shows an analysis of the molecules released by SF and AC in the OA knee joint, corroborating the importance of the presence of both cell populations in the initiation and progression of the disease. In addition, our results support the anti-inflammatory and anti-catabolic properties of VIP in rheumatic diseases. VIP-modulated proteins reported in this study would provide potential new targets for OA treatment.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/article/10.3390/ijms22126441/s1.
Author Contributions
Conceptualization, S.P.-G., V.C. and R.P.G.; methodology, S.P.-G., V.C. and R.P.G.; software, S.P.-G. and V.C.; validation, V.C.; formal analysis, S.P.-G., V.C., R.V.-R. and D.C.; investigation, S.P.-G., V.C., R.P.G.; resources, T.H.-G.; data curation, S.P.-G. and V.C.; writing—original draft preparation, S.P.-G., V.C. and R.P.G.; writing—review and editing, T.H.-G., I.G.-C., M.C., R.V.-R., D.C., C.M., Y.J.; supervision, R.P.G., Y.J. and F.J.B.; project administration, Y.J., F.J.B. and R.P.G.; funding acquisition, Y.J., F.J.B. and R.P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, grant numbers PI17/00027, PI20/00078, RD16/0012/0008 and RD16/0012/0002, co-financed by Fondo Europeo de Desarrollo Regional (FEDER).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Comité Autonómico de Ética da Investigación de Galicia (CAEIG) (protocol code 2013/107) that has been inscribed in the National Registry of Biobanks (Registro Nacional de Biobancos), Collections Section code: C.0000424.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the lack of an existing repository.
Acknowledgments
We are grateful to all patients and the collaborating clinicians for their participation in this study. We are grateful to the personnel of the INIBIC Proteomics Platform (PBR3-ProteoRed/ISCIII).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| AC | Articular chondrocytes |
| ADAMTS | A disintegrin and metalloproteinase with thrombospondin motifs |
| C1q | Complement C1q subcomponent |
| C1r | Complement C1r subcomponent |
| C1s | Complement C1s subcomponent |
| C3 | Complement C3 |
| CF | Complement factor |
| CHI3L1 | Chitinase-3-like protein 1 |
| CTSB | Cathepsin B |
| DCN | Decorin |
| ECM | Extracellular cellular matrix |
| Fn | Fibronectin |
| Fn-fs | Fibronectin fragments |
| GPCRs | G protein-coupled receptors |
| MMP | Matrix metalloproteinase |
| OA | Osteoarthritis |
| PTX3 | Pentraxin-related protein 3 |
| QSOX1 | Sulfhydryl oxidase 1 |
| RA | Rheumatoid arthritis |
| SF | Synovial fibroblasts |
| SILAC | Stable isotope labeling with amino acids |
| uPA | Urokinase-type plasminogen activator |
| VIP | Vasoactive intestinal peptide |
References
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef]
- Abramoff, B.; Caldera, F.E. Osteoarthritis: Pathology, Diagnosis, and Treatment Options. Med. Clin. N. Am. 2020, 104, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Belluzzi, E.; Olivotto, E.; Toso, G.; Cigolotti, A.; Pozzuoli, A.; Biz, C.; Trisolino, G.; Ruggieri, P.; Grigolo, B.; Ramonda, R.; et al. Conditioned media from human osteoarthritic synovium induces inflammation in a synoviocyte cell line. Connect. Tissue Res. 2018, 60, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; FitzGerald, U.; Murphy, J.M. Interplay of Inflammatory Mediators with Epigenetics and Cartilage Modifi-cations in Osteoarthritis. Front Bioeng. Biotechnol. 2018, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, J.W.; Berenbaum, F.; Lafeber, F.P. Osteoarthritis: An update with relevance for clinical practice. Lancet 2011, 377, 2115–2126. [Google Scholar] [CrossRef]
- Monfort, J. Artrosis: Fisiopatología, Diagnóstico y Tratamiento; Médica Panamericana: Madrid, Spain, 2010. [Google Scholar]
- Pérez-García, S.; Gutiérrez-Cañas, I.; Seoane, I.V.; Fernández, J.; Mellado, M.; Leceta, J.; Tío, L.; Romero, R.V.; Juarranz, Y.; Gomariz, R.P. Healthy and Osteoarthritic Synovial Fibroblasts Produce a Disintegrin and Metalloproteinase with Thrombospondin Motifs 4, 5, 7, and 12. Am. J. Pathol. 2016, 186, 2449–2461. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, S.; Carrión, M.; Jimeno, R.; Ortiz, A.M.; González-Álvaro, I.; Fernández, J.; Gomariz, R.P.; Juarranz, Y. Urokinase Plasminogen Activator System in Synovial Fibroblasts from Osteoarthritis Patients: Modulation by Inflammatory Mediators and Neuropeptides. J. Mol. Neurosci. 2013, 52, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, S.; Carrion, M.; Villanueva-Romero, R.; Hermida-Gomez, T.; Fernandez-Moreno, M.; Mellado, M.; Blanco, F.J.; Juarranz, Y.; Gomariz, R.P. Wnt and RUNX2 mediate cartilage breakdown by osteoarthritis synovial fibro-blast-derived ADAMTS-7 and -12. J. Cell Mol. Med. 2019, 23, 3974–3983. [Google Scholar] [CrossRef]
- Lefevre, S.; Meier, F.; Neumann, E.; Muller-Ladner, U. Role of Synovial Fibroblasts in Rheumatoid Arthritis. Curr. Pharm. Des. 2014, 21, 130–141. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123 Pt 24, 4195–4200. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Manou, D.; Karamanos, N.K. The extracellular matrix as a multitasking player in disease. FEBS J. 2019, 286, 2830–2869. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Takai, K.; Weaver, V.M.; Werb, Z. Extracellular Matrix Degradation and Remodeling in Development and Disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a005058. [Google Scholar] [CrossRef]
- Geurts, J.; Jurić, D.; Müller, M.; Schären, S.; Netzer, C. Novel Ex Vivo Human Osteochondral Explant Model of Knee and Spine Osteoarthritis Enables Assessment of Inflammatory and Drug Treatment Responses. Int. J. Mol. Sci. 2018, 19, 1314. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Sun, H.; Bunpetch, V.; Koh, Y.; Wen, Y.; Wu, D.; Ouyang, H. The regulation of cartilage extracellular matrix homeostasis in joint cartilage degeneration and regeneration. Biomaterials 2021, 268, 120555. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T. Cartilage destruction by matrix degradation products. Mod. Rheumatol. 2006, 16, 197–205. [Google Scholar] [CrossRef]
- Kapoor, M.; Martel-Pelletier, J.; Lajeunesse, D.; Pelletier, J.-P.; Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 2010, 7, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, S.; Carrion, M.; Gutierrez-Canas, I.; Villanueva-Romero, R.; Castro, D.; Martinez, C.; Gonzalez-Alvaro, I.; Blanco, F.J.; Juarranz, Y.; Gomariz, R.P. Profile of Matrix-Remodeling Proteinases in Osteoarthritis: Impact of Fibron-ectin. Cells 2019, 9, 40. [Google Scholar] [CrossRef]
- Grässel, S.; Muschter, D. Do Neuroendocrine Peptides and Their Receptors Qualify as Novel Therapeutic Targets in Osteoarthritis? Int. J. Mol. Sci. 2018, 19, 367. [Google Scholar] [CrossRef]
- Gomariz, R.P.; Juarranz, Y.; Carrión, M.; Pérez-García, S.; Romero, R.V.; González-Álvaro, I.; Gutiérrez-Cañas, I.; Lamana, A.; Martínez, C. An Overview of VPAC Receptors in Rheumatoid Arthritis: Biological Role and Clinical Significance. Front. Endocrinol. 2019, 10, 729. [Google Scholar] [CrossRef]
- Martínez, C.; Juarranz, Y.; Gutiérrez-Cañas, I.; Carrión, M.; Pérez-García, S.; Villanueva-Romero, R.; Castro, D.; Lamana, A.; Mellado, M.; González-Álvaro, I.; et al. A Clinical Approach for the Use of VIP Axis in Inflammatory and Autoimmune Diseases. Int. J. Mol. Sci. 2019, 21, 65. [Google Scholar] [CrossRef]
- Juarranz, Y.; Gutiérrez-Cañas, I.; Santiago, B.; Carrión, M.; Pablos, J.L.; Gomariz, R.P. Differential expression of vaso-active intestinal peptide and its functional receptors in human osteoarthritic and rheumatoid synovial fibroblasts. Arthritis Rheum 2008, 58, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Wang, H.; Li, Y.-S.; Luo, W. Role of vasoactive intestinal peptide in osteoarthritis. J. Biomed. Sci. 2016, 23, 63. [Google Scholar] [CrossRef]
- Jiang, W.; Gao, S.G.; Chen, X.G.; Xu, X.C.; Xu, M.; Luo, W.; Tu, M.; Zhang, F.J.; Zeng, C.; Lei, G.H. Expression of synovial fluid and articular cartilage VIP in human osteoarthritic knee: A new indicator of disease severity? Clin. Biochem. 2012, 45, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Calamia, V.; Lourido, L.; Fernández-Puente, P.; Mateos, J.; Rocha, B.; Montell, E.; Vergés, J.; Ruiz-Romero, C.; Blanco, F.J. Secretome analysis of chondroitin sulfate-treated chondrocytes reveals anti-angiogenic, anti-inflammatory and anti-catabolic properties. Arthritis Res. Ther. 2012, 14, R202. [Google Scholar] [CrossRef]
- Li, X.J.; Xu, M.; Zhao, X.Q.; Zhao, J.N.; Chen, F.F.; Yu, W.; Gao, D.Y.; Luo, B. Proteomic analysis of synovial fibro-blast-like synoviocytes from rheumatoid arthritis. Clini. Exp. Rheumatol. 2013, 31, 552–558. [Google Scholar]
- Sanchez, C.; Mazzucchelli, G.; Lambert, C.; Comblain, F.; Depauw, E.; Henrotin, Y. Comparison of secretome from osteoblasts derived from sclerotic versus non-sclerotic subchondral bone in OA: A pilot study. PLoS ONE 2018, 13, e0194591. [Google Scholar] [CrossRef]
- Klocke, R.; Levasseur, K.; Kitas, G.D.; Smith, J.P.; Hirsch, G. Cartilage turnover and intra-articular corticosteroid injec-tions in knee osteoarthritis. Rheumatol. Int. 2018, 38, 455–459. [Google Scholar] [CrossRef]
- Johansen, J.S.; Pedersen, A.N.; Schroll, M.; Jørgensen, T.; Pedersen, B.K.; Bruunsgaard, H. High serum YKL-40 level in a cohort of octogenarians is associated with increased risk of all-cause mortality. Clin. Exp. Immunol. 2007, 151, 260–266. [Google Scholar] [CrossRef]
- Recklies, A.D.; White, C.; Ling, H. The chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways. Biochem. J. 2002, 365, 119–126. [Google Scholar] [CrossRef]
- Kzhyshkowska, J.; Gratchev, A.; Goerdt, S. Human Chitinases and Chitinase-Like Proteins as Indicators for Inflammation and Cancer. Biomark. Insights 2007, 2, 128–146. [Google Scholar] [CrossRef]
- Mylin, A.K.; Abildgaard, N.; Johansen, J.S.; Heickendorff, L.; Kreiner, S.; Waage, A.; Turesson, I.; Gimsing, P.; Nordic Myeloma Study Group. Serum YKL-40: A new independent prognostic marker for skeletal complications in patients with multiple myeloma. Leuk. Lymphoma 2015, 56, 2650–2659. [Google Scholar] [CrossRef]
- Lee, C.G.; Da Silva, C.A.; Cruz, C.S.D.; Ahangari, F.; Ma, B.; Kang, M.-J.; He, C.-H.; Takyar, S.; Elias, J.A. Role of Chitin and Chitinase/Chitinase-Like Proteins in Inflammation, Tissue Remodeling, and Injury. Annu. Rev. Physiol. 2011, 73, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Lotz, M.J.; Martelpelletier, J.; Christiansen, C.F.; Brandi, M.L.; Bruyere, O.; Chapurlat, R.; Collette, J.; Cooper, C.L.; Giacovelli, G.; Kanis, J.A.; et al. Republished: Value of biomarkers in osteoarthritis: Current status and perspectives. Postgrad. Med. J. 2014, 90, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Väänänen, T.; Vuolteenaho, K.; Kautiainen, H.; Nieminen, R.; Mottonen, T.; Hannonen, P.; Korpela, M.; Kauppi, M.J.; Laiho, K.; Kaipiainen-Seppänen, O.; et al. Glycoprotein YKL-40: A potential biomarker of disease activity in rheumatoid arthritis during intensive treatment with csDMARDs and infliximab. Evidence from the randomised controlled NEO-RACo trial. PLoS ONE 2017, 12, e0183294. [Google Scholar] [CrossRef] [PubMed]
- Lafeber, F.; van Spil, W. Osteoarthritis year 2013 in review: Biomarkers; reflecting before moving forward, one step at a time. Osteoarthr. Cartil. 2013, 21, 1452–1464. [Google Scholar] [CrossRef]
- Živanović, S.; Rackov, L.P.; Vojvodic, D.; Vučetić, D. Human cartilage glycoprotein 39—Biomarker of joint damage in knee osteoarthritis. Int. Orthop. 2009, 33, 1165–1170. [Google Scholar] [CrossRef]
- Goldring, M.B.; Marcu, K.B. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res. Ther. 2009, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Otero, M.; Goldring, M.B. Cells of the synovium in rheumatoid arthritis. Chondrocytes. Arthritis Res. Ther. 2007, 9, 220. [Google Scholar] [CrossRef]
- Guan, J.; Zhang, L.; Liu, Z.; Feng, J.-S.; Wang, H.-J.; Chu, J.-G.; Song, Y.-Z.; Xie, L.; Ding, L.-B. Increased Synovial Fluid YKL-40 Levels are Linked with Symptomatic Severity in Knee Osteoarthritis Patients. Clin. Lab. 2015, 61, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Conrozier, T.; Carlier, M.-C.; Mathieu, P.; Colson, F.; Debard, A.L.; Richard, S.; Favret, H.; Bienvenu, J.; Vignon, E. Serum levels of YKL-40 and C reactive protein in patients with hip osteoarthritis and healthy subjects: A cross sectional study. Ann. Rheum. Dis. 2000, 59, 828–831. [Google Scholar] [CrossRef]
- Väänänen, T.; Koskinen, A.; Paukkeri, E.-L.; Hämäläinen, M.; Moilanen, T.; Moilanen, E.; Vuolteenaho, K. YKL-40 as a Novel Factor Associated with Inflammation and Catabolic Mechanisms in Osteoarthritic Joints. Mediat. Inflamm. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Salomon, J.; Matusiak, L.; Nowicka-Suszko, D.; Szepietowski, J.C. Chitinase-3-Like Protein 1 (YKL-40) Is a New Bi-omarker of Inflammation in Psoriasis. Mediat. Inflamm. 2017, 2017, 9538451. [Google Scholar] [CrossRef] [PubMed]
- Salomon, J.; Matusiak, Ł.; Nowicka-Suszko, D.; Szepietowski, J.C. Chitinase-3-like protein 1 (YKL-40) is a biomarker of severity of joint involvement in psoriatic arthritis. Adv. Dermatol. Allergol. 2018, 35, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Khan, R.; Gupta, N.; Zaheer, M.; Abbas, M.; Khan, S. Acute phase reactant, Pentraxin 3, as a novel marker for the diagnosis of rheumatoid arthritis. Clin. Chim. Acta 2018, 480, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Scimeca, M.; Salustri, A.; Bonanno, E.; Nardozi, D.; Rao, C.; Piccirilli, E.; Feola, M.; Tancredi, V.; Rinaldi, A.; Iolascon, G.; et al. Impairment of PTX3 expression in osteoblasts: A key element for osteoporosis. Cell Death Dis. 2017, 8, e3125. [Google Scholar] [CrossRef] [PubMed]
- Satomura, K.; Torigoshi, T.; Koga, T.; Maeda, Y.; Izumi, Y.; Jiuchi, Y.; Miyashita, T.; Yamasaki, S.; Kawakami, A.; Aiba, Y.; et al. Serum amyloid A (SAA) induces pentraxin 3 (PTX3) production in rheumatoid synoviocytes. Mod. Rheumatol. 2013, 23, 28–35. [Google Scholar] [CrossRef]
- Luchetti, M.M.; Piccinini, G.; Mantovani, A.; Peri, G.; Matteucci, C.; Pomponio, G.; Fratini, M.; Fraticelli, P.; Sambo, P.; di Loreto, C.; et al. Expression and production of the long pentraxin PTX3 in rheumatoid arthritis (RA). Clin. Exp. Immunol. 2000, 119, 196–202. [Google Scholar] [CrossRef]
- Di Rosa, M.; Szychlinska, M.A.; Tibullo, D.; Malaguarnera, L.; Musumeci, G. Expression of CHI3L1 and CHIT1 in os-teoarthritic rat cartilage model. A morphological study. Eur. J. Histochem. 2014, 58, 2423. [Google Scholar] [CrossRef]
- Gomariz, R.P.; Martinez, C.; Abad, C.; Leceta, J.; Delgado, M. Immunology of VIP: A review and therapeutical per-spectives. Curr. Pharm. Des. 2001, 7, 89–111. [Google Scholar] [CrossRef]
- Gomariz, R.P.; Juarranz, Y.; Abad, C.; Arranz, A.; Leceta, J.; Martinez, C. VIP-PACAP System in Immunity: New Insights for Multitarget Therapy. Ann. N. Y. Acad. Sci. 2006, 1070, 51–74. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.; Ganea, D. Vasoactive intestinal peptide: A neuropeptide with pleiotropic immune functions. Amino Acids 2013, 45, 25–39. [Google Scholar] [CrossRef]
- Galindo-Izquierdo, M.; Alvarez, J.L.P. Complement as a Therapeutic Target in Systemic Autoimmune Diseases. Cells 2021, 10, 148. [Google Scholar] [CrossRef]
- Silawal, S.; Triebel, J.; Bertsch, T.; Schulze-Tanzil, G. Osteoarthritis and the Complement Cascade. Clin. Med. Insights Arthritis Musculoskelet. Disord. 2018, 11, 1179544117751430. [Google Scholar] [CrossRef]
- Happonen, K.E.; Saxne, T.; Aspberg, A.; Morgelin, M.; Heinegard, D.; Blom, A.M. Regulation of complement by cartilage oligomeric matrix protein allows for a novel molecular diagnostic principle in rheumatoid arthritis. Arthritis Rheum. 2010, 62, 3574–3583. [Google Scholar] [CrossRef] [PubMed]
- Happonen, K.E.; Saxne, T.; Geborek, P.; Andersson, M.; Bengtsson, A.A.; Hesselstrand, R.; Heinegård, D.; Blom, A.M. Serum COMP-C3b complexes in rheumatic diseases and relation to anti-TNF-α treatment. Arthritis Res. Ther. 2012, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- Melin Furst, C.; Morgelin, M.; Vadstrup, K.; Heinegard, D.; Aspberg, A.; Blom, A.M. The C-type lectin of the aggrecan G3 domain activates complement. PLoS ONE 2013, 8, e61407. [Google Scholar] [CrossRef] [PubMed]
- Sjöberg, A.P.; Manderson, G.A.; Mörgelin, M.; Day, A.J.; Heinegård, D.; Blom, A.M. Short leucine-rich glycoproteins of the extracellular matrix display diverse patterns of complement interaction and activation. Mol. Immunol. 2009, 46, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Barilla, M.-L.; Carsons, S.E. Fibronectin fragments and their role in inflammatory arthritis. Semin. Arthritis Rheum. 2000, 29, 252–265. [Google Scholar] [CrossRef]
- Carsons, S.E.; Schwartzman, S.; Diamond, H.S.; Berkowitz, E. Interaction between fibronectin and C1q in rheumatoid synovial fluid and normal plasma. Clin. Exp. Immunol. 1988, 72, 37–42. [Google Scholar]
- Assirelli, E.; Pulsatelli, L.; Dolzani, P.; Mariani, E.; Lisignoli, G.; Addimanda, O.; Meliconi, R. Complement Expression and Activation in Osteoarthritis Joint Compartments. Front. Immunol. 2020, 11, 535010. [Google Scholar] [CrossRef]
- Gulati, P.; Lemercier, C.; Lappin, D.; Whaley, K.; Gue, D. Expression of the components and regulatory proteins of the classical pathway of complement in normal and diseased synovium. Rheumatol. Int. 1994, 14, 13–19. [Google Scholar] [CrossRef]
- Guc, D.; Gulati, P.; Lemercier, C.; Lappin, D.; Birnie, G.D.; Whaley, K. Expression of the components and regulatory proteins of the alternative complement pathway and the membrane attack complex in normal and diseased synovium. Rheumatol. Int. 1993, 13, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Bradley, K.; North, J.; Saunders, D.; Schwaeble, W.; Jeziorska, M.; Woolley, D.E.; Whaley, K. Synthesis of classical pathway complement components by chondrocytes. Immunology 1996, 88, 648–656. [Google Scholar] [PubMed]
- Lubbers, R.; van Schaarenburg, R.A.; Kwekkeboom, J.C.; Levarht, N.E.; Bakker, A.M.; Mahdad, R.; Monteagudo, S.; Cherifi, C.; Lories, R.J.; Toes, R.E.; et al. Complement component C1q is produced by isolated articular chondrocytes. Osteoarthr. Cartil. 2020, 28, 675–684. [Google Scholar] [CrossRef]
- Vicenti, G.; Bizzoca, D.; Carrozzo, M.; Solarino, G.; Moretti, B. Multi-omics analysis of synovial fluid: A promising ap-proach in the study of osteoarthritis. J. Biol. Regul. Homeost. Agents 2018, 32 (Suppl. S1), 9–13. [Google Scholar]
- Struglics, A.; Okroj, M.; Sward, P.; Frobell, R.; Saxne, T.; Lohmander, L.S.; Blom, A.M. The complement system is activated in synovial fluid from subjects with knee injury and from patients with osteoarthritis. Arthritis Res. Ther. 2016, 18, 223. [Google Scholar] [CrossRef]
- Rossi, V.; Bally, I.; Lacroix, M.; Arlaud, G.J.; Thielens, N.M. Classical Complement Pathway Components C1r and C1s: Purification from Human Serum and in Recombinant Form and Functional Characterization. Adv. Struct. Saf. Stud. 2013, 1100, 43–60. [Google Scholar] [CrossRef]
- Liao, W.; Li, Z.; Li, T.; Zhang, Q.; Zhang, H.; Wang, X. Proteomic analysis of synovial fluid in osteoarthritis using SWATH-mass spectrometry. Mol. Med. Rep. 2017, 17, 2827–2836. [Google Scholar] [CrossRef]
- Laurell, A.-B.; Mårtensson, U.; Sjöholm, A. Trimer and tetramer complexes containing C1 esterase inhibitor, C1r and C1s, in serum and synovial fluid of patients with rheumatic disease. J. Immunol. Methods 1990, 129, 55–61. [Google Scholar] [CrossRef]
- Sjöholm, A.G.; Berglund, K.; Johnson, U.; Laurell, A.-B.; Sturfelt, G. C1 Activation, with C1q in Excess of Functional C1 in Synovial Fluid from Patients with Rheumatoid Arthritis. Int. Arch. Allergy Immunol. 1986, 79, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, N.-E.; Fiedler, J.; Schrezenmeier, H.; Huber-Lang, M.; Brenner, R.E. Crucial Role of IL1beta and C3a in the In Vitro-Response of Multipotent Mesenchymal Stromal Cells to Inflammatory Mediators of Polytrauma. PLoS ONE 2015, 10, e0116772. [Google Scholar] [CrossRef]
- Ourradi, K.; Xu, Y.; De Seny, M.; Kirwan, J.; Blom, A.; Sharif, M. Development and validation of novel biomarker assays for osteoarthritis. PLoS ONE 2017, 12, e0181334. [Google Scholar] [CrossRef]
- de Seny, D.; Sharif, M.; Fillet, M.; Cobraiville, G.; Meuwis, M.A.; Maree, R.; Hauzeur, J.P.; Wehenkel, L.; Louis, E.; Merville, M.P.; et al. Discovery and biochemical characterisation of four novel biomarkers for osteoarthritis. Ann. Rheum. Dis. 2011, 70, 1144–1152. [Google Scholar] [CrossRef]
- Corigliano, A.; Preianò, M.; Terracciano, R.; Savino, R.; De Gori, M.; Galasso, O.; Gasparini, G. C3f is a potential tool for the staging of osteoarthritis. J. Boil. Regul. Homeost. Agents 2017, 31, 29–35. [Google Scholar]
- Cobraiville, G.; Fillet, M.; Sharif, M.; Ourradi, K.; Nys, G.; Malaise, M.G.; de Seny, D. Validation of a new method by nano-liquid chromatography on chip tandem mass spectrometry for combined quantitation of C3f and the V65 vitron-ectin fragment as biomarkers of diagnosis and severity of osteoarthritis. Talanta 2017, 169, 170–180. [Google Scholar] [CrossRef]
- Cheng, T.H.; Yoon, S.H.; Lee, P.; Dimaculangan, D.; Vikram Maheshwari, A.; Zhang, M. Knee synovial fluid com-plement C3-beta chain levels correlate with clinical symptoms of knee osteoarthritis. Int. J. Rheum. Dis. 2020, 23, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Thordardottir, S.; Vikingsdottir, T.; Bjarnadottir, H.; Jonsson, H., Jr.; Gudbjornsson, B. Activation of Complement Fol-lowing Total Hip Replacement. Scand. J. Immunol. 2016, 83, 219–224. [Google Scholar] [CrossRef]
- Abbink, J.J.; Kamp, A.M.; Nuijens, J.H.; Erenberg, A.J.; Swaak, A.J.; Hack, C.E. Relative contribution of contact and complement activation to inflammatory reactions in arthritic joints. Ann. Rheum. Dis. 1992, 51, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Gronau, T.; Kruger, K.; Prein, C.; Aszodi, A.; Gronau, I.; Iozzo, R.V.; Mooren, F.C.; Clausen-Schaumann, H.; Bertrand, J.; Pap, T.; et al. Forced exercise-induced osteoarthritis is attenuated in mice lacking the small leucine-rich proteoglycan decorin. Ann. Rheum. Dis. 2017, 76, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Guilak, F.; Nims, R.J.; Dicks, A.; Wu, C.-L.; Meulenbelt, I. Osteoarthritis as a disease of the cartilage pericellular matrix. Matrix Biol. 2018, 71–72, 40–50. [Google Scholar] [CrossRef]
- Cillero-Pastor, B.; Eijkel, G.B.; Kiss, A.; Blanco, F.J.; Heeren, R.M.A. Matrix-assisted laser desorption ionization-imaging mass spectrometry: A new methodology to study human osteoarthritic cartilage. Arthritis Rheum. 2013, 65, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Wiberg, C.; Klatt, A.R.; Wagener, R.; Paulsson, M.; Bateman, J.F.; Heinegård, D.; Mörgelin, M. Complexes of Matrilin-1 and Biglycan or Decorin Connect Collagen VI Microfibrils to Both Collagen II and Aggrecan. J. Biol. Chem. 2003, 278, 37698–37704. [Google Scholar] [CrossRef] [PubMed]
- Redini, F. Structure and regulation of articular cartilage proteoglycan expression. Pathol. Biol. 2001, 49, 364–375. [Google Scholar] [CrossRef]
- Knudson, C.B.; Knudson, W. Cartilage proteoglycans. Semin. Cell Dev. Biol. 2001, 12, 69–78. [Google Scholar] [CrossRef]
- Zaia, J.; Liu, B.; Boynton, R.; Barry, F. Structural analysis of cartilage proteoglycans and glycoproteins using ma-trix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 2000, 277, 94–103. [Google Scholar] [CrossRef]
- Tesche, F.; Miosge, N. New aspects of the pathogenesis of osteoarthritis: The role of fibroblast-like chondrocytes in late stages of the disease. Histol. Histopathol. 2005, 20, 329–337. [Google Scholar]
- Barreto, G.; Soininen, A.; Ylinen, P.; Sandelin, J.; Konttinen, Y.T.; Nordström, D.C.; Eklund, K.K. Soluble biglycan: A potential mediator of cartilage degradation in osteoarthritis. Arthritis Res. 2015, 17, 379. [Google Scholar] [CrossRef]
- Bock, H.; Michaeli, P.; Bode, C.; Schultz, W.; Kresse, H.; Herken, R.; Miosge, N. The small proteoglycans decorin and biglycan in human articular cartilage of late-stage osteoarthritis. Osteoarthr. Cartil. 2001, 9, 654–663. [Google Scholar] [CrossRef]
- Melrose, J.; Fuller, E.S.; Roughley, P.J.; Smith, M.M.; Kerr, B.; Hughes, C.E.; Caterson, B.; Little, C.B. Fragmentation of decorin, biglycan, lumican and keratocan is elevated in degenerate human meniscus, knee and hip articular cartilages compared with age-matched macroscopically normal and control tissues. Arthritis Res. Ther. 2008, 10, R79. [Google Scholar] [CrossRef] [PubMed]
- Young, A.A.; Smith, M.M.; Smith, S.M.; Cake, M.A.; Ghosh, P.; Read, R.A.; Melrose, J.; Sonnabend, D.H.; Roughley, P.J.; Little, C.B. Regional assessment of articular cartilage gene expression and small proteoglycan metabolism in an animal model of osteoarthritis. Arthritis Res. 2005, 7, R852–R861. [Google Scholar] [CrossRef]
- Arranz, A.; Gutiérrez-Cañas, I.; Carrión, M.; Juarranz, Y.; Pablos, J.L.; Martínez, C.; Gomariz, R.P. VIP reverses the expression profiling of TLR4-stimulated signaling pathway in rheumatoid arthritis synovial fibroblasts. Mol. Immunol. 2008, 45, 3065–3073. [Google Scholar] [CrossRef] [PubMed]
- Polgár, A.; Falus, A.; Koó, E.; Ujfalussy, I.; Seszták, M.; Szuts, I.; Konrád, K.; Hodinka, L.; Bene, E.; Mészáros, G.; et al. Elevated levels of synovial fluid antibodies reactive with the small proteoglycans biglycan and decorin in patients with rheumatoid arthritis or other joint diseases. Rheumatology 2003, 42, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Anders, H.J.; Schaefer, L. Beyond tissue injury-damage-associated molecular patterns, toll-like receptors, and inflam-masomes also drive regeneration and fibrosis. J. Am. Soc. Nephrol. 2014, 25, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Muneta, T.; Morito, T.; Mochizuki, T.; Sekiya, I. Autologous synovial fluid enhances migration of mesenchymal stem cells from synovium of osteoarthritis patients in tissue culture system. J. Orthop. Res. 2008, 26, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Rengel, Y.; Ospelt, C.; Gay, S. Proteinases in the joint: Clinical relevance of proteinases in joint destruction. Arthritis Res. Ther. 2007, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2012, 1824, 133–145. [Google Scholar] [CrossRef]
- Martel-Pelletier, J.; Welsch, D.J.; Pelletier, J.-P. Metalloproteases and inhibitors in arthritic diseases. Best Pr. Res. Clin. Rheumatol. 2001, 15, 805–829. [Google Scholar] [CrossRef]
- Ruettger, A.; Schueler, S.; Mollenhauer, J.A.; Wiederanders, B. Cathepsins B, K, and L are regulated by a defined collagen type II peptide via activation of classical protein kinase C and p38 MAP kinase in articular chondrocytes. J. Biol. Chem. 2008, 283, 1043–1051. [Google Scholar] [CrossRef]
- Požgan, U.; Caglič, D.; Rozman, B.; Nagase, H.; Turk, V.; Turk, B. Expression and activity profiling of selected cysteine cathepsins and matrix metalloproteinases in synovial fluids from patients with rheumatoid arthritis and osteoarthritis. Biol. Chem. 2010, 391, 571–579. [Google Scholar] [CrossRef]
- Clutterbuck, A.L.; Asplin, K.E.; Harris, P.; Allaway, D.; Mobasheri, A. Targeting matrix metalloproteinases in inflammatory conditions. Curr. Drug Targets 2009, 10, 1245–1254. [Google Scholar] [CrossRef]
- Lang, A.; Hörler, D.; Baici, A. The relative importance of cysteine peptidases in osteoarthritis. J. Rheumatol. 2000, 27, 1970–1979. [Google Scholar]
- Roughley, P.J. The degradation of cartilage proteoglycans by tissue proteinases. Proteoglycan heterogeneity and the pathway of proteolytic degradation. Biochem. J. 1977, 167, 639–646. [Google Scholar] [CrossRef]
- Maciewicz, R.A.; Wotton, S.F.; Etherington, D.J.; Duance, V.C. Susceptibility of the cartilage collagens types II, IX and XI to degradation by the cysteine proteinases, cathepsins B and L. FEBS Lett. 1990, 269, 189–193. [Google Scholar] [CrossRef]
- Kostoulas, G.; Lang, A.; Nagase, H.; Baici, A. Stimulation of angiogenesis through cathepsin B inactivation of the tissue inhibitors of matrix metalloproteinases. FEBS Lett. 1999, 455, 286–290. [Google Scholar] [CrossRef]
- Baici, A.; Lang, A.; Zwicky, R.; Müntener, K. Cathepsin B in Osteoarthritis: Uncontrolled Proteolysis in the Wrong Place. Semin. Arthritis Rheum. 2004, 34, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Zwicky, R.; Baici, A. Cytoskeletal architecture and cathepsin B trafficking in human articular chondrocytes. Histochem. Cell Biol. 2000, 114, 363–372. [Google Scholar] [CrossRef]
- Batshon, G.; Elayyan, J.; Qiq, O.; Reich, E.; Ben-Aderet, L.; Kandel, L.; Haze, A.; Steinmeyer, J.; Lefebvre, V.; Zhang, H.; et al. Serum NT/CT SIRT1 ratio reflects early osteoarthritis and chondrosenescence. Ann. Rheum. Dis. 2020, 79, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Mishiro, T.; Nakano, S.; Takahara, S.; Miki, M.; Nakamura, Y.; Yasuoka, S.; Nikawa, T.; Yasui, N. Relationship between cathepsin B and thrombin in rheumatoid arthritis. J. Rheumatol. 2004, 31, 1265–1273. [Google Scholar]
- Hashimoto, Y.; Kakegawa, H.; Narita, Y.; Hachiya, Y.; Hayakawa, T.; Kos, J.; Turk, V.; Katunuma, N. Significance of Cathepsin B Accumulation in Synovial Fluid of Rheumatoid Arthritis. Biochem. Biophys. Res. Commun. 2001, 283, 334–339. [Google Scholar] [CrossRef]
- Ikeda, Y.; Ikata, T.; Mishiro, T.; Nakano, S.; Ikebe, M.; Yasuoka, S. Cathepsins B and L in synovial fluids from patients with rheumatoid arthritis and the effect of cathepsin B on the activation of pro-urokinase. J. Med. Investig. 2000, 47, 61–75. [Google Scholar]
- Keyszer, G.; Redlich, A.; Haupl, T.; Zacher, J.; Sparmann, M.; Engethum, U.; Gay, S.; Burmester, G.R. Differential expression of cathepsins B and L compared with matrix metalloproteinases and their respective inhibitors in rheumatoid arthritis and osteoarthritis: A parallel investigation by semiquantitative reverse transcriptase-polymerase chain reaction and immunohistochemistry. Arthritis Rheum. 1998, 41, 1378–1387. [Google Scholar] [PubMed]
- Solau-Gervais, E.; Zerimech, F.; Lemaire, R.; Fontaine, C.; Huet, G.; Flipo, R.M. Cysteine and serine proteases of synovial tissue in rheumatoid arthritis and osteoarthritis. Scand. J. Rheumatol. 2007, 36, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.; Petrow, P.K.; Gaumann, A.; Keyszer, G.M.; Eysel, P.; Eckardt, A.; Bräuer, R.; Kriegsmann, J. Cathepsin B and its endogenous inhibitor cystatin C in rheumatoid arthritis synovium. J. Rheumatol. 2000, 27, 859–865. [Google Scholar]
- Bayliss, M.T.; Ali, S.Y. Studies on cathepsin B in human articular cartilage. Biochem. J. 1978, 171, 149–154. [Google Scholar] [CrossRef]
- Martel-Pelletier, J.; Cloutier, J.M. Cathepsin B and cysteine protease inhibitors in human osteoarthritis. J. Orthop. Res. 1990, 8, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Baici, A.; Horler, D.; Lang, A.; Merlin, C.; Kissling, R. Cathepsin B in osteoarthritis: Zonal variation of enzyme activity in human femoral head cartilage. Ann. Rheum. Dis. 1995, 54, 281–288. [Google Scholar] [CrossRef]
- Baici, A.; Lang, A.; Horler, D.; Kissling, R.; Merlin, C. Cathepsin B in osteoarthritis: Cytochemical and histochemical analysis of human femoral head cartilage. Ann. Rheum. Dis. 1995, 54, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Ben-Aderet, L.; Merquiol, E.; Fahham, D.; Kumar, A.; Reich, E.; Ben-Nun, Y.; Kandel, L.; Haze, A.; Liebergall, M.; Kosinska, M.K.; et al. Detecting cathepsin activity in human osteoar-thritis via activity-based probes. Arthritis Res. Ther. 2015, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Berardi, S.; Lang, A.; Kostoulas, G.; Vilei, E.M.; Baici, A. Alternative messenger RNA splicing and enzyme forms of cathepsin B in human osteoarthritic cartilage and cultured chondrocytes. Arthritis Rheum. 2001, 44, 1819–1831. [Google Scholar] [CrossRef]
- Zwicky, R.; Muntener, K.; Csucs, G.; Goldring, M.B.; Baici, A. Exploring the Role of 5′ Alternative Splicing and of the 3′-Untranslated Region of Cathepsin B mRNA. Biol. Chem. 2003, 384, 1007–1018. [Google Scholar] [CrossRef]
- Murphy, G.; Knauper, V.; Atkinson, S.; Butler, G.; English, W.; Hutton, M.; Stracke, J.; Clark, I. Matrix metalloproteinases in arthritic disease. Arthritis Res. 2002, 4 (Suppl. S3), S39–S49. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Lee, M.H. What are the roles of metalloproteinases in cartilage and bone damage? Ann. Rheum. Dis. 2005, 64 (Suppl. S4), iv44–iv47. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Asp. Med. 2008, 29, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Kevorkian, L.; Young, D.A.; Darrah, C.; Donell, S.T.; Shepstone, L.; Porter, S.; Brockbank, S.M.; Edwards, D.R.; Parker, A.E.; Clark, I.M. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004, 50, 131–141. [Google Scholar] [CrossRef]
- Lipari, L.; Gerbino, A. Expression of Gelatinases (MMP-2, MMP-9) in Human Articular Cartilage. Int. J. Immunopathol. Pharmacol. 2013, 26, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Duerr, S.; Stremme, S.; Soeder, S.; Bau, B.; Aigner, T. MMP-2/gelatinase A is a gene product of human adult articular chondrocytes and is increased in osteoarthritic cartilage. Clin. Exp. Rheumatol. 2004, 22, 603–608. [Google Scholar] [PubMed]
- Hulejova, H.; Baresova, V.; Klezl, Z.; Polanska, M.; Adam, M.; Senolt, L. Increased level of cytokines and matrix metal-loproteinases in osteoarthritic subchondral bone. Cytokine 2007, 38, 151–156. [Google Scholar] [CrossRef]
- Zeng, G.; Chen, A.; Li, W.; Song, J.; Gao, C. High MMP-1, MMP-2, and MMP-9 protein levels in osteoarthritis. Genet. Mol. Res. 2015, 14, 14811–14822. [Google Scholar] [CrossRef]
- Alunno, A.; Falcinelli, E.; Luccioli, F.; Petito, E.; Bartoloni, E.; Momi, S.; Mirabelli, G.; Mancini, G.B.; Gerli, R.; Gresele, P. Platelets Contribute to the Accumulation of Matrix Metalloproteinase Type 2 in Synovial Fluid in Osteoarthritis. Thromb. Haemost. 2017, 117, 2116–2124. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.B.; Setton, L.A.; Bell, R.D.; Easley, M.E.; Huebner, J.L.; Stabler, T.; Kraus, V.B.; Leimer, E.M.; Olson, S.A.; Nettles, D.L. Inflammatory Cytokines and Matrix Metalloproteinases in the Synovial Fluid After Intra-articular Ankle Fracture. Foot Ankle Int. 2015, 36, 1264–1271. [Google Scholar] [CrossRef]
- Sandya, S.; Achan, M.A.; Sudhakaran, P.R. Multiple matrix metalloproteinases in type II collagen induced arthritis. Indian J. Clin. Biochem. 2009, 24, 42–48. [Google Scholar] [CrossRef]
- Yoshida, K.; Takatsuka, S.; Hatada, E.; Nakamura, H.; Tanaka, A.; Ueki, K.; Nakagawa, K.; Okada, Y.; Yamamoto, E.; Fukuda, R. Expression of matrix metalloproteinases and aggrecanase in the synovial fluids of patients with symptomatic temporomandibular disorders. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2006, 102, 22–27. [Google Scholar] [CrossRef]
- Yang, C.-C.; Lin, C.-Y.; Wang, H.-S.; Lyu, S.-R. Matrix Metalloproteases and Tissue Inhibitors of Metalloproteinases in Medial Plica and Pannus-like Tissue Contribute to Knee Osteoarthritis Progression. PLoS ONE 2013, 8, e79662. [Google Scholar] [CrossRef]
- Xue, M.; McKelvey, K.; Shen, K.; Minhas, N.; March, L.; Park, S.-Y.; Jackson, C. Endogenous MMP-9 and not MMP-2 promotes rheumatoid synovial fibroblast survival, inflammation and cartilage degradation. Rheumatology 2014, 53, 2270–2279. [Google Scholar] [CrossRef]
- Hot, A.; Zrioual, S.; Lenief, V.; Miossec, P. IL-17 and tumour necrosis factor α combination induces a HIF-1α-dependent invasive phenotype in synoviocytes. Ann. Rheum. Dis. 2012, 71, 1393–1401. [Google Scholar] [CrossRef]
- Wang, C.; Chi, Q.; Xu, C.; Xu, K.; Zhang, Y.; Liu, Y.; Yang, L.; Sung, K.L. Expression of LOXs and MMP-1, 2, 3 by ACL Fibroblasts and Synoviocytes Impact of Coculture and TNF-α. J. Knee Surg. 2019, 32, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Prasadam, I.; Crawford, R.; Xiao, Y. Aggravation of ADAMTS and matrix metalloproteinase production and role of ERK1/2 pathway in the interaction of osteoarthritic subchondral bone osteoblasts and articular cartilage chondrocytes—Possible pathogenic role in osteoarthritis. J. Rheumatol. 2012, 39, 621–634. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, W.; Xian, G.; Pan, B.; Ye, Y.; Gu, M.; Ma, Y.; Zhang, Z.; Sheng, P. Identification of abnormally methylated-differentially expressed genes and pathways in osteoarthritis: A comprehensive bioinformatic study. Clin. Rheumatol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Wang, C.; Wan, S.; Yao, Z.; Zhang, Y.; Liu, J.; Zhang, C. Identification of Diagnostic Biomarkers of Osteoarthritis Based on Multi-Chip Integrated Analysis and Machine Learning. DNA Cell Biol. 2020, 39, 2245–2256. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.-Y.; Yang, M.; Guo, J.; Zhang, C.; Lin, L.-L.; Liu, Y.; Wei, R.-X. Identification of the Biomarkers and Pathological Process of Osteoarthritis: Weighted Gene Co-expression Network Analysis. Front. Physiol. 2019, 10, 275. [Google Scholar] [CrossRef] [PubMed]
- Werner, N.C.; Stoker, A.M.; Bozynski, C.C.; Keeney, J.A.; Cook, J.L. Characterizing correlations among disease severity measures in osteochondral tissues from osteoarthritic knees. J. Orthop. Res. 2021, 39, 1103–1112. [Google Scholar] [CrossRef]
- Thorson, C.; Galicia, K.; Burleson, A.; Bouchard, O.; Hoppensteadt, D.; Fareed, J.; Hopkinson, W. Matrix Metalloproteinases and Their Inhibitors and Proteoglycan 4 in Patients Undergoing Total Joint Arthroplasty. Clin. Appl. Thromb. Hemost. 2019, 25, 1076029619828113. [Google Scholar] [CrossRef] [PubMed]
- Vacas, E.; Bajo, A.M.; Schally, A.V.; Sánchez-Chapado, M.; Prieto, J.C.; Carmena, M.J. Vasoactive intestinal peptide induces oxidative stress and suppresses metastatic potential in human clear cell renal cell carcinoma. Mol. Cell. Endocrinol. 2013, 365, 212–222. [Google Scholar] [CrossRef] [PubMed]
- García, S.P.; Carrión, M.; Gutiérrez-Cañas, I.; González-Álvaro, I.; Gomáriz, R.M.P.; Juarranz, Y. VIP and CRF reduce ADAMTS expression and function in osteoarthritis synovial fibroblasts. J. Cell. Mol. Med. 2016, 20, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.; Abad, C.; Martinez, C.; Leceta, J.; Gomariz, R.P. Vasoactive intestinal peptide prevents experimental arthritis by downregulating both autoimmune and inflammatory components of the disease. Nat. Med. 2001, 7, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Juarranz, Y.; Abad, C.; Martinez, C.; Arranz, A.; Gutierrez-Cañas, I.; Rosignoli, F.; Gomariz, R.P.; Leceta, J. Protective effect of vasoactive intestinal peptide on bone destruction in the collagen-induced arthritis model of rheumatoid arthritis. Arthritis Res. 2005, 7, R1034–R1045. [Google Scholar] [CrossRef]
- Martínez, C.; Ortiz, A.M.; Juarranz, Y.; Lamana, A.; Seoane, I.V.; Leceta, J.; García-Vicuña, R.; Gomariz, R.P.; González-Álvaro, I. Serum Levels of Vasoactive Intestinal Peptide as a Prognostic Marker in Early Arthritis. PLoS ONE 2014, 9, e85248. [Google Scholar] [CrossRef]
- Seoane, I.V.; Martínez, C.; García-Vicuña, R.; Ortiz, A.M.; Juarranz, Y.; Talayero, V.C.; González-Álvaro, I.; Gomariz, R.P.; Lamana, A. Vasoactive intestinal peptide gene polymorphisms, associated with its serum levels, predict treatment requirements in early rheumatoid arthritis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Schuelert, N.; McDougall, J. Electrophysiological evidence that the vasoactive intestinal peptide receptor antagonist VIP6–28 reduces nociception in an animal model of osteoarthritis. Osteoarthr. Cartil. 2006, 14, 1155–1162. [Google Scholar] [CrossRef]
- Rahman, S.; Dobson, P.R.; Bunning, R.A.; Russell, R.G.; Brown, B.L. The regulation of connective tissue metabolism by vasoactive intestinal polypeptide. Regul. Pept. 1992, 37, 111–121. [Google Scholar] [CrossRef]
- Altman, R.; Asch, E.; Bloch, D.; Bole, G.; Borenstein, D.; Brandt, K.; Christy, W.; Cooke, T.D.; Greenwald, R.; Hochberg, M.; et al. Development of criteria for the classification and reporting of osteoarthritis: Classification of osteoarthritis of the knee. Arthritis Rheum. 1986, 29, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Calamia, V.; Ruiz-Romero, C.; Rocha, B.; Fernandez-Puente, P.; Mateos, J.; Montell, E.; Verges, J.; Blanco, F.J. Pharmacoproteomic study of the effects of chondroitin and glucosamine sulfate on human articular chondrocytes. Arthritis Res. Ther. 2010, 12, R138. [Google Scholar] [CrossRef] [PubMed]
- Calamia, V.; Rocha, B.; Mateos, J.; Fernandez-Puente, P.; Ruiz-Romero, C.; Blanco, F.J. Metabolic labeling of chondro-cytes for the quantitative analysis of the interleukin-1-beta-mediated modulation of their intracellular and extracellular proteomes. J. Proteome Res. 2011, 10, 3701–3711. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).