Abstract
Congenital diaphragmatic hernia (CDH) is a relatively common major life-threatening birth defect that results in significant mortality and morbidity depending primarily on lung hypoplasia, persistent pulmonary hypertension, and cardiac dysfunction. Despite its clinical relevance, CDH multifactorial etiology is still not completely understood. We reviewed current knowledge on normal diaphragm development and summarized genetic mutations and related pathways as well as cellular mechanisms involved in CDH. Our literature analysis showed that the discovery of harmful de novo variants in the fetus could constitute an important tool for the medical team during pregnancy, counselling, and childbirth. A better insight into the mechanisms regulating diaphragm development and genetic causes leading to CDH appeared essential to the development of new therapeutic strategies and evidence-based genetic counselling to parents. Integrated sequencing, development, and bioinformatics strategies could direct future functional studies on CDH; could be applied to cohorts and consortia for CDH and other birth defects; and could pave the way for potential therapies by providing molecular targets for drug discovery.
1. Introduction
Congenital diaphragmatic hernia (CDH) consists of a life-threatening developmental defect in variable size in the fetal diaphragm that allows abdominal viscera to herniate into the chest [1]. CDH occurs in approximately 1 in 3000 live births [2]. Despite improvements in survival with advanced diagnostic techniques along with medical and surgical care, the average mortality rate worldwide is 50%, depending primarily on pulmonary hypoplasia, pulmonary hypertension, and heart failure [3,4]. Long-term morbidity among survivors is common [5], including longer postnatal hospital stays of affected neonates, poor growth, developmental delay, gastroesophageal reflux, and chronic oxygen dependence [6].
Congenital diaphragmatic defects are variable in size encompassing diaphragm agenesis; well-circumscribed defects or “holes”; and, less often, a thinning or undermuscularization of diaphragmatic tissue. Prenatal ultrasound detection is successful in 50% of CDH cases at a mean gestational age of 24 weeks; fetal magnetic resonance imaging (MRI), fetal echocardiography, and three-dimensional ultrasound imaging also play a role in the study of congenital diaphragmatic hernia, including diagnosis, severity stratification, and prognostic prediction [7,8]. The vast majority of neonates with CDH present with cardiorespiratory distress within the first hours or days of life; however, about 5% to 25% of diaphragmatic hernias present beyond the neonatal period (late-presenting CDH), and clinical manifestations may include respiratory or gastrointestinal symptoms, or a combination of both [9,10,11].
Despite its high impact on neonatal health, pathogenesis and etiology of CDH remain poorly understood. CDH is thought to be multi-factorial, with genetic, environmental, and nutritional factors playing a role [7,12,13]. The involvement of multiple genetic factors is suggested by recent advances in our understanding of the genetic pathways regulating normal diaphragm development and genetic mutations leading to CDH. New insights have been produced after the development and availability of new genetic testing procedures including whole genome sequencing (WGS) and whole exome sequencing (WES) [14,15]. Monogenic syndromic disorders, single-gene mutations, multiple chromosomal abnormalities such as deletions, and aneuploidies have been reported to be associated to CDH [16,17].
This review of the literature provides an overview of current knowledge of normal diaphragm embryogenesis and a summary of genetic mutations as well as related pathways and cellular mechanisms involved in CDH whose knowledge is essential to the development of new therapeutic strategies and evidence-based genetic counselling to parents. Systematic searches were performed in PubMed, Embase, Cochrane Library, Scopus, Google Scholar, and ClinicalTrials.gov accessed on 30 March 2021. Language was restricted to English. Search terms included congenital diaphragmatic hernia (CDH), diaphragm development and embryology, molecular genetic pathways, genetics, and genetic testing. Case reports, case series, original research studies, review articles, letters to the editor, randomized controlled trials (RCTs), non-RCTs, and cohort studies (prospective or retrospective) published were included.
2. Diaphragm Development
2.1. Diaphragm Basic Anatomy
The diaphragm muscle is a dome-shaped musculotendinous structure [18]. Because of distinct attachment sites, the muscular part of diaphragm is bilaterally divided into sternal, costal, and lumbar parts. Muscular fibers originate from the internal and the external arcuate ligaments, from the lateral lower six ribs on each side and from the sternum. The muscle fibers from these attachments converge in a three-leaf shaped central tendon, forming the crest of the dome. The right lateral leaf is the largest, the left lateral one is the smallest, and the anterior leaf is intermediate in size and is fused with the diaphragmatic surface of the pericardium.
The diaphragm is a passageway for structures from the thorax to the abdomen: the major orifices are the aortic hiatus, vena caval foramen, and the oesophageal hiatus. The phrenic nerve provides motor and sensory innervation to the diaphragm. The right and left phrenic nerves originate in the spinal cervical roots (C3, C4, and C5), and each phrenic nerve innervates half of the diaphragm. Both nerves always divide into a variable number of macroscopic branches, from two to seven, varying in size and thickness [18]. Three main branches are described: anterior, lateral, and posterior ones. The anterior branches run anteromedially toward the sternum, the anterolateral branches run laterally anterior to the lateral edges of the central tendon, while the posterior branches divide into two rami: posterolateral and posterior [19,20] (Figure 1).
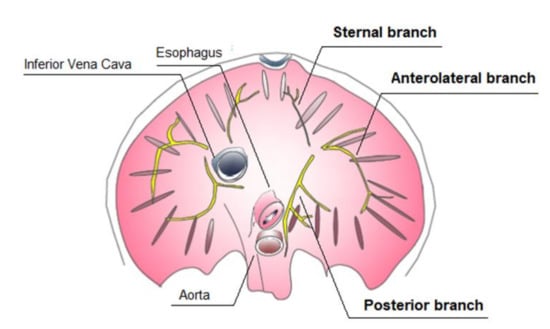
Figure 1.
Inferior view of diaphragm: intramuscular distribution of the phrenic nerve.
The presence of an accessory phrenic nerve arising from the fifth or fifth and sixth cervical nerves has been noted [21].
2.2. Anatomic Findings
CDH is classified according to the location of the defect in the diaphragm (Figure 2). Posterolateral diaphragmatic hernias, referred to as Bochdalek hernias, are the most common hernia type (70–75%), with the majority occurring on the left side (85%), and less frequently on the right side (13%) or bilaterally (2%). Anterior defects also known as Morgagni–Larrey hernias (23–28%) and central hernias (2–7%) are the other types [1,22,23].
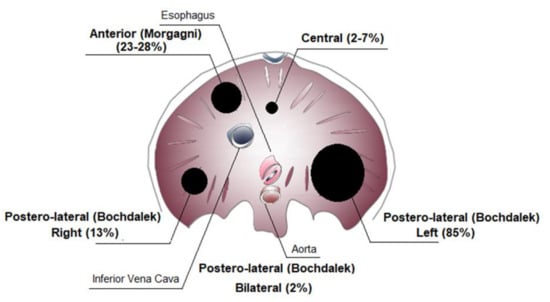
Figure 2.
Congenital diaphragmatic hernia (CDH) classification.
The size of the defect can range from small to the most extreme variant consisting of complete agenesis [24].
2.3. Diaphragm Embryogenesis
Defects in diaphragm development lead to CDH. Recently, progress has been made in understanding the genetic pathways regulating diaphragm development and the genetic mutations leading to CDH. Some CDH-associated genetic mutations and related defects in different molecular pathways can directly affect diaphragm development and of other organs, such as the lungs and the hearth [25,26]. Transgenic mouse models’ studies have helped to understand normal diaphragm development [27]. The mature diaphragm derives from multiple embryonic sources: the septum transversum, the pleuroperitoneal folds (PPFs), muscular ingrowths from body wall, and the dorsal mesentery of esophagus (Figure 3).
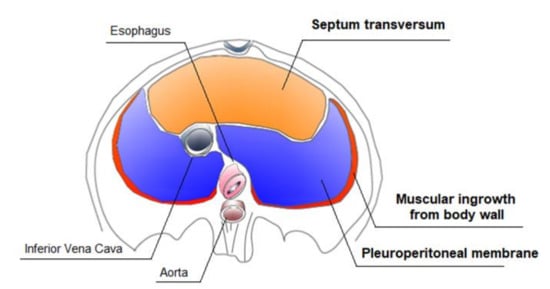
Figure 3.
Development of the diaphragm.
The largest contributors are the pleuroperitoneal folds (PPFs), two transient pyramidal structures appearing at the beginning of the fifth week of pregnancy and situated between the thoracic and abdominal cavities. The PPFs expand dorsally and ventrally, merging with the septum transversum and mesentery of the esophagus, giving rise to the diaphragm’s muscle connective tissue and central tendon in the seventh week of pregnancy, definitively separating the thoracic cavity from the from the abdominal cavity. An additional rim originating from the body wall forms the most peripheral part of the diaphragm [27]. The muscle progenitors migrate from cervical somites to the developing diaphragm into the PPFs [28,29]. After reaching the pleuroperitoneal folds, the muscle progenitors undergo the myogenic program controlled by the network of myogenic regulatory transcription factors (MRF), including Myf5, MyoD, and myogenin [30,31]. Defects in pre-muscular cell migration, differentiation, and proliferation lead to the formation of an abnormal diaphragm musculature [32].
Similar to the muscle progenitors, the phrenic nerve axons originating from the cervical segments of the neural tube reach the PPFs via a ventromedial pathway. Subsequently, the phrenic nerve splits into three branches and elongates in close association with the dorsal and central expansion of the muscle [29,33]. Correct primary and secondary phrenic nerve branching and motoneuron axon targeting through the formation of neuromuscular junctions with differentiated myofibers during development are critical for diaphragmatic function [34,35].
PPF-derived signals regulate migration of muscle and neural progenitors to the developing diaphragm: outgrowth of the phrenic nerve may be guided by neural cell adhesion molecule (NCAM) and low-affinity nerve growth factor receptor (NGFR) expressed along the path from the neural tube to the pleuroperitoneal folds. In addition, hepatocyte growth factor (HGF), the ligand for the Met receptor, is expressed along the migratory path for both the muscle progenitors and the nerves [33,36].
3. Prognostic Factors
3.1. Prenatal Prognostic Factors
CDH is a pathology diagnosed in prenatal age [37]. Prenatal diagnosis of CDH is possible as early as 12 weeks of gestation during first trimester ultrasound screen; CDH ultrasound detection is successful in 50% of cases at a mean gestational age of 24 weeks. Fetal magnetic resonance imaging (MRI), three-dimensional ultrasound imaging, and fetal echocardiography also play a role in prenatal CDH detection and severity stratification [8,22,38]. The main determinants of CDH outcomes are the presence of associated abnormalities, particularly heart disease, extent of pulmonary hypoplasia, and liver position (intra-abdominal or intrathoracic) [8].
The prognosis of isolated CDH is better than CDH associated with multiple abnormalities; a higher survival rate has also been demonstrated for the former [39,40]. Liver position is a survival index: liver herniation (liver up) is associated with a worse prognosis. Metkus et al. reported a survival rate of 100% in CDH cases without hepatic hernia (liver-down) as compared to 56% with liver herniation (liver-down) [41]. Mullassery et al. have described a decrease in survival from 73.7% to 45.4% with liver herniation; liver herniation as a marker for survival showed a sensitivity of 73%, a specificity of 54%, a positive predictive value of 54%, and a negative predictive value of 73% [42].
Prenatal prognostic indicators also include gestational age at diagnosis [43,44], stomach position [45,46,47], polyhydramnios [43], lung size [41], mediastinal shift [48], lung-to-head ratio (LHR) [41,49,50,51], and the preferable observed to expected normal mean for gestation lung-to-head ratio (O/E LHR) [52,53,54,55]. Survival is less than 50% with an O/E LHR of less than 25% and exceeds 80% with an O/E LHR greater than 40%.
As regards stomach position in prediction of survival, Basta et al. established degrees of herniation on ultrasound examination: grade 1, stomach not visualized; grade 2, stomach visualized anteriorly, next to apex of heart, with no structure in between stomach and sternum; grade 3, stomach visualized along from apex of heart and abdominal structures anteriorly; or grade 4, which is the same as grade 3 with stomach posterior to level of atrioventricular heart valves [46].
Lastly, additional indexes derived from imaging modalities other than two-dimensional ultrasound, such as three-dimensional ultrasound, Doppler ultrasonographic analysis of fetal pulmonary vasculature, fetal echocardiography, and magnetic resonance, have been found to be risk predictors of pulmonary hypertension, extra corporeal membrane oxygenation (ECMO) need, and prognostic factors for survival [8,56,57].
3.2. Postnatal Prognostic Factors
Very often, prenatal prognostic factors are insufficient to predict postnatal risk. Many neonates are born without a prenatal diagnosis. Prenatal imaging can often be limited. It appears that the birth weight, Apgar score at 5 min > 7, the severity of respiratory failure (measured by the highest values of fraction of inspired oxygen (FiO2), mean arterial pressure (MAP), Oxygenation Index (OI), and alveolar–arterial oxygen gradient (AaDO2)), and primary pulmonary hypertension (PPH) act as independent factors in influencing the survival rate of patients with CDH [58]. Congenital Diaphragmatic Hernia Study Group (CDHSG) predicted survival, CDHSG defect size, Willford Hall/Santa Rosa clinical prediction formula, Brindle score, and SNAP-II are validated postnatal prediction tools. CDHSG predictive survival is applied in the immediate postnatal period and is based on the Apgar score at 5 min and birth weight to generate a probability of survival [51]. CDHSG defect size is based on the size of the defect that is a marker significantly correlated with survival, need for patch repair, and other adverse outcomes [59,60]. The Willford Hall/Santa Rosa clinical prediction formula uses the difference between the highest value of partial pressure of arterial oxygen (PaO2) and the highest value of partial pressure of carbon dioxide (PCO2) in the first 24 h of life to generate a score of survival [51]. The variables of the Brindle score are instead very low birth weight, absent or low Apgar score at the fifth minute, echocardiographic evidence of pulmonary hypertension, or cardiac or chromosomal abnormalities, and are defined as a mortality score [61]. SNAP-II is a survival score calculated in the first 24–48 h of life on the basis of mean blood pressure, lowest temperature, PO2/FiO2 ratio, lowest serum pH, seizure activity, and urine output [51,62].
Persistent pulmonary hypertension and pulmonary hypoplasia are believed to be responsible for the high morbidity [63,64]. The degree of pulmonary hypoplasia is almost impossible to assess before and immediately after birth, and in most cases, the degree of pulmonary hypoplasia is an important determining factor in the outcome. In addition, pulmonary hypertension (PHT) causes an increase in the right ventricle (RV) after loading, progressively compromising the RV function. Dysfunction and dilation of the RV, in turn, lead to left ventricular (LV) dysfunction, potentially contributing to adverse clinical outcomes [7,65]. Aggarwal et al. in fact demonstrated that LV dysfunction was associated with death and negative outcomes, justifying incorporation of echocardiographic indices as prognostic markers of CDH [66].
Neonates with left-sided CDH had a significantly lower left ventricular mass assessed by echocardiography than ones with other causes of persistent pulmonary hypertension of the newborn (PPHN). The reduced left ventricular mass contributes to functional LV hypoplasia and may result in increased left atrial pressure, pulmonary venous hypertension, and a reduced left ventricular output [67,68]. Yamoto et al. revealed an association of pulmonary circulation parameters predictive of poor CDH prognosis, including the following echocardiographic parameters: right-to-left (R-L) shunt, right pulmonary artery (RPA) diameter, left pulmonary artery (LPA) diameter and left ventricular dimension at diastole (LVDd) between patients who survived up to 90 days and those who died. Smaller RPA diameter and LVDd were good predictors of mortality in CDH [69].
4. Classification of Congenital Diaphragmatic Hernia (CDH)
Isolated and Non-Isolated Congenital Diaphragmatic Hernia (CDH)
CDH hernia may occur in isolation (isolated CDH) in approximately 50% of cases or in association with additional congenital anomalies for the remainder (non-isolated CDH or CDH +), either as part of a genetic syndrome, a chromosome abnormality, or a nonsyndromic collection of major congenital malformations (Figure 4) [16].
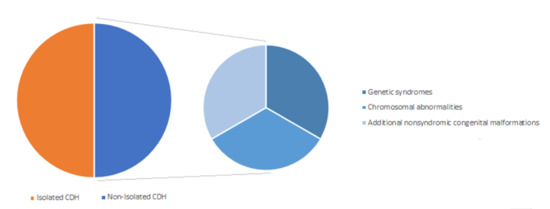
Figure 4.
Classification of congenital diaphragmatic hernia (CDH).
Pulmonary hypoplasia, intestinal malrotation, and left hearth hypoplasia may coexist with isolated CDH and are usually considered part of a sequence. Nonsyndromic major congenital malformations associated with non-isolated CDH may involve the cardiovascular (27.5%), urogenital (17.7%), musculoskeletal (15.7%), and the central nervous system (9.8%) [70].
Many patients with CDH are the only family members affected, and therefore this condition has been typically interpreted as sporadic. The majority of CDH cases are sporadic and appear to be multifactorial. The etiology of most cases remains unknown; however, there is increasing evidence that genetic factors may play a role in the development of CDH.
Genetic contributions to CDH are heterogeneous, identified from animal models, conventional karyotype, chromosome microarray, and next-generation sequencing technology, including whole exome sequencing (WES) and whole genome sequencing (WGS) [71].
Chromosomal aberrations have been discovered as an important etiology for CDH, reported in about 10% of CDH cases and detected by routine karyotyping and chromosome microarray. Complete and mosaic aneuploidies, copy number variant (CNV) deletions, insertions, duplications, and translocations are included in this percentage. Nearly all chromosomes may be affected by structural anomalies [72,73,74,75].
5. Genetic Contribution to Congenital Diaphragmatic Hernia (CDH)
5.1. Aneuploidies
Aneuploidy is a major chromosomal anomaly in which chromosome number is abnormal.
Different aneuploid conditions are associated with CDH: trisomy 18, trisomy 13, trisomy 21, and Turner syndrome (45, X) are the most frequent aneuploidies reported in association.
5.1.1. Trisomy 21—Down Syndrome
Congenital Morgagni’s hernia (CMH), also called congenital Morgagni–Larrey’s hernia, is rare when compared with other types of congenital diaphragmatic hernia, accounting for about 23–28% of all types of CDH [1,22,23].
Congenital Morgagni’s hernia (CMH) can be associated with trisomy 21 with a variable reported incidence ranging from 14% to 50% [76,77,78,79].
The age of presentation varies from the neonatal period to adulthood [80,81,82,83].
5.1.2. Trisomy 18—Edwards Syndrome
Trisomy 18 is the most prevalent aneuploidy associated with CDH cases. CDH occurs more often in male fetuses than in trisomy 18 female fetuses [84].
5.1.3. Trisomy 13—Patau Syndrome
Association of CDH with trisomy 13 (Patau syndrome) has been rarely described [85,86].
5.1.4. Turner syndrome (45, X) and Trisomy X (46, XXX)
Turner syndrome (45, X) and Trisomy X (46, XXX) have been rarely described in association with CDH [87,88].
5.1.5. Tetrasomy 12p—Mosaic Isochromosome 12p Syndrome—Pallister–Killian Syndrome (OMIM 601803)
Pallister–Killian syndrome is caused by mosaicism for tetrasomy of chromosome 12p [89,90,91]. It is characterized by seizures; mental retardation; skin abnormalities; and coarse facial features, including prominent forehead, hypertelorism, short nose, anteverted nostrils, and flat occiput [92,93,94]. A high rate of association between Pallister–Killian syndrome and congenital diaphragmatic hernia has been described in the literature [95,96].
5.2. Copy Number Variants (CNVs)
5.2.1. Deletion 1q41-q42 (OMIM 612530)
Clinical features of patients diagnosed with deletion of 1q41-q42 region include dysmorphic features (deep-set eyes, frontal bossing, broad nasal tip, anteverted nares, and depressed nasal bridge), microcephaly, seizures, and neurodevelopmental disabilities, as well as, less commonly, cleft palate, clubfeet, and congenital diaphragmatic hernia [97,98]. Deletion in this region involves the candidate genes H2.0-like homeobox (HLX) and dispatched RND transporter family member 1 (DISP1) [99,100,101,102].
5.2.2. Duplication 1q25q31.2
Few CDH cases associated with duplication of 1q25q31.2 have been reported in literature [103].
5.2.3. Deletion 3q22
Deletion in this region involves the candidate genes for cellular retinol binding protein 1 (RBP1) and cellular retinol binding protein 2 (RBP2), which play a role in retinol signaling pathway, essential for embryonic development [104,105]. CDH cases associated with this deletion have been reported in the literature [106,107].
5.2.4. Deletion 4p16 (OMIM 194190)
Deletion of chromosomal region 4p16, also known as Wolf–Hirschhorn syndrome, is characterized by pre- and postnatal growth retardation; microcephaly; “Greek helmet” facies; mental retardation; seizures; and/or epilepsy and closure defects including cleft lip or palate, cardiac septal defects, and coloboma of the eye [108,109,110]. Although not a common finding, CDH has been described in association with Wolf–Hirschhorn syndrome [111,112,113,114].
5.2.5. Deletion 6p25
CDH has been described in individuals with 6p25 deletion [115].
5.2.6. Deletion 8q23 (OMIM 610187)
Few CDH cases associated with deletion on chromosome 8q23 have been reported in the literature. This chromosomal region harbors zinc finger protein and FOG family member 2 (ZFPM2, FOG2) genes [116]. Fog2 is required for normal diaphragm and lung development in mice and humans [25]. Variants and deletions in ZFMP2 have been described in patients with CDH [117].
5.2.7. Deletion 8p23 (OMIM 222400)
Deletions of 8p23.1 of variable size have been reported in association with complex congenital heart anomalies, CDH, facial dysmorphisms, and neurodevelopmental disabilities [118,119,120,121,122,123]. Deletion in this region involves the candidate gene GATA4, encoding GATA4 zinc finger DNA-binding protein involved in heart, lung, and diaphragm development [26,124].
5.2.8. Duplication of 8p21-p23.1
Duplication of 8p21-p23.1 has been described in association with CDH [125,126,127].
5.2.9. Deletion 9p24-pter
Terminal deletions of this region have been described in patients with nonisolated CDH [128,129].
5.2.10. Deletion 11p13
Deletion of chromosomal region 11p13, harboring the Wilms tumor 1 gene (WT1), has been described in association with CDH cases [130,131]. WT1 plays a crucial role in diaphragm development [132].
5.2.11. Duplication 11q23.3-qter
Duplication of 11q23.3-qter has been reported in several cases of CDH [133,134].
5.2.12. Deletion 15q26
Non-isolated CDH cases have been described in association with deletion of the distal long arm of chromosome 15 [135]. NR2F2, also known as chick ovalbumin upstream promoter-transcription factor II (COUP-TFII), harbored in this chromosomal region, is believed to play a role in retinoic acid metabolism and diaphragm development [135,136].
5.3. Monogenic Syndromes
Well-characterized monogenic syndromes have been associated with CDH, accounting for approximately 3–10% of all CDH cases [74,137]. Associated syndromes include both X-linked and autosomal-dominant and -recessive inheritance [138]. No one genetic cause accounts for more than 1–2% of CDH cases. Below, we summarize the most known monogenic syndrome causes of CDH. Both de novo and inherited variants contribute to CDH [138].
5.3.1. Simpson–Golabi–Behmel Syndrome (OMIM 312870)
Simpson–Golabi–Behmel syndrome (SBGS) is a X-linked recessive disorder due to mutations in glypican-3 (GPC3) [139]. It is characterized by prenatal and postnatal macrosomia; coarse facial features; hypertelorism; macroglossia; macrocephaly; skeletal; cardiac and renal abnormalities; thoracoabdominal wall defects; and, commonly, mild to severe intellectual disability. CDH has been reported in 24% of individuals with Simpson–Golabi–Behmel syndrome, accounting for about 34% of all types of gastrointestinal and abdominal wall malformations described in association with this syndrome [140,141].
5.3.2. Craniofrontonasal Syndrome (OMIM 304110)
Craniofrontonasal syndrome (CFNS) is an X-linked dominant disorder caused by a variant in Ephrin B1 (efnb1). It is characterized by craniosynostosis, hypertelorism, broad nasal tip, grooved nails of the hallux and thumb, syndactyly, and skeletal abnormalities [142]. Male patients show a milder phenotype than females [143]. CDH has been described in both genders [142,143,144].
5.3.3. Myotubular Myopathy 1 (OMIM 310400)
Myotubular myopathy 1 (MTM1) is an X-linked recessive centronuclear myopathy due to loss of function mutations in the myotubularin 1 (MTM1) at Xq28 [145]. Diaphragm eventration and diaphragm ventilator-dependent dysfunction have been described [146].
5.3.4. Opitz G/BBB Syndrome (OMIM 300000)
Opitz G/BBB syndrome is characterized by facial anomalies, genitourinary abnormalities, and laryngotracheoesophageal defects caused by loss of function of MID1 gene [147]. CDH has been described in association with Opitz G/BBB syndrome [148].
5.3.5. Lowe Syndrome (OMIM 309000)
Lowe syndrome, also called oculocerebrorenal syndrome, is a multisystemic disorder characterized by the triad of proximal renal tubular dysfunction, congenital cataracts, and intellectual disability. It is caused by variants in inositol polyphosphate 5-phosphatase ocrl-1 [149,150]. CDH has been described in association with this syndrome [151].
5.3.6. Focal Dermal Hypoplasia—Goltz Syndrome (OMIM 305600)
Focal dermal hypoplasia or Goltz syndrome is caused by variants in porcupine O-acyltransferase (PORCN) gene [152]. It is characterized by developmental skin malformations and digital, ocular, and dental abnormalities [153,154]. CDH has been reported in in association with Goltz syndrome [155,156].
5.3.7. MIDAS Syndrome (OMIM 309801)
MIDAS syndrome is characterized by unilateral or bilateral microphthalmia and linear areas of aplastic skin limited to the face and neck [157]. Additional features, including CDH, have been described in association with this syndrome [158].
5.3.8. Cornelia de Lange Syndrome (OMIM 122470)
Cornelia de Lange syndrome, also called Brachmann de Lange syndrome, is caused by heterozygous variants in NIBL gene at chromosome 5p13.1, encoding for the NIPBL protein [159]. Distinctive clinical features are prenatal and postnatal growth restriction, facial dysmorphisms, developmental delay, malformations of the upper extremities, heart defects, and gastrointestinal and genitourinary malformations [160]. CDH is a recognized clinical feature of Cornelia de Lange syndrome [161,162,163]. In one large review of 426 patients diagnosed with Cornelia de Lange syndrome, CDH has been identified as the cause of death in 10% of cases [164].
5.3.9. Denys–Drash Syndrome (OMIM 194080)
Denys–Drash syndrome is caused by variants in the tumor-suppressor gene wt1. It is characterized by male pseudohermaphroditism, nephrotic syndrome leading to end-stage renal disease, and increased risk of development of Wilms’ tumor [165]. To date, only three CDH cases associated with Denys–Drash syndrome have been reported in literature [166,167,168]. WT1 plays a crucial role in diaphragm morphogenesis as confirmed by the development of congenital diaphragmatic hernia in wt1-null mouse embryos. However, the mechanisms regulated by WT1 are unknown [132].
5.3.10. Marfan Syndrome (OMIM 154700)
Marfan syndrome is a heritable disorder of fibrous connective tissue with a great clinical variability. It is a multisystemic disease with ocular, skeletal, cardiovascular, and pulmonary involvement. Marfan syndrome results from mutations in the fbn1 gene encoding fibrillin-1, an extracellular matrix protein [169,170]. To date, over 3000 FBN1 gene variants have been identified [171].
Diaphragmatic abnormalities are rare features in Marfan syndrome. Severe neonatal presentations have been described [172,173], and approximately 20% of patients with early onset Marfan syndrome have a diaphragmatic eventration [174].
5.3.11. CHARGE Syndrome (OMIM 214800)
CHARGE syndrome results from variants within the chromodomain helicase DNA-binding protein 7 gene (CHD7) located on 8q12 [175,176]. CHARGE is an acronym for coloboma, heart defects, choanal atresia, retardation (growth and/or development retardation), genitourinary malformation, and ear abnormalities [177,178]. The diagnosis is based on major and minor criteria, as published by Blake et al. and modified by Verloes [179,180]. CDH has been rarely described in CHARGE syndrome [181,182].
5.3.12. Fryns Syndrome (OMIM 229850)
Fryns syndrome has been reported to be the most common autosomal recessive syndrome associated with CDH, accounting for 1.3%–10% of all CDH cases [137,183,184,185]. Recessive variants in the phosphatidyl inositol glycan biosynthesis type N (PIGN) gene are the genetic basis of disease [186,187].
Diagnostic criteria for Fryns syndrome include diaphragmatic defect, characteristic facial appearance, distal digital hypoplasia, pulmonary hypoplasia, at least one characteristic associated anomaly, and family history consistent with autosomal recessive inheritance [188]. The syndrome encompasses a broad spectrum of diaphragmatic developmental defects including diaphragmatic hernia in any location, eventration, agenesis or significant hypoplasia of the diaphragm [189,190].
5.3.13. Donnai–Barrow Syndrome (OMIM 222448)
Donnai–Barrow syndrome is a rare autosomal recessive entity characterized by typical facial dysmorphism (hypertelorism, bulging eyes, down slanting palpebral fissures, posteriorly rotated ears, widow’s peak), large anterior fontanel, high-grade myopia, low molecular weight proteinuria, sensorineural deafness, corpus callosum anomalies, and congenital diaphragmatic hernia [191,192].
The syndrome results from variants in the low-density lipoprotein receptor-related protein 2 gene (LRP2) encoding megalin, a multi-ligand transmembrane endocytic receptor critical for reuptake of numerous ligands, including lipoproteins, sterols, vitamin-binding proteins, and hormones [193,194]. Megalin has a role in cell signaling by interacting with vitamin A (retinol), critical for diaphragm development [195]. Donnai–Barrow syndrome is associated with CDH in >50% of patients [196,197].
5.3.14. Matthew–Wood Syndrome (OMIM 601186, 615524)
Matthew–Wood syndrome, also known as Spear syndrome or microphthalmic syndrome 9, is caused by homozygous variants in stra6 (stimulated by retinoic acid 6) gene encoding for a multitransmembrane domain protein acting as retinol-binding protein (RBP) receptor. RBP is the principal physiological carrier of retinol (vitamin A) [198]. RBP receptor binds to RBP and mediates vitamin A uptake from vitamin A-loaded RBP (holo-RBP) [199]. From this, it follows the importance of stra6 expression and the role of vitamin A during embryonic development.
Matthew–Wood disease is a multisystemic syndrome characterized by pulmonary hypoplasia/aplasia, cardiac malformations, anophthalmia, and diaphragmatic hernia/eventration [200,201,202].
5.3.15. Multiple Vertebral Segmentation Defects
CDH has been reported in association with disorders of vertebral segmentation, variously called Jarcho–Levin syndrome, spondylothoracic dysostosis, and spondylocostal dysostosis [203,204,205,206]. Multiple contiguous vertebral abnormalities (such as hemivertebrae, block vertebrae, or unsegmented bars), rib anomalies (such as absent or fused ribs), and shortened trunk are common features of these disorders [207].
Exact distinction among these disorders is missing. Multiple vertebral segmentation defects can be inherited in an autosomal-dominant or an autosomal-recessive pattern; to date, variants have been described as being related to delta-like 3 (DLL3), lunatic fringe (LFNG), and mesoderm posterior 2 (MESP2) in the Notch signaling pathway, playing a role in somite formation [208]. DLL3 is the most commonly mutated, inherited in an autosomal recessive pattern [209].
5.4. Most Frequent Genes identified by Next-Generation Sequencing and Related Pathways
The text below and Table 1 shows an overview of the most frequent candidate genes and signaling pathways for CDH.

Table 1.
Overview of candidate genes and signaling pathways for congenital diaphragmatic hernia (CDH).
5.4.1. GATA4
GATA4 gene is localized on chromosomal region 8p23.1 and codes for GATA4 zinc finger DNA-binding protein. GATA4 is a component of the retinoic pathway required for normal diaphragm, heart, and lung development, as confirmed by mutant mice lacking GATA4 suffering from defective morphogenesis [26,124,210,211]. Chromosomal deletions of 8p23.1 of variable size have been described in association with congenital diaphragmatic hernia [118,119,120,121,122,123].
De novo GATA4 variants identified by WES have also been described in association with sporadic and familial cases of CDH [212].
5.4.2. GATA6
GATA6 is a zinc finger transcription factor that plays a role in visceral endoderm differentiation and organogenesis [213,214]. GATA6 is essential for pulmonary, diaphragm, and pericardium development [215,216,217,218,219,220]. GATA6 is expressed in murine primordial septum transversum mesenchyme and lateral PPFs [15,221]. Early embryonic lethality has been observed in mice nullizygous for Gata6 [222].
De novo and inherited variants in GATA6 have been identified in CDH patients. Yu et al. used WES and identified de novo variants in GATA6 in patients with CDH and congenital heart disease [223].
5.4.3. FOG2 (ZFPM2)
FOG2, also known as ZFPM2, gene is located on chromosomal region 8q23 and codes for zinc finger protein ZFPM2, FOG family member 2, which modulates the activity of GATA transcription factors. ZFPM2 protein interacts primarily with gata4, which in turn modulates embryonic development. FOG2 is necessary for both diaphragm and lung development, as underscored in mice nullizygous for Fog2 gene affected with diaphragmatic eventration and pulmonary hypoplasia [25].
Inherited and de novo variants in ZFPM2 gene have been described in association with familial and sporadic CDH cases [116,117,224].
5.4.4. COUP-TFII (NRF2)
Chick ovalbumin upstream promoter-transcription factor II (COUP-TFII), also known as NRF2 (nuclear receptor subfamily 2, group F, member 2), has been mapped to chromosome 15q26 and is believed to play a role in retinoic acid metabolism and diaphragm development [135,136,225,226].
COUP-TFII has also been associated with other congenital anomalies, including pancreatic agenesis and congenital heart disease [227].
COUP-TFII is expressed in the developing diaphragm, in PPFs, and in the transverse septum. COUP-TFII is also expressed in the developing lung mesenchyme, suggesting a possible etiological link with lung hypoplasia frequently associated with diaphragmatic defects [226,228].
High et al. have identified by WES that mutations in the coding region of COUP-TFII, alone or in combination with other contributing factors, could be a rare cause of CDH. Larger deletions are associated with complex phenotypes, smaller deletions with isolated CDH, or CDH with congenital heart anomalies [227].
5.4.5. SIN3A
SIN3A is a corepressor regulating genes transcription through interaction with retinoic acid receptors (RARs) [229].
SIN3A gene mutations have been described in human CDH cases. SIN3A is required for normal lung and diaphragm development, as confirmed by Sin3a mutant mice [230].
5.4.6. MYRF (MRF)
Myelin regulatory factor (MYRF), also known as myelin gene regulatory factor (MRF), is a membrane-associated transcription factor that is highly expressed in developing heart and diaphragm. MYRF mutations have been described in association with congenital heart defects (hypoplastic left heart syndrome, scimitar syndrome, septal defects, and valvular anomalies), genitourinary anomalies (ambiguous genitalia, hypospadias, and cryptorchidism), congenital diaphragmatic hernia, and pulmonary hypoplasia [231,232].
6. Implications of Congenital Diaphragmatic Hernia (CDH) Diagnosis
CDH can occur as an isolated defect or complex defect associated with other congenital anomalies. Chromosomal abnormalities, affecting the entire chromosome or partial aneuploidies, are the most common genetic causes recognized for CDH [16,233,234,235,236]. Karyotype, chromosomal microarray, and WES are the techniques used in the prenatal period capable of identifying about 30% of the causes of non-isolated CDH [74,237]. Overall, 10% of chromosomal abnormalities causative for CDH are identified by chromosomal microarray analysis [74]. WES/WGS represents the best method to assess the contribution of de novo mutations.
The purpose of identifying the cause is to discover and clarify the risk of recurrence. It is essentially a careful family history in the identification of the cause of CDH. Trio studies (proband, mother, and father) have been carried out to identify de novo and recessive mutations, and their importance as tools for identifying undiagnosed genetic conditions is emerging. Serious undiagnosed developmental disorders are based on de novo harmful mutations in developmentally important genes [238]. Most patients with CDH have no family history of CDH, leading to the hypothesis that de novo variants are an important etiological mechanism [239].
However, WES and WGS should be reserved to particular cases because of the high costs and difficult to identify new candidate genes. Approximately 10–22% of CDH isolated and complex patients have a de novo sequence variant that can be identified through the use of WES/WGS [237,240,241,242]. Recent advances in WES and WGS have had an advantage in facilitating detection of de novo (as well as those inherited) single-nucleotide variants (SNVs), short insertions and deletions (indels), and copy number variations (CNVs). This has significant implications for genetic counseling for parents and risk stratification of recurrence. These techniques also represent a useful tool for the diagnosis of new syndromes but also for the counseling and follow-up of newborns born with apparently isolated congenital defects.
7. Conclusions
CDH is a relatively common major life-threatening birth defect that results in significant mortality and morbidity depending primarily on lung hypoplasia, persistent pulmonary hypertension, and cardiac dysfunction. Despite its clinical relevance, CDH multifactorial etiology is still not completely understood. Genetic contributions to CDH appeared heterogeneous. Advances in genomics, coupled with functional studies in animal models, are increasingly identifying the causes of CDH in both familial and sporadic cases [243,244,245]. Through these approaches, we are beginning to elucidate the mechanisms and molecular pathways that are responsible for diaphragm and lung development abnormalities in CDH patients. A key challenge will be to understand which molecular pathways are most commonly disrupted and contribute to diaphragm and lung defects in CDH. An additional challenge will be to understand what causes the phenotypic variability and different clinical outcomes of CDH patients who share the same genetic mutation. A better insight into the mechanisms regulating diaphragm development and genetic causes leading to CDH appeared essential to the development of a personalized approach, new therapeutic strategies, and evidence-based genetic counselling to parents. The discovery of harmful de novo variants in the fetus could constitute an important tool for the medical team (gynecologists, neonatologists, geneticists, anesthetists, and pediatric surgeons) during pregnancy. Integrated sequencing, development, and bioinformatics strategies could direct future functional studies on CDH, could be applied to cohorts and consortia for CDH and other birth defects, and could pave the way for potential therapies by providing molecular targets for drug discovery. Due to genetic predispositions for CDH, there is the possibility of recurrence in later pregnancies [246]. This information should be shared with parents to help them make an informed choice between expectant management and prenatal referral for elective delivery; termination of pregnancy; or, in selected patients, fetal intervention. Presence of a human element in medical communication increases parents’ confidence in healthcare professionals and enhances their emotional resilience and preparedness [246].
Author Contributions
G.C. wrote the first draft of the manuscript. C.C. and F.G. performed the literature analysis. S.P. gave her scientific contribution. S.E. critically revised the text and made substantial scientific contributions. All authors have read and agreed to the published version of the manuscript.
Funding
Ri.Cli.Ped.—University of Parma, Parma, Italy (PED-2021-03).
Institutional Review Board Statement
Not applicable in a review article.
Informed Consent Statement
Not applicable in a review article.
Data Availability Statement
Not applicable in a review article.
Conflicts of Interest
The authors declare no competing interests.
References
- Greer, J.J. Current concepts on the pathogenesis and etiology of congenital diaphragmatic hernia. Respir. Physiol. Neurobiol. 2013, 189, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Grivell, R.M.; Andersen, C.; Dodd, J.M. Prenatal interventions for congenital diaphragmatic hernia for improving outcomes. Cochrane Database Syst. Rev. 2015, 11, CD008925. [Google Scholar] [CrossRef] [PubMed]
- Mohseni-Bod, H.; Bohn, D. Pulmonary hypertension in congenital diaphragmatic hernia. Semin. Pediatr. Surg. 2007, 16, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Coughlin, M.A.; Werner, N.L.; Gajarski, R.; Gadepalli, S.; Hirschl, R.; Barks, J.; Treadwell, M.C.; Ladino-Torres, M.; Kreutzman, J.; Mychaliska, G.B. Prenatally diagnosed severe CDH: Mortality and morbidity remain high. J. Pediatr. Surg. 2016, 51, 1091–1095. [Google Scholar] [CrossRef]
- Harrison, M.R.; Bjordal, R.I.; Langmark, F.; Knutrud, O. Congenital diaphragmatic hernia: The hidden mortality. J. Pediatr. Surg. 1978, 13, 227–230. [Google Scholar] [CrossRef]
- Nobuhara, K.K.; Lund, D.P.; Mitchell, J.; Kharasch, V.; Wilson, J.M. Long-term Outlook for Survivors of Congenital Diaphragmatic Hernia. Clin. Perinatol. 1996, 23, 873–887. [Google Scholar] [CrossRef]
- Deprest, J.A.; Nicolaides, K.H.; Benachi, A.; Gratacos, E.; Ryan, G.; Persico, N.; Sago, H.; Johnson, A.; Wielgoś, M.; Berg, C.; et al. TOTAL Trial for Severe Hypoplasia Investigators.Randomized Trial of Fetal Surgery for Severe Left Diaphragmatic Hernia. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef]
- Graham, G.; Devine, P.C. Antenatal Diagnosis of Congenital Diaphragmatic Hernia. Semin. Perinatol. 2005, 29, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Bagłaj, M. Late-presenting congenital diaphragmatic hernia in children: A clinical spectrum. Pediatr. Surg. Int. 2004, 20, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Chao, P.-H.; Chuang, J.-H.; Lee, S.-Y.; Huang, H.-C. Late-presenting congenital diaphragmatic hernia in childhood. Acta Paediatr. 2010, 100, 425–428. [Google Scholar] [CrossRef]
- Kitano, Y.; Lally, K.P.; Lally, P. A Congenital Diaphragmatic Hernia Study Group. Late-presenting congenital diaphragmatic hernia. J. Pediatr. Surg. 2005, 40, 1839–1843. [Google Scholar] [CrossRef]
- Beurskens, L.W.J.E.; Tibboel, D.; Lindemans, J.; Duvekot, J.J.; Cohen-Overbeek, T.E.; Veenma, D.C.M.; De Klein, A.; Greer, J.J.; Steegers-Theunissen, R.P.M. Retinol Status of Newborn Infants Is Associated with Congenital Diaphragmatic Hernia. Pediatry 2010, 126, 712–720. [Google Scholar] [CrossRef]
- Beurskens, L.W.J.E.; Tibboel, D.; Steegers-Theunissen, R.P. Role of nutrition, lifestyle factors, and genes in the pathogenesis of congenital diaphragmatic hernia: Human and animal studies. Nutr. Rev. 2009, 67, 719–730. [Google Scholar] [CrossRef]
- Longoni, M.; High, F.A.; Russell, M.K.; Kashani, A.; Tracy, A.A.; Coletti, C.M.; Hila, R.; Shamia, A.; Wells, J.; Ackerman, K.G.; et al. Molecular pathogenesis of congenital diaphragmatic hernia revealed by exome sequencing, developmental data, and bioinformatics. Proc. Natl. Acad. Sci. USA 2014, 111, 12450–12455. [Google Scholar] [CrossRef]
- Russell, M.K.; Longoni, M.; Wells, J.; Maalouf, F.; Tracy, A.A.; Loscertales, M.; Ackerman, K.G.; Pober, B.R.; Lage, K.; Bult, C.J.; et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc. Natl. Acad. Sci. USA 2012, 109, 2978–2983. [Google Scholar] [CrossRef]
- Pober, B.R. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am. J. Med. Genet. Part. C: Semin. Med. Genet. 2007, 145C, 158–171. [Google Scholar] [CrossRef]
- Pober, B.R. Genetic aspects of human congenital diaphragmatic hernia. Clin. Genet. 2008, 74, 1–15. [Google Scholar] [CrossRef]
- Botha, G.S.M. The Anatomy of Phrenic Nerve Termination and the Motor Innervation of the Diaphragm. Thorax 1957, 12, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Shane Tubbs, R.; Rizk, E.; Shoja, M.M.; Loukas, M.; Barbaro, N.; Spinner, R.J. Volume 1: History, Embryology, Anatomy, Imaging and Diagnosis. In Nerves and Nerve Injuries; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- An, X.; Yue, B.; Lee, J.-H.; Lee, M.-S.; Lin, C.; Han, S.-H. Intramuscular distribution of the phrenic nerve in human diaphragm as shown by Sihler staining. Muscle Nerve 2012, 45, 522–526. [Google Scholar] [CrossRef]
- Bergman, R.A.; Thompson, S.A.; Saadeh, F.A. Compendium of Human Anatomic Variation; Urban Schwarz: Baltimore, MD, USA, 1988; Volume 1, pp. 138–139. [Google Scholar]
- Chandrasekharan, P.K.; Rawat, M.; Madappa, R.; Rothstein, D.H.; Lakshminrusimha, S. Congenital Diaphragmatic hernia—A review. Matern. Health Neonatol. Perinatol. 2017, 3, 1–16. [Google Scholar] [CrossRef]
- Veenma, D.; de Klein, A.; Tibboel, D. Developmental and genetic aspects of congenital diaphragmatic hernia. Pediatr. Pulmonol. 2012, 47, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Baglaj, M.; Spicer, R.; Ashworth, M. Unilateral agenesis of the diaphragm: A separate entity or an extremely large defect? Pediatr. Surg. Int. 1999, 15, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, K.G.; Herron, B.J.; Vargas, S.O.; Huang, H.; Tevosian, S.G.; Kochilas, L.; Rao, C.; Pober, B.R.; Babiuk, R.P.; Epstein, J.A.; et al. Fog2 Is Required for Normal Diaphragm and Lung Development in Mice and Humans. PLoS Genet. 2005, 1, e10–e65. [Google Scholar] [CrossRef] [PubMed]
- Jay, P.Y.; Bielinska, M.; Erlich, J.; Mannisto, S.; Pu, W.; Heikinheimo, M.; Wilson, D.B. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev. Biol. 2007, 301, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Sefton, E.M.; Gallardo, M.; Kardon, G. Developmental origin and morphogenesis of the diaphragm, an essential mammalian muscle. Dev. Biol. 2018, 440, 64–73. [Google Scholar] [CrossRef]
- Merrell, A.J.; Ellis, B.J.; Fox, Z.D.; Lawson, J.A.; Weiss, J.A.; Kardon, G. Muscle connective tissue controls development of the diaphragm and is a source of congenital diaphragmatic hernias. Nat. Genet. 2015, 47, 496–504. [Google Scholar] [CrossRef]
- Babiuk, R.P.; Zhang, W.; Clugston, R.; Allan, D.W.; Greer, J.J. Embryological origins and development of the rat diaphragm. J. Comp. Neurol. 2003, 455, 477–487. [Google Scholar] [CrossRef]
- Chal, J.; Pourquié, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef]
- Buckingham, M.; Bajard, L.; Chang, T.; Daubas, P.; Hadchouel, J.; Meilhac, S.; Montarras, D.; Rocancourt, D.; Relaix, F. The formation of skeletal muscle: From somite to limb. J. Anat. 2003, 202, 59–68. [Google Scholar] [CrossRef]
- Merrell, A.J.; Kardon, G. Development of the diaphragm—A skeletal muscle essential for mammalian respiration. FEBS J. 2013, 280, 4026–4035. [Google Scholar] [CrossRef]
- Allan, D.W.; Greer, J.J. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J. Comp. Neurol. 1997, 382, 459–468. [Google Scholar] [CrossRef]
- Uetani, N.; Chagnon, M.J.; Kennedy, T.E.; Iwakura, Y.; Tremblay, M.L. Mammalian Motoneuron Axon Targeting Requires Receptor Protein Tyrosine Phosphatases σ and δ. J. Neurosci. 2006, 26, 5872–5880. [Google Scholar] [CrossRef]
- Philippidou, P.; Walsh, C.M.; Aubin, J.; Jeannotte, L.; Dasen, J.S. Sustained Hox5 gene activity is required for respiratory motor neuron development. Nat. Neurosci. 2012, 15, 1636–1644. [Google Scholar] [CrossRef]
- Dietrich, S.; Abou-Rebyeh, F.; Brohmann, H.; Bladt, F.; Sonnenberg-Riethmacher, E.; Yamaai, T.; Lumsden, A.; Brand-Saberi, B.; Birchmeier, C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development 1999, 126, 1621–1629. [Google Scholar] [CrossRef]
- Reiss, I.; Schaible, T.; Hout, L.V.D.; Capolupo, I.; Allegaert, K.; van Heijst, A.; Silva, M.G.; Greenough, A.; Tibboel, D. For the CDH EURO consortium Standardized Postnatal Management of Infants with Congenital Diaphragmatic Hernia in Europe: The CDH EURO Consortium Consensus. Neonatology 2010, 98, 354–364. [Google Scholar] [CrossRef]
- Sepulveda, W.; Wong, A.E.; Casasbuenas, A.; Solari, A.; Alcalde, J.L. Congenital diaphragmatic hernia in a first-trimester ultrasound aneuploidy screening program. Prenat. Diagn. 2008, 28, 531–534. [Google Scholar] [CrossRef]
- Tennant, P.W.; Pearce, M.S.; Bythell, M.; Rankin, J. 20-year survival of children born with congenital anomalies: A population-based study. Lancet 2010, 375, 649–656. [Google Scholar] [CrossRef]
- McGivern, M.R.; Best, K.; Rankin, J.; Wellesley, D.; Greenlees, R.; Addor, M.-C.; Arriola, L.; De Walle, H.; Barisic, I.; Beres, J.; et al. Epidemiology of congenital diaphragmatic hernia in Europe: A register-based study. Arch. Dis. Child. Fetal Neonatal Ed. 2014, 100, F137–F144. [Google Scholar] [CrossRef]
- Metkus, A.P.; Filly, R.A.; Stringer, M.D.; Harrison, M.R.; Adzick, N. Sonographic predictors of survival in fetal diaphragmatic hernia. J. Pediatr. Surg. 1996, 31, 148–152. [Google Scholar] [CrossRef]
- Mullassery, D.; Ba’Ath, M.E.; Jesudason, E.C.; Losty, P.D. Value of liver herniation in prediction of outcome in fetal congenital diaphragmatic hernia: A systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2010, 35, 609–614. [Google Scholar] [CrossRef]
- Adzick, N.S.; Vacanti, J.P.; Lillehei, C.W.; O’Rourke, P.P.; Crone, R.K.; Wilson, J.M. Fetal diaphragmatic hernia: Ultrasound diagnosis and clinical outcome in 38 cases. J. Pediatr. Surg. 1989, 24, 654–658. [Google Scholar] [CrossRef]
- Stringer, M.D.; Goldstein, R.B.; Filly, R.A.; Howell, L.J.; Sola, A.; Adzick, N.; Harrison, M.R. Fetal diaphragmatic hernia without visceral herniation. J. Pediatr. Surg. 1995, 30, 1264–1266. [Google Scholar] [CrossRef]
- Hatch, E.I.; Kendall, J.; Blumhagen, J. Stomach position as an in utero predictor of neonatal outcome in left-sided diaphragmatic hernia. J. Pediatr. Surg. 1992, 27, 778–779. [Google Scholar] [CrossRef]
- Basta, A.M.; Lusk, L.A.; Keller, R.L.; Filly, R.A. Fetal Stomach Position Predicts Neonatal Outcomes in Isolated Left-Sided Congenital Diaphragmatic Hernia. Fetal Diagn. Ther. 2016, 39, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Cordier, A.-G.; Jani, J.C.; Cannie, M.M.; Rodó, C.; Fabietti, I.; Persico, N.; Saada, J.; Carreras, E.; Senat, M.-V.; Benachi, A. Stomach position in prediction of survival in left-sided congenital diaphragmatic hernia with or without fetoscopic endoluminal tracheal occlusion. Ultrasound Obstet. Gynecol. 2014, 46, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Dommergues, M.; Louis-Sylvestre, C.; Mandelbrot, L.; Oury, J.F.; Herlicoviez, M.; Body, G.; Gamerre, M.; Dumez, Y. Congenital diaphragmatic hernia: Can prenatal ultrasonography predict outcome? Am. J. Obstet. Gynecol. 1996, 174, 1377–1381. [Google Scholar] [CrossRef]
- Lipshutz, G.S.; Albanese, C.T.; Feldstein, V.A.; Jennings, R.W.; Housley, H.T.; Beech, R.; Farrell, J.A.; Harrison, M.R. Prospective analysis of lung-to-head ratio predicts survival for patients with prenatally diagnosed congenital diaphragmatic hernia. J. Pediatr. Surg. 1997, 32, 1634–1636. [Google Scholar] [CrossRef]
- Jani, J.; Keller, R.L.; Benachi, A.; Nicolaides, K.H.; Favre, R.; Gratacos, E.; Laudy, J.; Eisenberg, V.; Eggink, A.; Vaast, P.; et al. Prenatal prediction of survival in isolated left-sided diaphragmatic hernia. Ultrasound Obstet. Gynecol. 2005, 27, 18–22. [Google Scholar] [CrossRef]
- Gentili, A.; Pasini, L.; Iannella, E.; Landuzzi, V.; Lima, M.; Reggiani, M.L.B.; Baroncini, S. Predictive outcome indexes in neonatal Congenital Diaphragmatic Hernia. J. Matern. Neonatal Med. 2014, 28, 1–6. [Google Scholar] [CrossRef]
- Ba’Ath, M.E.; Jesudason, E.C.; Losty, P.D. How useful is the lung-to-head ratio in predicting outcome in the fetus with congenital diaphragmatic hernia? A systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2007, 30, 897–906. [Google Scholar] [CrossRef]
- Jani, J.; Nicolaides, K.H.; Keller, R.L.; Benachi, A.; Peralta, C.F.A.; Favre, R.; Moreno, O.; Tibboel, D.; Lipitz, S.; Eggink, A.; et al. Observed to expected lung area to head circumference ratio in the prediction of survival in fetuses with isolated diaphragmatic hernia. Ultrasound Obstet. Gynecol. 2007, 30, 67–71. [Google Scholar] [CrossRef]
- Deprest, J.; Flemmer, A.W.; Gratacos, E.; Nicolaides, K. Antenatal prediction of lung volume and in-utero treatment by fetal endoscopic tracheal occlusion in severe isolated congenital diaphragmatic hernia. Semin. Fetal Neonatal. Med. 2009, 14, 8–13. [Google Scholar] [CrossRef]
- Snoek, K.G.; Peters, N.C.J.; Van Rosmalen, J.; van Heijst, A.; Eggink, A.J.; Sikkel, E.; Wijnen, R.M.; Ijsselstijn, H.; Cohen-Overbeek, T.; Titia, E.; et al. The validity of the observed-to-expected lung-to-head ratio in congenital diaphragmatic hernia in an era of standardized neonatal treatment, a multicenter study. Prenat. Diagn. 2017, 37, 658–665. [Google Scholar] [CrossRef]
- Weidner, M.; Hagelstein, C.; Debus, A.; Walleyo, A.; Weiss, C.; Schoenberg, S.O.; Schaible, T.; Büsing, K.A.; Kehl, S.; Neff, K.W. MRI-Based Ratio of Fetal Lung Volume to Fetal Body Volume as a New Prognostic Marker in Congenital Diaphragmatic Hernia. Am. J. Roentgenol. 2014, 202, 1330–1336. [Google Scholar] [CrossRef]
- Ruano, R.; Aubry, M.-C.; Barthe, B.; Mitanchez, D.; Dumez, Y.; Benachi, A. Quantitative analysis of fetal pulmonary vasculature by 3-dimensional power Doppler ultrasonography in isolated congenital diaphragmatic hernia. Am. J. Obstet. Gynecol. 2006, 195, 1720–1728. [Google Scholar] [CrossRef]
- Lupo, E.; Castoldi, F.M.L.R.; Maestri, L.; Rustico, M.; Dani, C.; Lista, G. Outcome of congenital diaphragmatic hernia: Analysis of implicated factors. Minerva Pediatr. 2013, 65. [Google Scholar]
- Werner, N.L.; Coughlin, M.; Kunisaki, S.M.; Hirschl, R.; Ladino-Torres, M.; Berman, D.; Kreutzman, J.; Mychaliska, G.B. Prenatal and postnatal markers of severity in congenital diaphragmatic hernia have similar prognostic ability. Prenat. Diagn. 2015, 36, 107–111. [Google Scholar] [CrossRef]
- The Congenital Diaphragmatic Hernia Study Group*. Defect Size Determines Survival in Infants with Congenital Diaphragmatic Hernia. Pediatrics 2007, 120, e651–e657. [Google Scholar] [CrossRef]
- Brindle, M.E.; Cook, E.F.; Tibboel, D.; Lally, P.A.; Lally, K.P.; on behalf of the Congenital Diaphragmatic Hernia Study Group. A Clinical Prediction Rule for the Severity of Congenital Diaphragmatic Hernias in Newborns. Pediatry 2014, 134, e413–e419. [Google Scholar] [CrossRef]
- Baird, R.; Macnab, Y.C.; Skarsgard, E. Mortality prediction in congenital diaphragmatic hernia. J. Pediatr. Surg. 2008, 43, 783–787. [Google Scholar] [CrossRef]
- Langham, M.R., Jr.; Kays, D.W.; Ledbetter, D.J.; Frentzen, B.; Sanford, L.L.; Richards, D.S. Congenital diaphragmatic hernia. Epidemi-ology and outcome. Clin. Perinatol. 1996, 23, 671. [Google Scholar] [CrossRef] [PubMed]
- Downard, C.D.; Jaksic, T.; Garza, J.J.; Dzakovic, A.; Nemes, L.; Jennings, R.W.; Wilson, J.M. Analysis of an improved survival rate for congenital diaphragmatic hernia. J. Pediatr. Surg. 2003, 38, 729–732. [Google Scholar] [CrossRef]
- Chin, K.M.; Kim, N.H.S.; Rubin, L.J. The right ventricle in pulmonary hypertension. Coron. Artery Dis. 2005, 16, 13–18. [Google Scholar] [CrossRef]
- Aggarwal, S.; Stockmann, P.; Klein, M.D.; Natarajan, G. Echocardiographic measures of ventricular function and pulmonary artery size: Prognostic markers of congenital diaphragmatic hernia? J. Perinatol. 2011, 31, 561–566. [Google Scholar] [CrossRef]
- Byrne, F.A.; Keller, R.L.; Meadows, J.; Miniati, D.; Brook, M.M.; Silverman, N.H.; Moon-Grady, A.J. Severe left diaphragmatic hernia limits size of fetal left heart more than does right diaphragmatic hernia. Ultrasound Obstet. Gynecol. 2015, 46, 688–694. [Google Scholar] [CrossRef]
- Kinsella, J.P.; Ivy, D.D.; Abman, S.H. Pulmonary Vasodilator Therapy in Congenital Diaphragmatic Hernia: Acute, Late, and Chronic Pulmonary Hypertension. Semin. Perinatol. 2005, 29, 123–128. [Google Scholar] [CrossRef]
- Yamoto, M.; Inamura, N.; Terui, K.; Nagata, K.; Kanamori, Y.; Hayakawa, M.; Tazuke, Y.; Yokoi, A.; Takayasu, H.; Okuyama, H.; et al. Echocardiographic predictors of poor prognosis in congenital diaphragmatic hernia. J. Pediatr. Surg. 2016, 51, 1926–1930. [Google Scholar] [CrossRef]
- Stoll, C.; Alembik, Y.; Dott, B.; Roth, M.-P. Associated malformations in cases with congenital diaphragmatic hernia. Genet. Couns. 2008, 19, 331–339. [Google Scholar]
- Yu, L.; Hernan, R.R.; Wynn, J.; Chung, W.K. The influence of genetics in congenital diaphragmatic hernia. Semin. Perinatol. 2020, 44, 151169. [Google Scholar] [CrossRef]
- Tibboel, D.; Gaag, A.V. Etiologic and Genetic Factors in Congenital Diaphragmatic Hernia. Clin. Perinatol. 1996, 23, 689–699. [Google Scholar] [CrossRef]
- Holder, A.; Klaassens, M.; Tibboel, D.; de Klein, A.; Lee, B.; Scott, D. Genetic Factors in Congenital Diaphragmatic Hernia. Am. J. Hum. Genet. 2007, 80, 825–845. [Google Scholar] [CrossRef] [PubMed]
- Garne, E.; Haeusler, M.; Barisic, I.; Gjergja, R.; Stoll, C.; Clementi, M. Congenital diaphragmatic hernia: Evaluation of prenatal diagnosis in 20 European regions. Ultrasound Obstet. Gynecol. 2002, 19, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Lurie, I.W. Where to look for the genes related to diaphragmatic hernia? Genet. Couns. 2003, 14, 12725592. [Google Scholar]
- Honoré, L.H.; Torfs, C.P.; Curry, C.J.R. Possible association between the hernia of Morgagni and trisomy 21. Am. J. Med. Genet. 1993, 47, 255–256. [Google Scholar] [CrossRef]
- Pokorny, W.J.; McGill, C.W.; Harberg, F.J. Morgagni hernias during infancy: Presentation and associated anomalies. J. Pediatr. Surg. 1984, 19, 394–397. [Google Scholar] [CrossRef]
- Jetley, N.K.; Al-Assiri, A.H.; Al-Helal, A.S.; Ali, A.M.A.-B. Down’s syndrome as a factor in the diagnosis, management, and outcome in patients of Morgagni hernia. J. Pediatr. Surg. 2011, 46, 636–639. [Google Scholar] [CrossRef]
- Cigdem, M.K.; Onen, A.; Okur, H.; Otcu, S. Associated malformations in Morgagni hernia. Pediatr. Surg. Int. 2007, 23, 1101–1103. [Google Scholar] [CrossRef]
- Harris, G.J.; Soper, R.T.; Kimura, K.K. Foramen of morgagni hernia in identical twins: Is this an inheritable defect? J. Pediatr. Surg. 1993, 28, 177–178. [Google Scholar] [CrossRef]
- Snyder, W.H., Jr.; Greaney, E.M., Jr. Congenital diaphragmatic hernia; 77 consecutive cases. Surgery 1965, 57, 576. [Google Scholar] [PubMed]
- Quah, B.S.; Menon, B.S. Down syndrome associated with a retroperitoneal teratoma and Morgagni hernia. Clin. Genet. 2008, 50, 232–234. [Google Scholar] [CrossRef]
- Kubiak, R.; Platen, C.; Schmid, E.; Gruber, R.; Ludwig, K.-H.; Rauh, W. Delayed appearance of bilateral morgagni herniae in a child with Down’s syndrome. Pediatr. Surg. Int. 1998, 13, 600–601. [Google Scholar] [CrossRef]
- Chen, C.-P. Omphalocele and congenital diaphragmatic hernia associated with fetal trisomy 18. Prenat. Diagn. 2005, 25, 421–423. [Google Scholar] [CrossRef]
- PK, A.J.; Yk, P.K.S. Congenital diaphragmatic hernia in a case of patau syndrome: A rare association. J. Neonatal. Surg. 2015, 1, 4. [Google Scholar]
- Sahin, S.; Kutman, K.H.; Bozkurt, O.; Canpolat, F.E.; Uras, N.; Oguz, S.S.; Topcu, V.; Ozdemir, O.; Dilmen, U. A trisomy 13 case prsenting with congenita diaphragmatic hernia and microphthalmia. Genet. Couns. 2015, 26, 263. [Google Scholar] [PubMed]
- Ruano, R.; Bunduki, V.; Silva, M.M.; Yoshizaki, C.T.; Tanuri, U.; Macksoud, J.G.; Zugaib, M. Prenatal diagnosis and perinatal outcome of 38 cases with congenital diaphragmatic hernia: 8-year experience of a tertiary Brazilian center. Clinics 2006, 61, 197–202. [Google Scholar] [CrossRef]
- Zaiss, I.; Kehl, S.; Link, K.; Neff, W.; Schaible, T.; Sütterlin, M.; Siemer, J. Associated Malformations in Congenital Diaphragmatic Hernia. Am. J. Perinatol. 2010, 28, 211–218. [Google Scholar] [CrossRef]
- Pallister, P.D.; Meisner, L.F.; Elejalde, B.R.; Francke, U.; Herrmann, J.; Spranger, J.; Tiddy, W.; Inhorn, S.L.; Opitz, J.M. The pallister mosaic syndrome. Birth Defect. Orig. Artic. Ser. 1977, 13, 103. [Google Scholar] [PubMed]
- Peltomäki, P.; Knuutila, S.; Ritvanen, A.; Kaitila, I.; De La Chapelle, A. Pallister-Killian syndrome: Cytogenetic and molecular studies. Clin. Genet. 2008, 31, 399–405. [Google Scholar] [CrossRef]
- Warburton, D.; Anyane-Yeboa, K.; Francke, U.; Reynolds, J.F. Mosaic tetrasomy 12p: Four new cases, and confirmation of the chromosomal origin of the supernumerary chromosome in one of the original Pallister-Mosaic syndrome cases. Am. J. Med. Genet. 1987, 27, 275–283. [Google Scholar] [CrossRef]
- Schinzel, A. Tetrasomy 12p (Pallister-Killian syndrome). J. Med. Genet. 1991, 28, 122–125. [Google Scholar] [CrossRef]
- Blyth, M.; Maloney, V.; Beal, S.; Collinson, M.; Huang, S.; Crolla, J.; Temple, I.K.; Baralle, D. Pallister-Killian syndrome: A study of 22 British patients. J. Med. Genet. 2015, 52, 454–464. [Google Scholar] [CrossRef]
- Thakur, S.; Gupta, R.; Tiwari, B.; Singh, N.; Saxena, K. Pallister-Killian syndrome: Review of fetal phenotype. Clin. Genet. 2018, 95, 79–84. [Google Scholar] [CrossRef]
- Salzano, E.; Raible, S.; Kaur, M.; Wilkens, A.; Sperti, G.; Tilton, R.; Bettini, L.; Rocca, A.; Cocchi, G.; Selicorni, A.; et al. Prenatal profile of Pallister-Killian syndrome: Retrospective analysis of 114 pregnancies, literature review and approach to prenatal diagnosis. Am. J. Med. Genet. Part. A 2018, 176, 2575–2586. [Google Scholar] [CrossRef]
- Wilkens, A.; Liu, H.; Park, K.; Campbell, L.B.; Jackson, M.; Kostanecka, A.; Pipan, M.; Izumi, K.; Pallister, P.; Krantz, I.D. Novel clinical manifestations in Pallister-Killian syndrome: Comprehensive evaluation of 59 affected individuals and review of previously reported cases. Am. J. Med. Genet. Part. A 2012, 158A, 3002–3017. [Google Scholar] [CrossRef]
- Shaffer, L.G.; Theisen, A.; Bejjani, B.A.; Ballif, B.C.; Aylsworth, A.S.; Lim, C.; McDonald, M.; Ellison, J.W.; Kostiner, D.; Saitta, S.; et al. The discovery of microdeletion syndromes in the post-genomic era: Review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet. Med. 2007, 9, 607–616. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Lacassie, Y.; El-Khechen, D.; Escobar, L.F.; Reggin, J.; Heuer, C.; Chen, E.; Jenkins, L.S.; Collins, A.T.; Zinner, S.; et al. New cases and refinement of the critical region in the 1q41q42 microdeletion syndrome. Eur. J. Med. Genet. 2011, 54, 42–49. [Google Scholar] [CrossRef]
- Slavotinek, A.M.; Moshrefi, A.; Jiminez, N.L.; Chao, R.; Mendell, A.; Shaw, G.M.; Pennacchio, L.A.; Bates, M.D. Sequence variants in theHLXgene at chromosome 1q41-1q42 in patients with diaphragmatic hernia. Clin. Genet. 2009, 75, 429–439. [Google Scholar] [CrossRef]
- Hentsch, B.; Lyons, I.; Li, R.; Hartley, L.; Lints, T.J.; Adams, J.M.; Harvey, R.P. Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev. 1996, 10, 70–79. [Google Scholar] [CrossRef]
- Farrell, S.A.; Sodhi, S.; Marshall, C.R.; Guerin, A.; Slavotinek, A.; Paton, T.; Chong, K.; Sirkin, W.L.; Scherer, S.W.; Bérubé-Simard, F.-A.; et al. HLX is a candidate gene for a pattern of anomalies associated with congenital diaphragmatic hernia, short bowel, and asplenia. Am. J. Med. Genet. Part. A 2017, 173, 3070–3074. [Google Scholar] [CrossRef]
- Kantarci, S.; Ackerman, K.G.; Russell, M.K.; Longoni, M.; Sougnez, C.; Noonan, K.M.; Hatchwell, E.; Zhang, X.; Vanmarcke, R.P.; Anyane-Yeboa, K.; et al. Characterization of the chromosome 1q41q42.12 region, and the candidate gene DISP1, in patients with CDH. Am. J. Med. Genet. Part. A 2010, 152A, 2493–2504. [Google Scholar] [CrossRef]
- Clark, R.D.; Fenner-Gonzales, M. Apparent Fryns syndrome in a boy with a tandem duplication of 1q24-31. Am. J. Med. Genet. 1989, 34, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.E.X.; Lu, J.; Tso, P.; Blaner, W.S.; Levin, M.S.; Li, E. Increased Neonatal Mortality in Mice Lacking Cellular Retinol-binding Protein II. J. Biol. Chem. 2002, 277, 36617–36623. [Google Scholar] [CrossRef]
- Greer, J.J.; Babiuk, R.P.; Thébaud, B. Etiology of Congenital Diaphragmatic Hernia: The Retinoid Hypothesis. Pediatr. Res. 2003, 53, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, J.; Brown, J.; Masters, K.G.; Wright, C.; English, C.J. Blepharophimosis sequence and diaphragmatic hernia associated with interstitial deletion of chromosome 3 (46,XY,del(3)(q21q23)). J. Med. Genet. 1994, 31, 647–648. [Google Scholar] [CrossRef][Green Version]
- Dillon, E.; Renwick, M.; Wright, C. Congenital diaphragmatic herniation: Antenatal detection and outcome. Br. J. Radiol. 2000, 73, 360–365. [Google Scholar] [CrossRef]
- Hirschhorn, K.; Cooper, H.L.; Firschein, I.L. Deletion of short arms of chromosome 4–5 in a child with defects of midline fusion. Qual. Life Res. 1965, 1, 479–482. [Google Scholar] [CrossRef]
- Wolf, U.; Reinwein, H.; Porsch, R.; Schröter, R.; Baitsch, H. Deficiency on the short arms of a chromosome No. 4. Humangenetik 1965, 1, 397–413. [Google Scholar]
- Battaglia, A.; Filippi, T.; Carey, J.C. Update on the clinical features and natural history of Wolf-Hirschhorn (4p-) syndrome: Experience with 87 patients and recommendations for routine health supervision. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2008, 148C, 246–251. [Google Scholar] [CrossRef]
- Casaccia, G.; Mobili, L.; Braguglia, A.; Santoro, F.; Bagolan, P. Distal 4p microdeletion in a case of Wolf-Hirschhorn syndrome with congenital diaphragmatic hernia. Birth Defects Res. Part. A Clin. Mol. Teratol. 2006, 76, 210–213. [Google Scholar] [CrossRef]
- Van Dooren, M.; Brooks, A.; Hoogeboom, A.; Hoonaard, T.V.D.; De Klein, J.; Wouters, C.; Tibboel, D. Early diagnosis of Wolf-Hirschhorn syndrome triggered by a life-threatening event: Congenital diaphragmatic hernia. Am. J. Med. Genet. 2004, 127A, 194–196. [Google Scholar] [CrossRef]
- Tachdjian, G.; Fondacci, G.; TAPlA, S.; Huten, Y.; Blot, P.; Nessmann, C. The Wolf-Hirschhorn syndrome in fetuses. Clin. Genet. 2008, 42, 281–287. [Google Scholar] [CrossRef]
- Sergi, C.; Schulze, B.R.; Hager, H.D.; Beedgen, B.; Zilow, E.; Linderkamp, O.; Otto, H.F.; Tariverdian, G. Wolf-Hirschhorn syndrome: Case report and review of the chromosomal aberrations associated with diaphragmatic defects. Pathology 1998, 90, 285–293. [Google Scholar]
- Batanian, J.R.; Grange, D.K.; Fleming, R.; Gadre, B.; Wetzel, J. Two unbalanced translocations involving a common 6p25 region in two XY female patients. Clin. Genet. 2001, 59, 52–57. [Google Scholar] [CrossRef]
- Temple, I.K.; Barber, J.C.; James, R.S.; Burge, D. Diaphragmatic herniae and translocations involving 8q22 in two patients. J. Med. Genet. 1994, 31, 735–737. [Google Scholar] [CrossRef][Green Version]
- Longoni, M.; Russell, M.; High, F.; Darvishi, K.; Maalouf, F.; Kashani, A.; Tracy, A.; Coletti, C.; Loscertales, M.; Lage, K.; et al. Prevalence and penetrance ofZFPM2mutations and deletions causing congenital diaphragmatic hernia. Clin. Genet. 2015, 87, 362–367. [Google Scholar] [CrossRef]
- Longoni, M.; Lage, K.; Russell, M.K.; Loscertales, M.; Abdul-Rahman, O.A.; Baynam, G.; Bleyl, S.B.; Brady, P.D.; Breckpot, J.; Chen, C.P.; et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am. J. Med. Genet. Part. A 2012, 158A, 3148–3158. [Google Scholar] [CrossRef]
- Wat, M.J.; Shchelochkov, O.A.; Holder, A.M.; Breman, A.M.; Dagli, A.; Bacino, C.; Scaglia, F.; Zori, R.T.; Cheung, S.W.; Scott, D.A.; et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am. J. Med. Genet. Part. A 2009, 149A, 1661–1677. [Google Scholar] [CrossRef]
- Shimokawa, O.; Miyake, N.; Yoshimura, T.; Sosonkina, N.; Harada, N.; Mizuguchi, T.; Kondoh, S.; Kishino, T.; Ohta, T.; Remco, V.; et al. Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am. J. Med. Genet. Part. A 2005, 136A, 49–51. [Google Scholar] [CrossRef]
- Pecile, V.; Petroni, M.G.; Fertz, M.C.; Filippi, G. Deficiency of distal 8p -: Report of two cases and review of the literature. Clin. Genet. 2008, 37, 271–278. [Google Scholar] [CrossRef]
- Faivre, L.; Morichon-Delvallez, N.; Viot, G.; Narcy, F.; Loison, S.; Mandelbrot, L.; Aubry, M.C.; Raclin, V.; Edery, P.; Munnich, A.; et al. Prenatal diagnosis of an 8p23.1 deletion in a fetus with a diaphragmatic hernia and review of the literature. Prenat. Diagn. 1998, 18, 1055–1060. [Google Scholar] [CrossRef]
- Borys, D.; Taxy, J.B. Congenital Diaphragmatic Hernia and Chromosomal Anomalies: Autopsy Study. Pediatr. Dev. Pathol. 2004, 7, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, K.G.; Wang, J.; Luo, L.; Fujiwara, Y.; Orkin, S.H.; Beier, D.R. Gata4Is Necessary for Normal Pulmonary Lobar Development. Am. J. Respir. Cell Mol. Biol. 2007, 36, 391–397. [Google Scholar] [CrossRef] [PubMed]
- MorenoFuenmayor, H.; Meilinger, K.L.; Rucknagel, D.L.; Mohrenweiser, H.L.; Chu, E.H.Y.; Optiz, J.M. Duplication 8p syndrome: Studies in a family with a reciprocal translocation between chromosomes 8 and 12. Am. J. Med. Genet. 1980, 7, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Moog, U.; Engelen, J.J.M.; Albrechts, J.C.M.; Baars, L.G.M.; de Die-Smulders, C.E.M. Familial dup(8)(p12p21.1): Mild phenotypic effect and review of partial 8p duplications. Am. J. Med. Genet. 2000, 94, 306–310. [Google Scholar] [CrossRef]
- Ringer, K.; Rogers, J.; Pasztor, L.M. Inversion duplication of chromosome 8 with diaphragmatic hernia. Am. J. Human Genet. Suppl. 1995, 57, A124107. [Google Scholar]
- Liberfarb, R.M.; Atkins, L.; Holmes, L.B. A clinical syndrome associated with 5p duplication and 9p deletion. Ann. Génét. 1980, 23, 26–30. [Google Scholar]
- Donnenfeld, A.E.; Campbell, T.J.; Byers, J.; Librizzi, R.J.; Weiner, S. Tissue-specific mosaicism among fetuses with prenatally diagnosed diaphragmatic hernia. Am. J. Obstet. Gynecol. 1993, 169, 1017–1021. [Google Scholar] [CrossRef]
- Scott, D.; Cooper, M.; Stankiewicz, P.; Patel, A.; Potocki, L.; Cheung, S. Congenital diaphragmatic hernia in WAGR syndrome. Am. J. Med. Genet. Part. A 2005, 134A, 430–433. [Google Scholar] [CrossRef]
- Gustavson, K.-H.; Anneren, G.; Wranne, L. Two cases of 11p13 interstitial deletion and unusual clinical features. Clin. Genet. 2008, 26, 247–249. [Google Scholar] [CrossRef]
- Carmona, R.; Cañete, A.; Cano, E.; Ariza, L.; Rojas, A.; Muñoz-Chápuli, R. Conditional deletion of WT1 in the septum transversum mesenchyme causes congenital diaphragmatic hernia in mice. eLife 2016, 5, e16009. [Google Scholar] [CrossRef]
- Klaassens, M.; Scott, D.; van Dooren, M.; Hochstenbach, R.; Eussen, H.; Cai, W.; Galjaard, R.; Wouters, C.; Poot, M.; Laudy, J.; et al. Congenital diaphragmatic hernia associated with duplication of 11q23-qter. Am. J. Med. Genet. Part. A 2006, 140A, 1580–1586. [Google Scholar] [CrossRef]
- Park, J.P.; McDermet, M.K.; Doody, A.M.; Moeschler, J.B.; Marin-Padilla, J.M.; Wurster-Hill, D.H. Familial t(11;13)(q21;q14) and the duplication 11q, 13q phenotype. Am. J. Med. Genet. 1993, 45, 46–48. [Google Scholar] [CrossRef]
- Klaassens, M.; Van Dooren, M.; Eussen, H.J.; Douben, H.; Dekker, A.T.D.; Lee, C.; Donahoe, P.K.; Galjaard, R.J.; Goemaere, N.; De Krijger, R.R.; et al. Congenital diaphragmatic hernia and chromosome 15q26: Determination of a Candidate Region by Use of Fluorescent In Situ Hybridization and Array-Based COmparative Genomic Hybridization. Am. J. Hum. Genet. 2005, 76, 877–882. [Google Scholar] [CrossRef]
- Marguet, F.; Vezain, M.; Marcorelles, P.; Audebert-Bellanger, S.; Cassinari, K.; Drouot, N.; Chambon, P.; Gonzalez, B.J.; Horowitz, A.; Laquerriere, A.; et al. Neuropathological hallmarks of fetal hydrocephalus linked to CCDC88C pathogenic variants. Acta Neuropathol. Commun. 2021, 9, 104. [Google Scholar] [CrossRef]
- Enns, G.M.; Cox, V.A.; Goldstein, R.B.; Gibbs, D.L.; Harrison, M.R.; Golabi, M. Congenital diaphragmatic defects and associated syn-dromes, malformations, and chromosome anomalies: A retrospective study of 60 patients and literature review. Am. J. Med. Genet. 1998, 79, 215. [Google Scholar] [CrossRef] [PubMed]
- Kardon, G.; Ackerman, K.G.; McCulley, D.J.; Shen, Y.; Wynn, J.; Shang, L.; Bogenschutz, E.; Sun, X.; Chung, W.K. Congenital diaphragmatic hernias: From genes to mechanisms to therapies. Dis. Model. Mech. 2017, 10, 955–970. [Google Scholar] [CrossRef]
- Pilia, G.; Hughes-Benzie, R.M.; MacKenzie, A.E.; Baybayan, P.; Chen, E.Y.; Huber, R.; Neri, G.; Cao, A.; Forabosco, A.; Schlessinger, D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 1996, 12, 241–247. [Google Scholar] [CrossRef]
- Cottereau, E.; Mortemousque, I.; Moizard, M.-P.; Bürglen, L.; Lacombe, D.; Gilbert-Dussardier, B.; Sigaudy, S.; Boute, O.; David, A.; Faivre, L.; et al. Phenotypic Spectrum of Simpson-Golabi-Behmel Syndrome in a Series of 42 Cases with a Mutation in GPC 3 and Review of the Literature. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2013, 163, 92–105. [Google Scholar] [CrossRef]
- Chen, E.; Johnson, J.P.; Cox, V.A.; Golabi, M. Simpson-Golabi-Behmel syndrome: Congenital diaphragmatic hernia and radiologic findings in two patients and follow-up of a previously reported case. Am. J. Med. Genet. 1993, 46, 574–578. [Google Scholar] [CrossRef]
- Twigg, S.R.F.; Kan, R.; Babbs, C.; Bochukova, E.G.; Robertson, S.P.; Wall, S.A.; Morriss-Kay, G.M.; Wilkie, A.O.M. Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8652–8657. [Google Scholar] [CrossRef]
- Vasudevan, P.C.; Twigg, S.R.F.; Mulliken, J.B.; Cook, J.A.; Quarrell, O.W.J.; Wilkie, A.O.M. Expanding the phenotype of craniofrontonasal syndrome: Two unrelated boys with EFNB1 mutations and congenital diaphragmatic hernia. Eur. J. Hum. Genet. 2006, 14, 884–887. [Google Scholar] [CrossRef]
- Brooks, A.S.; Van Dooren, M.; Hoogeboom, J.; Gischler, S.; Willems, P.J.; Tibboel, D. Congenital diaphragmatic hernia in a female patient with craniofrontonasal syndrome. Clin. Dysmorphol. 2002, 11, 151–153. [Google Scholar] [CrossRef]
- Laporte, J.; Hu, L.J.; Kretz, C.; Mandel, J.-L.; Kioschis, P.; Coy, J.F.; Klauck, S.M.; Poustka, A.; Dahl, N. A gene mutated in X–linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 1996, 13, 175–182. [Google Scholar] [CrossRef]
- Grogan, P.M.; Tanner, S.M.; Orstavik, K.H.; Knudsen, G.; Saperstein, D.S.; Vogel, H.; Barohn, R.J.; Herbelin, L.L.; McVey, A.L.; Katz, J.S. Myopathy with skeletal asymmetry and hemidiaphragm elevation is caused by myotubularin mutations. Neurology 2005, 64, 1638–1640. [Google Scholar] [CrossRef]
- Quaderi, N.A.; Schweiger, S.; Gaudenz, K.; Franco, B.; Rugarli, E.; Berger, W.; Feldman, G.J.; Volta, M.; Andolfi, G.; Gilgenkrantz, S.; et al. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp. Nat. Genet. 1997, 17, 285–291. [Google Scholar] [CrossRef]
- Taylor, J.; Aftimos, S. Congenital diaphragmatic hernia is a feature of Opitz G/BBB syndrome. Clin. Dysmorphol. 2010, 19, 225–226. [Google Scholar] [CrossRef]
- Lowe, C.; Terrey, M.; MacLachlan, E.A. Organic-aciduria, decreased renal ammonia production, hydrophthalmos, and mental retardationa clinical entity. Arch. Pediatr. Adolesc. Med. 1952, 83, 164. [Google Scholar] [CrossRef]
- Loi, M. Lowe syndrome. Orphanet. J. Rare Dis. 2006, 1, 16. [Google Scholar] [CrossRef]
- Race, H.J.; Elhadi, N.; Holder, S.E. Congenital diaphragmatic hernia in Lowe syndrome: A rare association? Clin. Dysmorphol. 2010, 19, 226. [Google Scholar] [CrossRef]
- Wang, X.; Sutton, V.R.; Peraza-Llanes, J.O.; Yu, Z.; Rosetta, R.; Kou, Y.-C.; Eble, T.N.; Patel, A.; Thaller, C.; Fang, P.; et al. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat. Genet. 2007, 39, 836–838. [Google Scholar] [CrossRef] [PubMed]
- Goltz, R.W.; Peterson, W.C.; Gorlin, R.J.; Ravits, H.G. Focal Dermal Hypoplasia. Arch. Dermatol. 1962, 86, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jin, X.; Zhao, X.; Liu, D.; Hu, T.; Li, W.; Jiang, L.; Dan, H.; Zeng, X.; Chen, Q. Focal dermal hypoplasia: Updates. Oral Dis. 2013, 20, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Smigiel, R.; Jakubiak, A.; Lombardi, M.P.; Jaworski, W.; Slezak, R.; Patkowski, D.; Hennekam, R.C. Co-occurrence of severe Goltz-Gorlin syndrome and pentalogy of Cantrell—Case report and review of the literature. Am. J. Med. Genet. Part. A 2011, 155, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.M.; Lombardi, M.P.; Van Essen, A.J.; Wakeling, E.; Castle, B.; Temple, I.K.; Kumar, V.K.A.; Writzl, K.; Hennekam, R.C.M. Phenotype and genotype in 17 patients with Goltz-Gorlin syndrome. J. Med. Genet. 2009, 46, 716–720. [Google Scholar] [CrossRef]
- Wimplinger, I.; Morleo, M.; Rosenberger, G.; Iaconis, D.; Orth, U.; Meinecke, P.; Lerer, I.; Ballabio, A.; Gal, A.; Franco, B.; et al. Mutations of the Mitochondrial Holocytochrome c–Type Synthase in X-Linked Dominant Microphthalmia with Linear Skin Defects Syndrome. Am. J. Hum. Genet. 2006, 79, 878–889. [Google Scholar] [CrossRef]
- Veyver, I.V.D. Microphthalmia with linear skin defects (MLS), Aicardi, and Goltz syndromes: Are they related X-linked dominant male-lethal disorders? Cytogenet. Genome Res. 2002, 99, 289–296. [Google Scholar] [CrossRef]
- Krantz, I.D.; McCallum, J.; De Scipio, C.; Kaur, M.; Gillis, L.A.; Yaeger, D.; Jukofsky, L.; Wasserman, N.; Bottani, A.; Morris, C.A.; et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 2004, 36, 631–635. [Google Scholar] [CrossRef]
- Ireland, M.; Donnai, D.; Burn, J. Brachmann-de Lange syndrome. Delineation of the clinical phenotype. Am. J. Med. Genet. 1993, 47, 959–964. [Google Scholar] [CrossRef]
- Martínez-Frías, M.L.; Bermejo, E.; Félix, V.; Jiménez, N.; Gómez-Ullate, J.; López, J.A.; Aparicio, P.; Ayala, A.; Gairi, J.M.; Galán, E.; et al. Síndrome de Brachmann de Lange en nuestro medio: Características clínicas y epidemiológicas [Brachmann-de-Lange syndrome in our population: Clinical and epidemiological characteristics]. An. Esp. Pediatr. 1998, 48, 293–298. (In Spanish) [Google Scholar]
- Marino, T.; Wheeler, P.G.; Simpson, L.L.; Craigo, S.D.; Bianchi, D.W. Fetal diaphragmatic hernia and upper limb anomalies suggest Brachmann-de Lange syndrome. Prenat. Diagn. 2002, 22, 144–147. [Google Scholar] [CrossRef]
- Jelsema, R.D.; Isada, N.B.; Kazzi, N.J.; Sargent, K.; Harrison, M.R.; Johnson, M.P.; Evans, M.I. Prenatal diagnosis of congenital diaphragmatic hernia not amenable to prenatal or neonatal repair: Brachmann-de Lange syndrome. Am. J. Med. Genet. 1993, 47, 1022–1023. [Google Scholar] [CrossRef]
- Schrier, S.A.; Sherer, I.; Deardorff, M.A.; Clark, D.; Audette, L.; Gillis, L.; Kline, A.D.; Ernst, L.; Loomes, K.; Krantz, I.D.; et al. Causes of death and autopsy findings in a large study cohort of individuals with Cornelia de Lange syndrome and review of the literature. Am. J. Med. Genet. Part. A 2011, 155, 3007–3024. [Google Scholar] [CrossRef]
- Pelletier, J.; Bruening, W.; Kashtan, C.E.; Mauer, S.M.; Manivel, J.C.; Striegel, J.E.; Houghton, D.C.; Junien, C.; Habib, R.; Fouser, L. Germline mutations in the Wilms’ tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell 1991, 67, 437–447. [Google Scholar] [CrossRef]
- Devriendt, K.; DeLoof, E.; Moerman, P.; Legius, E.; Vanhole, C.; De Zegher, F.; Proesmans, W.; Devlieger, H. Diaphragmatic hernia in Denys-Drash syndrome. Am. J. Med. Genet. 1995, 57, 97–101. [Google Scholar] [CrossRef]
- Cho, H.Y.; Lee, B.S.; Kang, C.H.; Kim, W.-H.; Ha, I.S.; Cheong, H.I.; Choi, Y. Hydrothorax in a patient with Denys-Drash syndrome associated with a diaphragmatic defect. Pediatr. Nephrol. 2006, 21, 1909–1912. [Google Scholar] [CrossRef]
- Antonius, T.; van Bon, B.; Eggink, A.; van der Burgt, I.; Noordam, K.; van Heijst, A. Denys–Drash syndrome and congenital diaphragmatic hernia: Another case with the 1097G > A(Arg366His) mutation. Am. J. Med. Genet. Part. A 2008, 146A, 496–499. [Google Scholar] [CrossRef]
- Faivre, L.; Collod-Beroud, G.; Loeys, B.; Child, A.; Binquet, C.; Gautier, E.; Callewaert, B.; Arbustini, E.; Mayer, K.; Arslan-Kirchner, M.; et al. Effect of Mutation Type and Location on Clinical Outcome in 1,013 Probands with Marfan Syndrome or Related Phenotypes and FBN1 Mutations: An International Study. Am. J. Hum. Genet. 2007, 81, 454–466. [Google Scholar] [CrossRef]
- Aubart, M.; Gazal, S.; Arnaud, P.; Benarroch, L.; Gross, M.-S.; Buratti, J.; Boland, A.; Meyer, V.; Zouali, H.; Hanna, N.; et al. Association of modifiers and other genetic factors explain Marfan syndrome clinical variability. Eur. J. Hum. Genet. 2018, 26, 1759–1772. [Google Scholar] [CrossRef]
- Universal Mutation Database (UMD) FBN. Available online: http://www.umd.be/FBN1/;http://www.umd.be/FBN1/4DACTION/w_mutations (accessed on 27 November 2020).
- Jacobs, A.M.; Toudjarska, I.; Racine, A.; Tsipouras, P.; Kilpatrick, M.W.; Shanske, A. A Recurring FBN1 Gene Mutation in Neonatal Marfan Syndrome. Arch. Pediatr. Adolesc. Med. 2002, 156, 1081–1085. [Google Scholar] [CrossRef]
- Revencu, N.; Quenum, G.; Detaille, T.; Verellen, G.; De Paepe, A.; Verellen-Dumoulin, C. Congenital diaphragmatic eventration and bilateral uretero-hydronephrosis in a patient with neonatal Marfan syndrome caused by a mutation in exon 25 of the FBN1 gene and review of the literature. Eur. J. Nucl. Med. Mol. Imaging 2003, 163, 33–37. [Google Scholar] [CrossRef]
- Veiga-Fernández, A.; Prieto, L.J.; Álvarez, T.; Ruiz, Y.; Pérez, R.; Gámez, F.; Abad, V.O.; Yllana, F.; De León-Luis, J. Perinatal diagnosis and management of early-onset Marfan syndrome: Case report and systematic review. J. Matern. Neonatal Med. 2019, 33, 2493–2504. [Google Scholar] [CrossRef]
- Bergman, J.E.; Janssen, N.; Hoefsloot, L.H.; Jongmans, M.C.; Hofstra, R.M.; Van Ravenswaaij-Arts, C.M.A. CHD7 mutations and CHARGE syndrome: The clinical implications of an expanding phenotype. J. Med. Genet. 2011, 48, 334–342. [Google Scholar] [CrossRef]
- Janssen, N.; Bergman, J.E.H.; Swertz, M.; Tranebjaerg, L.; Lodahl, M.; Schoots, J.; Hofstra, R.M.W.; van Ravenswaaij-Arts, C.M.A.; Hoefsloot, L.H. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum. Mutat. 2012, 33, 1149–1160. [Google Scholar] [CrossRef]
- Pagon, R.A.; Graham, J.M.; Zonana, J.; Yong, S.-L. Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J. Pediatr. 1981, 99, 223–227. [Google Scholar] [CrossRef]
- Hall, B.D. Choanal atresia and associated multiple anomalies. J. Pediatr. 1979, 95, 395–398. [Google Scholar] [CrossRef]
- Blake, K.D.; Davenport, S.L.H.; Hall, B.D.; Hefner, M.A.; Pagon, R.A.; Williams, M.S.; Lin, A.E.; Graham, J.J.M. CHARGE Association: An Update and Review for the Primary Pediatrician. Clin. Pediatr. 1998, 37, 159–173. [Google Scholar] [CrossRef]
- Verloes, A. Updated diagnostic criteria for CHARGE syndrome: A proposal. Am. J. Med. Genet. Part. A 2005, 133A, 306–308. [Google Scholar] [CrossRef]
- Casaccia, G.; Digilio, M.C.; Seymandi, P.L.; Bagolan, P. Congenital diaphragmatic hernia in CHARGE syndrome. Pediatr. Surg. Int. 2007, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Jongmans, M.C.J.; Admiraal, R.J.; Van Der Donk, K.P.; Vissers, L.E.L.M.; Baas, A.F.; Kapusta, L.; Van Hagen, J.M.; Donnai, D.; De Ravel, T.J.; Veltman, J.; et al. CHARGE syndrome: The phenotypic spectrum of mutations in the CHD7 gene. J. Med. Genet. 2005, 43, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Philip, N.; Gambarelli, D.; Guys, J.M.; Camboulives, J.; Ayme, S. Epidemiological study of congenital diaphragmatic defects with special reference to aetiology. Eur. J. Nucl. Med. Mol. Imaging 1991, 150, 726–729. [Google Scholar] [CrossRef] [PubMed]
- Dillon, E.; Renwick, M. Antenatal detection of congenital diaphragmatic hernias: The northern region experience. Clin. Radiol. 1993, 48, 264–267. [Google Scholar] [CrossRef]
- Neville, H.L.; Jaksic, T.; Wilson, J.M.; Lally, P.A.; Hardin, W.D.; Hirschl, R.B.; Langham, M.R.; Lally, K.P. Fryns syndrome in children with congenital diaphragmatic hernia. J. Pediatr. Surg. 2002, 37, 1685–1687. [Google Scholar] [CrossRef]
- Brady, P.; Moerman, P.; De Catte, L.; Deprest, J.; Devriendt, K.; Vermeesch, J. Exome sequencing identifies a recessive PIGN splice site mutation as a cause of syndromic Congenital Diaphragmatic Hernia. Eur. J. Med. Genet. 2014, 57, 487–493. [Google Scholar] [CrossRef]
- McInerney-Leo, A.M.; Harris, J.E.; Gattas, M.; Peach, E.E.; Sinnott, S.; Dudding-Byth, T.; Rajagopalan, S.; Barnett, C.P.; Anderson, L.K.; Wheeler, L.; et al. Fryns Syndrome Associated with Recessive Mutations in PIGN in two Separate Families. Hum. Mutat. 2016, 37, 695–702. [Google Scholar] [CrossRef]
- Lin, A.E.; Pober, B.R.; Mullen, M.P.; Slavotinek, A.M. Cardiovascular malformations in Fryns syndrome: Is there a pathogenic role for neural crest cells? Am. J. Med. Genet. Part. A 2005, 139, 186–193. [Google Scholar] [CrossRef]
- Fryns, J.P.; Moerman, F.; Goddeeris, P.; Bossuyt, C.; Berghe, H.V.D. A new lethal syndrome with cloudy corneae, diaphragmatic defects and distal limb deformities. Qual. Life Res. 1979, 50, 65–70. [Google Scholar] [CrossRef]
- Goddeeris, P.; Fryns, J.P.; van den Berghe, H. Diaphragmatic defects, craniofacial dysmorphism, cleft palate and distal limb de-formities—A new lethal syndrome. J. Genet. Hum. 1980, 28, 57. [Google Scholar] [PubMed]
- Donnai, D.; Barrow, M. Diaphragmatic hernia, exomphalos, absent corpus callosum, hypertelorism, myopia, and sensorineural deafness: A newly recognized autosomal recessive disorder? Am. J. Med. Genet. 1993, 47, 679–682. [Google Scholar] [CrossRef]
- Chassaing, N.; Lacombe, D.; Carles, D.; Calvas, P.; Saura, R.; Bieth, E. Donnai-Barrow syndrome: Four additional patients. Am. J. Med. Genet. 2003, 121A, 258–262. [Google Scholar] [CrossRef]
- Kantarci, S.; Al-Gazali, L.; Hill, R.S.; Donnai, D.; Black, G.C.M.; Bieth, E.; Chassaing, N.; Lacombe, D.; Devriendt, K.; Teebi, A.; et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat. Genet. 2007, 39, 957–959. [Google Scholar] [CrossRef]
- Raila, J.; Willnow, T.E.; Schweigert, F. Megalin-Mediated Reuptake of Retinol in the Kidneys of Mice Is Essential for Vitamin A Homeostasis. J. Nutr. 2005, 135, 2512–2516. [Google Scholar] [CrossRef]
- Kling, D.E.; Schnitzer, J.J. Vitamin A deficiency (VAD), teratogenic, and surgical models of congenital diaphragmatic hernia (CDH). Am. J. Med. Genet. Part. C Semin. Med. Genet. 2007, 145C, 139–157. [Google Scholar] [CrossRef]
- Khalifa, O.; Al-Sahlawi, Z.; Imtiaz, F.; Ramzan, K.; Allam, R.; Al-Mostafa, A.; Abdel-Fattah, M.; Abuharb, G.; Nester, M.; Verloes, A.; et al. Variable expression pattern in Donnai-Barrow syndrome: Report of two novel LRP2 mutations and review of the literature. Eur. J. Med. Genet. 2015, 58, 293–299. [Google Scholar] [CrossRef]
- Kantarci, S.; Donahoe, P.K. Congenital diaphragmatic hernia (CDH) etiology as revealed by pathway genetics. Am. J. Med. Genet. Part. C: Semin. Med. Genet. 2007, 145C, 217–226. [Google Scholar] [CrossRef]
- Goodman, D.S. The Retinoids; Sporn, M.B., Boberts, A.B., Goodman, D.S., Eds.; Academic Press: New York, NY, USA, 1984. [Google Scholar]
- Kawaguchi, R.; Yu, J.; Honda, J.; Hu, J.; Whitelegge, J.; Ping, P.; Wiita, P.; Bok, D.; Sun, H. A Membrane Receptor for Retinol Binding Protein Mediates Cellular Uptake of Vitamin A. Science 2007, 315, 820–825. [Google Scholar] [CrossRef]
- Pasutto, F.; Sticht, H.; Hammersen, G.; Gillessen-Kaesbach, G.; FitzPatrick, D.R.; Nürnberg, G.; Brasch, F.; Schirmer-Zimmermann, H.; Tolmie, J.L.; Chitayat, D.; et al. Mutations in STRA6 Cause a Broad Spectrum of Malformations Including Anophthalmia, Congenital Heart Defects, Diaphragmatic Hernia, Alveolar Capillary Dysplasia, Lung Hypoplasia, and Mental Retardation. Am. J. Hum. Genet. 2007, 80, 550–560. [Google Scholar] [CrossRef]
- Andijani, A.A.; Shajira, E.S.; Abushaheen, A.; Al-Matary, A. Microphthalmia Syndrome 9: Case Report of a Newborn Baby with Pulmonary Hypoplasia, Diaphragmatic Eventration, Microphthalmia, Cardiac Defect and Severe Primary Pulmonary Hypertension. Am. J. Case Rep. 2019, 20, 354–360. [Google Scholar] [CrossRef]
- Srour, M.; Chitayat, D.; Caron, V.; Chassaing, N.; Bitoun, P.; Patry, L.; Cordier, M.-P.; Capo-Chichi, J.-M.; Francannet, C.; Calvas, P.; et al. Recessive and Dominant Mutations in Retinoic Acid Receptor Beta in Cases with Microphthalmia and Diaphragmatic Hernia. Am. J. Hum. Genet. 2013, 93, 765–772. [Google Scholar] [CrossRef]
- Park, Y.; Gong, G.; Choe, G.; Yu, E.; Kim, K.S.; Lee, I. Jarcho-Levin syndrome: A report of an autopsy case with cytogenetic analysis. J. Korean Med. Sci. 1993, 8, 471–475. [Google Scholar] [CrossRef]
- Lam, Y.H.; Eik-Nes, S.H.; Tang, M.H.Y.; Lee, C.P.; Nicholls, J.M. Prenatal sonographic features of spondylocostal dysostosis and diaphragmatic hernia in the first trimester. Ultrasound Obstet. Gynecol. 1999, 13, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Day, R.; Fryer, A. Diaphragmatic hernia and preaxial polydactyly in spondylothoracic dysplasia. Clin. Dysmorphol. 2003, 12, 277–278. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, L.M.; García-García, I.; Correa-Rivas, M.S.; García-Fragoso, L. Pulmonary hypoplasia in Jarcho-Levin syndrome. Puerto Rico Health Sci. J. 2004, 23, 65–67. [Google Scholar]
- Cornier, A.; Ramírez, N.; Arroyo, S.; Acevedo, J.; García, L.; Carlo, S.; Korf, B. Phenotype characterization and natural history of spondylothoracic dysplasia syndrome: A series of 27 new cases. Am. J. Med. Genet. Part. A 2004, 128A, 120–126. [Google Scholar] [CrossRef]
- Turnpenny, P.D.; Alman, B.; Cornier, A.S.; Giampietro, P.F.; Offiah, A.; Tassy, O.; Pourquie, O.; Kusumi, K.; Dunwoodie, S. Abnormal vertebral segmentation and the notch signaling pathway in man. Dev. Dyn. 2007, 236, 1456–1474. [Google Scholar] [CrossRef]
- Bulman, M.P.; Kusumi, K.; Frayling, T.M.; McKeown, C.; Garrett, C.; Lander, E.S.; Krumlauf, R.; Hattersley, A.T.; Ellard, S.; Turnpenny, P.D. Mutations in the human Delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat. Genet. 2000, 24, 438–441. [Google Scholar] [CrossRef]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef]
- Yu, L.; Wynn, J.; Cheung, Y.H.; Shen, Y.; Mychaliska, G.B.; Crombleholme, T.M.; Azarow, K.S.; Lim, F.Y.; Chung, D.H.; Potoka, D.; et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Qual. Life Res. 2012, 132, 285–292. [Google Scholar] [CrossRef]
- Cai, K.Q.; Capo-Chichi, D.C.; Rula, M.E.; Yang, D.-H.; Xu, X.-X. Dynamic GATA6 expression in primitive endoderm formation and maturation in early mouse embryogenesis. Dev. Dyn. 2008, 237, 2820–2829. [Google Scholar] [CrossRef]
- Allen, H.L.; The International Pancreatic Agenesis Consortium; Flanagan, S.; Shaw-Smith, C.; De Franco, E.; Akerman, I.; Caswell, R.; Ferrer, J.; Hattersley, A.T.; Ellard, S. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat. Genet. 2011, 44, 20–22. [Google Scholar] [CrossRef]
- Keijzer, R.; van Tuyl, M.; Meijers, C.; Post, M.; Tibboel, D.; Grosveld, F.; Koutsourakis, M. The transcription factor GATA6 is essential for branching morphogenesis and epithelial cell differentiation during fetal pulmonary development. Development 2001, 128, 503–511. [Google Scholar] [CrossRef]
- Zhang, Y.; Goss, A.M.; Cohen, E.D.; Kadzik, R.; Lepore, J.J.; Muthukumaraswamy, K.; Yang, J.; DeMayo, F.; Whitsett, J.A.; Parmacek, M.S.; et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat. Genet. 2008, 40, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Kodo, K.; Nishizawa, T.; Furutani, M.; Arai, S.; Yamamura, E.; Joo, K.; Takahashi, T.; Matsuoka, R.; Yamagishi, H. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 13933–13938. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, M.; Shield, J.; Acerini, C.; Walker, J.; Ellard, S.; Marchand, M.; Polak, M.; Vaxillaire, M.; Crolla, J.; Bunyan, D.; et al. Pancreatic hypoplasia presenting with neonatal diabetes mellitus in association with congenital heart defect and developmental delay. Am. J. Med. Genet. Part. A 2010, 152A, 340–346. [Google Scholar] [CrossRef]
- Lin, X.; Huo, Z.; Liu, X.; Zhang, Y.; Li, L.; Zhao, H.; Yan, B.; Liu, Y.; Yang, Y.; Chen, Y.-H. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J. Hum. Genet. 2010, 55, 662–667. [Google Scholar] [CrossRef]
- Maitra, M.; Koenig, S.N.; Srivastava, D.; Garg, V. Identification of GATA6 Sequence Variants in Patients with Congenital Heart Defects. Pediatr. Res. 2010, 68, 281–285. [Google Scholar] [CrossRef]
- Zhao, R.; Watt, A.J.; Li, J.; Luebke-Wheeler, J.; Morrisey, E.E.; Duncan, S.A. GATA6 Is Essential for Embryonic Development of the Liver but Dispensable for Early Heart Formation. Mol. Cell. Biol. 2005, 25, 2622–2631. [Google Scholar] [CrossRef]
- Morrisey, E.E.; Tang, Z.; Sigrist, K.; Lu, M.M.; Jiang, F.; Ip, H.S.; Parmacek, M.S. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998, 12, 3579–3590. [Google Scholar] [CrossRef]
- Yu, L.; Bennett, J.T.; Wynn, J.; Carvill, G.L.; Cheung, Y.H.; Shen, Y.; Mychaliska, G.B.; Azarow, K.S.; Crombleholme, T.M.; Chung, D.H.; et al. Whole exome sequencing identifies de novo mutations inGATA6associated with congenital diaphragmatic hernia. J. Med. Genet. 2014, 51, 197–202. [Google Scholar] [CrossRef]
- Brady, P.D.; Van Houdt, J.; Callewaert, B.; Deprest, J.; Devriendt, K.; Vermeesch, J.R. Exome sequencing identifies ZFPM2 as a cause of familial isolated congenital diaphragmatic hernia and possibly cardiovascular malformations. Eur. J. Med. Genet. 2014, 57, 247–252. [Google Scholar] [CrossRef]
- Qiu, Y.; Krishnan, V.; Pereira, F.A.; Tsai, S.Y.; Tsai, M.J. Chicken ovalbumin upstream promoter-transcription factors and their reg-ulation. J. Steroid. Biochem. Mol. Biol. 1996, 56, 81–85. [Google Scholar] [CrossRef]
- You, L.-R.; Takamoto, N.; Yu, C.-T.; Tanaka, T.; Kodama, T.; DeMayo, F.; Tsai, S.Y.; Tsai, M.-J. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc. Natl. Acad. Sci. USA 2005, 102, 16351–16356. [Google Scholar] [CrossRef] [PubMed]
- High, F.A.; Bhayani, P.; Wilson, J.M.J.M.; Bult, C.J.; Donahoe, P.K.P.K.; Longoni, M. De Novo Frameshift Mutation in COUP-TFII (NR2F2) in Human Congenital Diaphragmatic Hernia. Am. J. Med. Genet. Part. A 2016, 170, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Clugston, R.D.; Zhang, W.; Greer, J.J. Gene expression in the developing diaphragm: Significance for congenital diaphragmatic hernia. Am. J. Physiol. Cell. Mol. Physiol. 2008, 294, L665–L675. [Google Scholar] [CrossRef]
- Hong, S.-H.; David, G.; Wong, C.-W.; Dejean, A.; Privalsky, M.L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor (RAR) and PLZF-RAR oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 1997, 94, 9028–9033. [Google Scholar] [CrossRef]
- Aljohani, O.; Haldeman, S.; Rao, R. Abstracts from the 11th International Conference on Neonatal and Childhood Pulmonary Vascular Disease. Pulm. Circ. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Rossetti, L.Z.; Glinton, K.; Yuan, B.; Liu, P.; Pillai, N.; Mizerik, E.; Magoulas, P.; Rosenfeld, J.A.; Karaviti, L.; Sutton, V.R.; et al. Review of the phenotypic spectrum associated with haploinsufficiency of MYRF. Am. J. Med. Genet. Part. A 2019, 179, 1376–1382. [Google Scholar] [CrossRef]
- Qi, H.; Yu, L.; Zhou, X.; Wynn, J.; Zhao, H.; Guo, Y.; Zhu, N.; Kitaygorodsky, A.; Hernan, R.; Aspelund, G.; et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018, 14, e1007822. [Google Scholar] [CrossRef]
- Slavotinek, A.M. Single gene disorders associated with congenital diaphragmatic hernia. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2007, 145C, 172–183. [Google Scholar] [CrossRef]
- Brady, P.D.; Dekoninck, P.; Fryns, J.P.; Devriendt, K.; Deprest, J.; Vermeesch, J.R. Identification of dosage-sensitive genes in fetuses referred with severe isolated congenital diaphragmatic hernia. Prenat. Diagn. 2013, 33, 1283–1292. [Google Scholar] [CrossRef]
- Yu, L.; Wynn, J.; Ma, L.; Guha, S.; Mychaliska, G.B.; Crombleholme, T.M.; Azarow, K.S.; Lim, F.Y.; Chung, D.H.; Potoka, D.; et al. De novo copy number variants are associated with congenital diaphragmatic hernia. J. Med. Genet. 2012, 49, 650–659. [Google Scholar] [CrossRef]
- Beurskens, N.; Klaassens, M.; Rottier, R.; de Klein, A.; Tibboel, D. Linking animal models to human congenital diaphragmatic hernia. Birth Defects Res. Part. A: Clin. Mol. Teratol. 2007, 79, 565–572. [Google Scholar] [CrossRef]
- Longoni, M.; High, F.A.; Qi, H.; Joy, M.P.; Hila, R.; Coletti, C.M.; Wynn, J.; Loscertales, M.; Shan, L.; Bult, C.J.; et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Qual. Life Res. 2017, 136, 679–691. [Google Scholar] [CrossRef]
- The Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nat. Cell Biol. 2015, 519, 223–228. [Google Scholar] [CrossRef]
- Yu, L.; Sawle, A.; Wynn, J.; Aspelund, G.; Stolar, C.J.; Arkovitz, M.S.; Potoka, D.; Azarow, K.S.; Mychaliska, G.B.; Shen, Y.; et al. Increased burden ofde novopredicted deleterious variants in complex congenital diaphragmatic hernia. Hum. Mol. Genet. 2015, 24, 4764–4773. [Google Scholar] [CrossRef]
- Ackerman, K.G.; Greer, J.J. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2007, 145C, 109–116. [Google Scholar] [CrossRef]
- Best, S.; Wou, K.; Vora, N.; Van Der Veyver, I.B.; Wapner, R.; Chitty, L.S. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenat. Diagn. 2018, 38, 10–19. [Google Scholar] [CrossRef]
- Meier, N.; Bruder, E.; Lapaire, O.; Hoesli, I.; Kang, A.; Hench, J.; Hoeller, S.; De Geyter, J.; Miny, P.; Heinimann, K.; et al. Exome sequencing of fetal anomaly syndromes: Novel phenotype–genotype discoveries. Eur. J. Hum. Genet. 2019, 27, 730–737. [Google Scholar] [CrossRef]
- Qiao, L.; Wynn, J.; Yu, L.; Ms, R.H.; Zhou, X.; Duron, V.; Aspelund, G.; Farkouh-Karoleski, C.; Zygumunt, A.; Krishnan, U.S.; et al. Likely damaging de novo variants in congenital diaphragmatic hernia patients are associated with worse clinical outcomes. Genet. Med. 2020, 22, 2020–2028. [Google Scholar] [CrossRef]
- Scott, T.M.; Campbell, I.M.; Hernandez-Garcia, A.; Lalani, S.R.; Liu, P.; Shaw, C.A.; Rosenfeld, J.A.; Scott, D.A. Clinical exome sequencing data reveal high diagnostic yields for congenital diaphragmatic hernia plus (CDH+) and new phenotypic expansions involving CDH. J. Med. Genet. 2021. [Google Scholar] [CrossRef]
- Nakamura, H.; Doi, T.; Puri, P.; Friedmacher, F. Transgenic animal models of congenital diaphragmatic hernia: A comprehensive overview of candidate genes and signaling pathways. Pediatr. Surg. Int. 2020, 36, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.M.; Debeer, A.; De Coppi, P.; Devriendt, K.; Crombag, N.; Hubble, T.; Power, B.; Benachi, A.; Deprest, J. What should we tell parents? Congenital diaphragmatic hernia. Prenat. Diagn. 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).