Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs
Abstract
1. Introduction
2. Results
2.1. VPS13D Cognate Mutants, but Not VPS13B or C, Disrupt a Subset of VPS13 Functions in Yeast
2.2. Vps13N2428S Localizes Normally to the Prospore Membrane
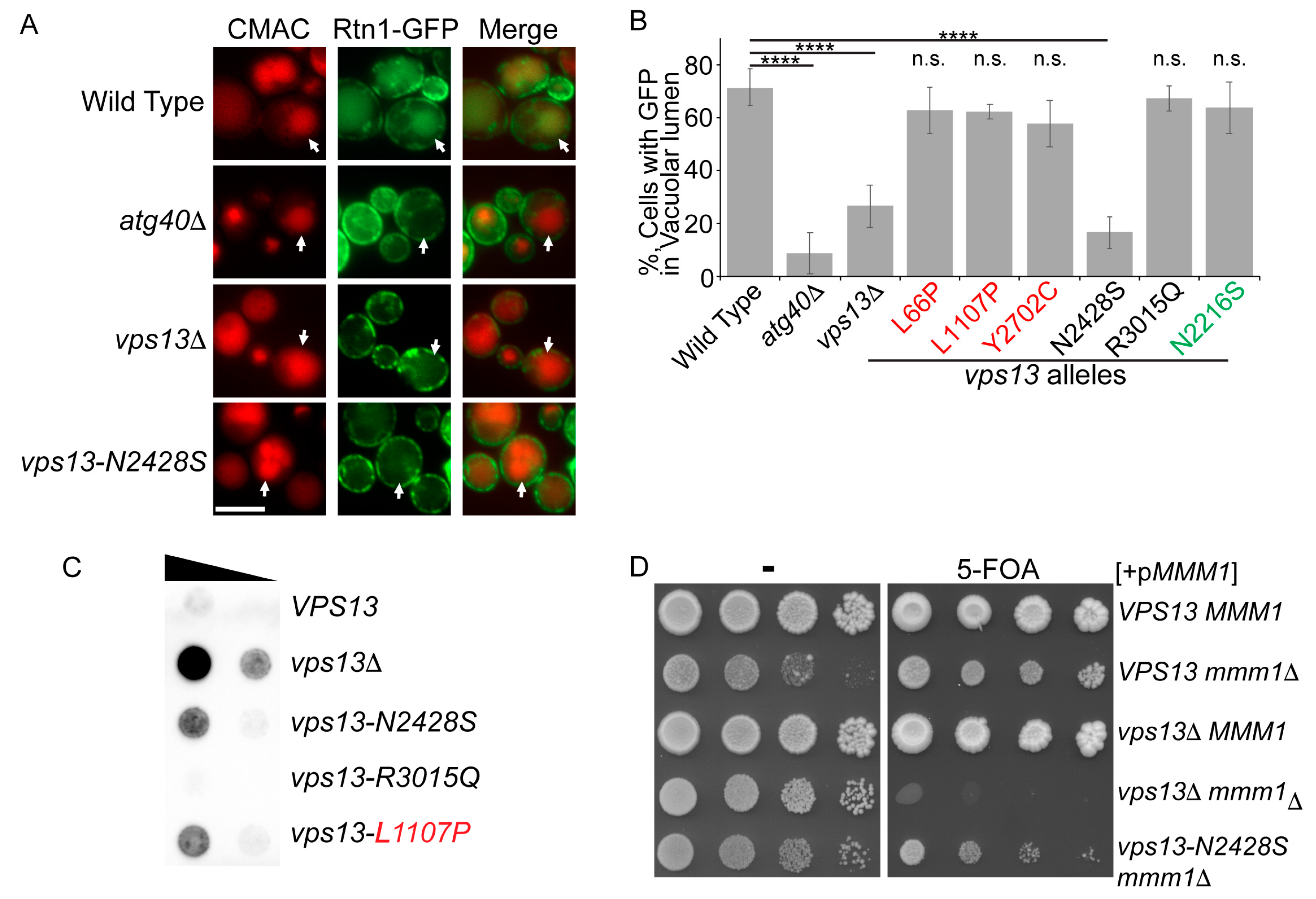
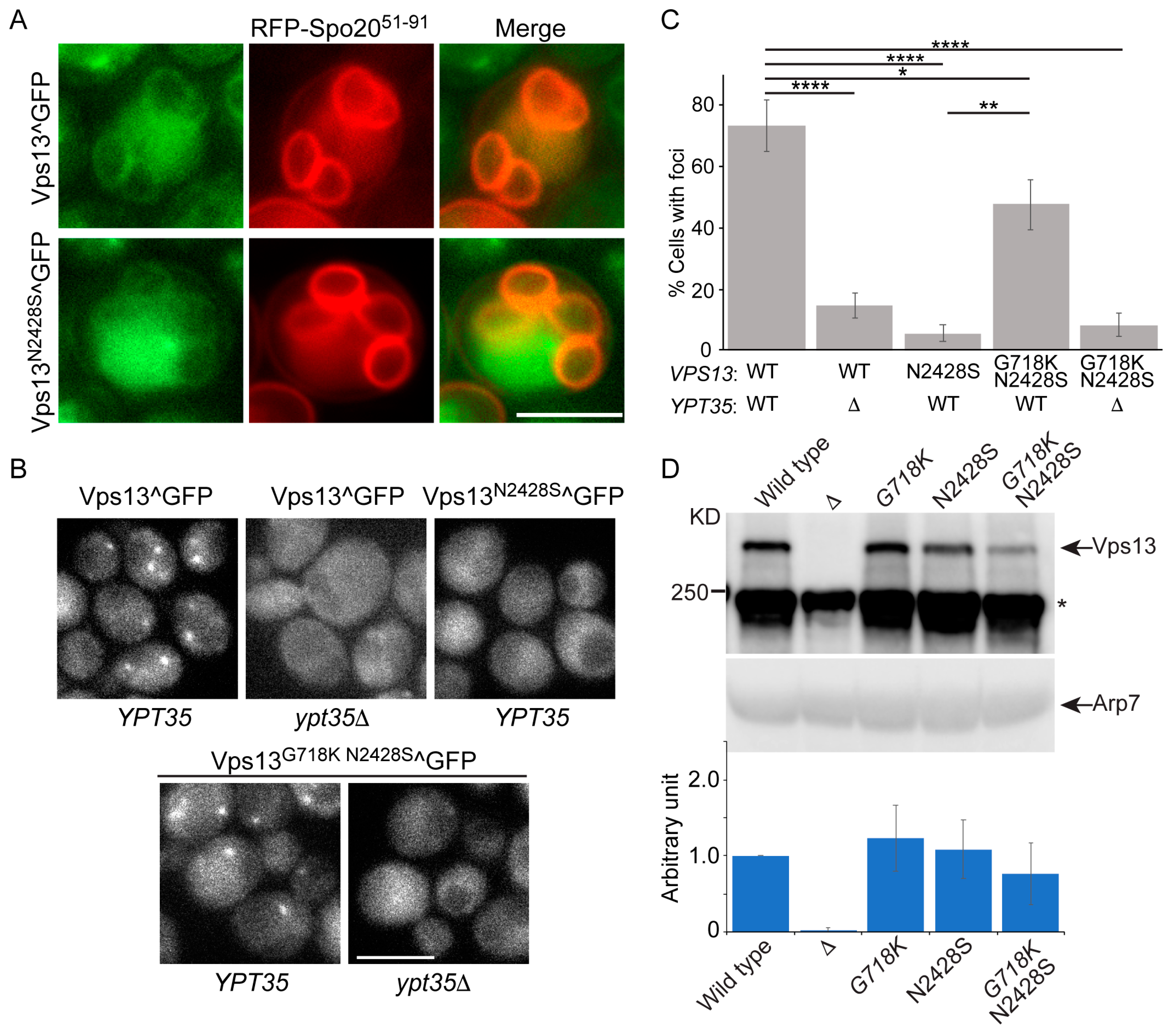
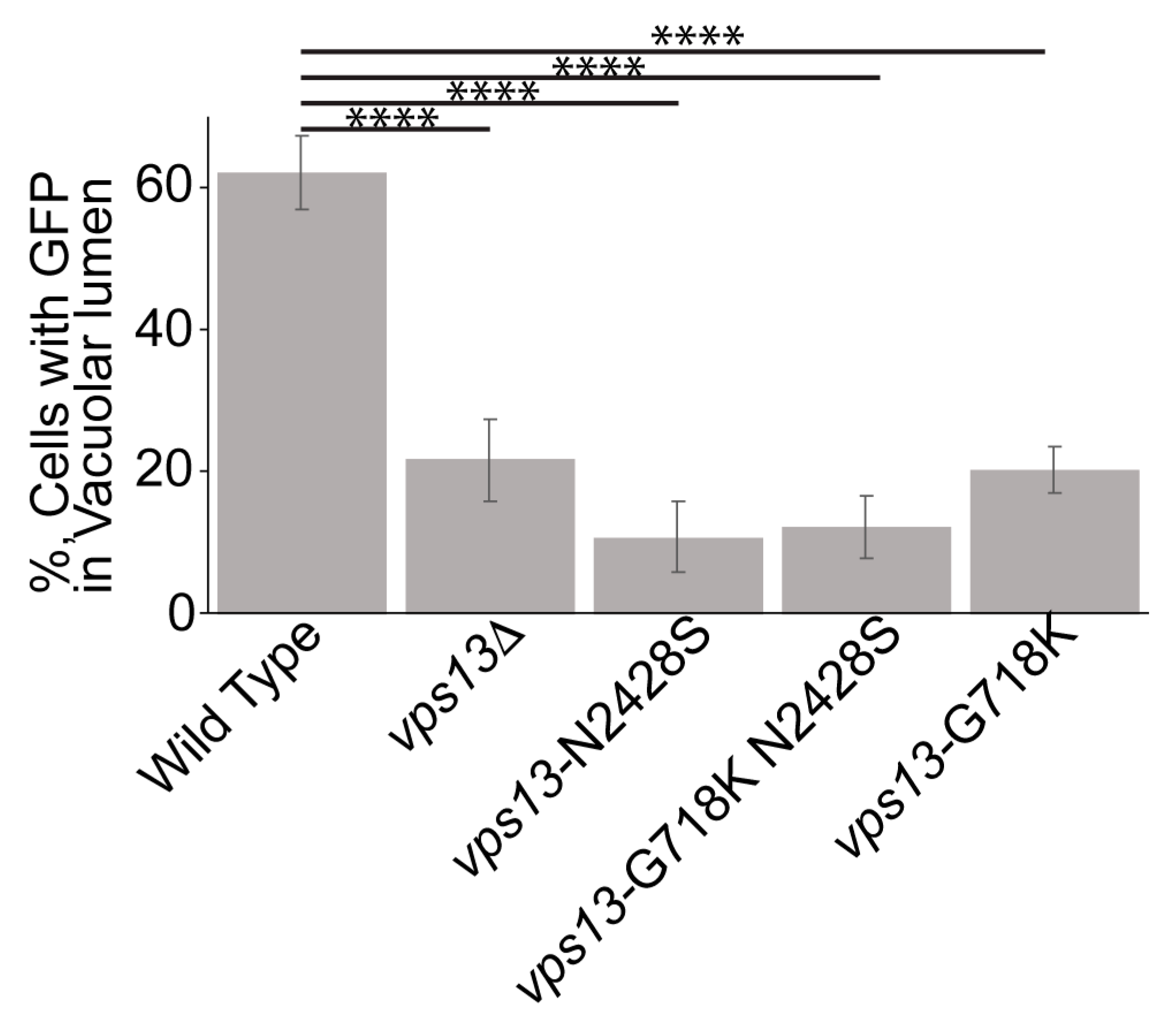
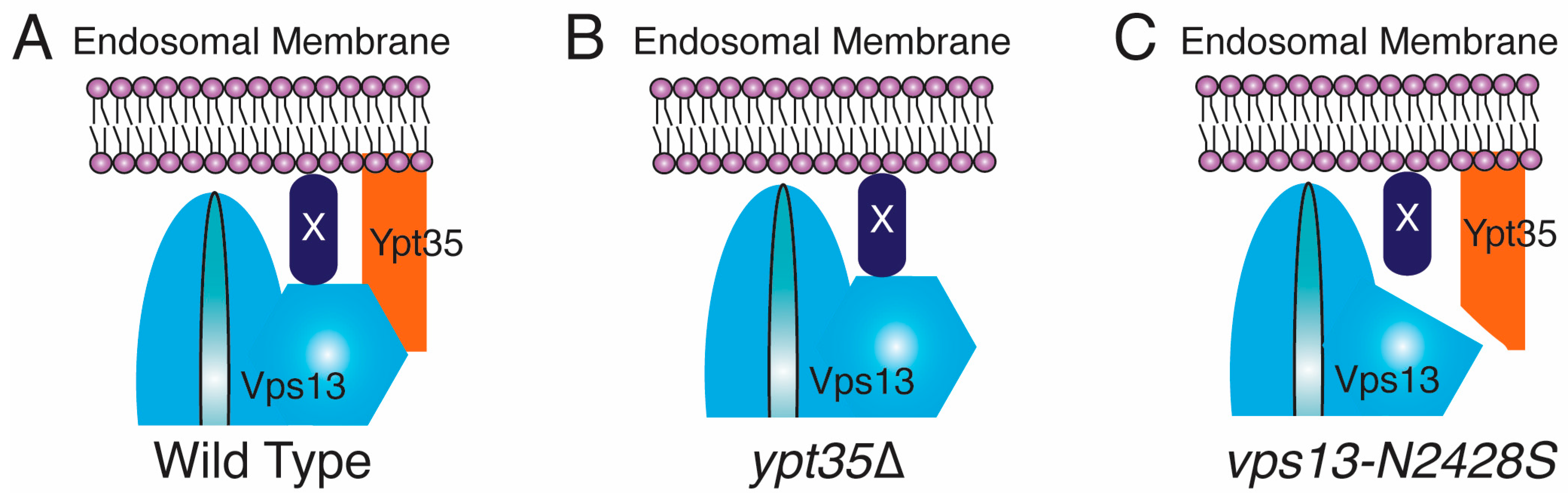
2.3. An Intragenic Mutation Restores Endosomal Localization to Vps13N2428S and Partially Suppresses the CPY Sorting Defect
2.4. The vps13-G718K Mutant Alone Is Defective in ER-phagy
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Media
4.2. Mitochondrial Function Assay
4.3. Vps13 Antibodies
4.4. Western Blot Analysis
4.5. CPY Secretion Assay
4.6. ER-phagy Assay
4.7. Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dziurdzik, S.K.; Conibear, E. The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites. Int. J. Mol. Sci. 2021, 22, 2905. [Google Scholar] [CrossRef]
- Gauthier, J.; Meijer, I.A.; Lessel, D.; Mencacci, N.E.; Krainc, D.; Hempel, M.; Tsiakas, K.; Prokisch, H.; Rossignol, E.; Helm, M.H.; et al. Recessive mutations in VPS13D cause childhood onset movement disorders. Ann. Neurol. 2018, 83, 1089–1095. [Google Scholar] [CrossRef]
- Kolehmainen, J.; Black, G.C.; Saarinen, A.; Chandler, K.; Clayton-Smith, J.; Traskelin, A.L.; Perveen, R.; Kivitie-Kallio, S.; Norio, R.; Warburg, M.; et al. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am. J. Hum. Genet. 2003, 72, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.M.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef]
- Rampoldi, L.; Dobson-Stone, C.; Rubio, J.P.; Danek, A.; Chalmers, R.M.; Wood, N.W.; Verellen, C.; Ferrer, X.; Malandrini, A.; Fabrizi, G.M.; et al. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 2001, 28, 119–120. [Google Scholar] [CrossRef]
- Seong, E.; Insolera, R.; Dulovic, M.; Kamsteeg, E.J.; Trinh, J.; Bruggemann, N.; Sandford, E.; Li, S.; Ozel, A.B.; Li, J.Z.; et al. Mutations in VPS13D lead to a new recessive ataxia with spasticity and mitochondrial defects. Ann. Neurol. 2018, 83, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Maruki, Y.; Nakamura, M.; Tomemori, Y.; Kamae, K.; Tanabe, H.; Yamashita, Y.; Matsuda, S.; Kaneko, S.; Sano, A. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet. 2001, 28, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Rzepnikowska, W.; Flis, K.; Munoz-Braceras, S.; Menezes, R.; Escalante, R.; Zoladek, T. Yeast and other lower eukaryotic organisms for studies of Vps13 proteins in health and disease. Traffic 2017, 18, 711–719. [Google Scholar] [CrossRef]
- Bankaitis, V.A.; Johnson, L.M.; Emr, S.D. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 1986, 83, 9075–9079. [Google Scholar] [CrossRef]
- Brickner, J.H.; Fuller, R.S. SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J. Cell Biol. 1997, 139, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Neiman, A.M. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 2012, 125, 3004–3011. [Google Scholar] [CrossRef]
- Lang, A.B.; John Peter, A.T.; Walter, P.; Kornmann, B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 2015, 210, 883–890. [Google Scholar] [CrossRef]
- Park, J.S.; Thorsness, M.K.; Policastro, R.; McGoldrick, L.; Hollingsworth, N.M.; Thorsness, P.E.; Neiman, A.M. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 2016, 27, 2435–2449. [Google Scholar] [CrossRef]
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc. Natl. Acad. Sci. USA 2020, 117, 18530–18539. [Google Scholar] [CrossRef] [PubMed]
- John Peter, A.T.; Herrmann, B.; Antunes, D.; Rapaport, D.; Dimmer, K.S.; Kornmann, B. Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 2017, 216, 3219–3229. [Google Scholar] [CrossRef]
- Helle, S.C.; Kanfer, G.; Kolar, K.; Lang, A.; Michel, A.H.; Kornmann, B. Organization and function of membrane contact sites. Biochim. Biophys. Acta 2013, 1833, 2526–2541. [Google Scholar] [CrossRef]
- Lahiri, S.; Toulmay, A.; Prinz, W.A. Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 2015, 33, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.A.; Reinisch, K.M. Inter-organelle lipid transfer: A channel model for Vps13 and chorein-N motif proteins. Curr. Opin. Cell Biol. 2020, 65, 66–71. [Google Scholar] [CrossRef]
- Li, P.; Lees, J.A.; Lusk, C.P.; Reinisch, K.M. Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Bean, B.D.M.; Dziurdzik, S.K.; Kolehmainen, K.L.; Fowler, C.M.S.; Kwong, W.K.; Grad, L.I.; Davey, M.; Schluter, C.; Conibear, E. Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J. Cell Biol. 2018, 217, 3593–3607. [Google Scholar] [CrossRef]
- Park, J.S.; Okumura, Y.; Tachikawa, H.; Neiman, A.M. SPO71 encodes a developmental stage-specific partner for VPS13 in Saccharomcyes cerevisiae. Eukaryot. Cell 2013, 12, 1530–1537. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Braceras, S.; Tornero-Ecija, A.R.; Vincent, O.; Escalante, R. VPS13A is closely associated with mitochondria and is required for efficient lysosomal degradation. Dis. Model. Mech. 2019, 12. [Google Scholar] [CrossRef]
- Wang, J.; Fang, N.; Xiong, J.; Du, Y.; Cao, Y.; Ji, W.K. An ESCRT-dependent step in fatty acid transfer from lipid droplets to mitochondria through VPS13D-TSG101 interactions. Nat. Commun. 2021, 12, 1252. [Google Scholar] [CrossRef] [PubMed]
- Yeshaw, W.M.; van der Zwaag, M.; Pinto, F.; Lahaye, L.L.; Faber, A.I.E.; Gomez-Sanchez, R.; Dolga, A.M.; Poland, C.; Monaco, A.P.; van IJzendoorn, S.C.D.; et al. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife 2019, 8, e43561. [Google Scholar] [CrossRef] [PubMed]
- Dobson-Stone, C.; Danek, A.; Rampoldi, L.; Hardie, R.J.; Chalmers, R.M.; Wood, N.W.; Bohlega, S.; Dotti, M.T.; Federico, A.; Shizuka, M.; et al. Mutational spectrum of the CHAC gene in patients with chorea-acanthocytosis. Eur. J. Hum. Genet. 2002, 10, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Neiman, A.M. XK is a partner for VPS13A: A moleular link between Chorea-Acanthocytosis and McLeod Syndrome. Mol. Biol. Cell 2020, 31, 2425–2436. [Google Scholar] [CrossRef]
- Dziurdzik, S.K.; Bean, B.D.M.; Davey, M.; Conibear, E. A VPS13D spastic ataxia mutation disrupts the conserved adaptor-binding site in yeast Vps13. Hum. Mol. Genet. 2020, 29, 635–648. [Google Scholar] [CrossRef]
- Rzepnikowska, W.; Flis, K.; Kaminska, J.; Grynberg, M.; Urbanek, A.; Ayscough, K.R.; Zoladek, T. Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate. Hum. Mol. Genet. 2017, 26, 1497–1510. [Google Scholar] [CrossRef]
- Liu, Z.; Liang, Y.; Ang, E.L.; Zhao, H. A New Era of Genome Integration-Simply Cut and Paste! ACS Synth Biol. 2017, 6, 601–609. [Google Scholar] [CrossRef]
- Holzen, T.M.; Shah, P.P.; Olivares, H.A.; Bishop, D.K. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006, 20, 2593–2604. [Google Scholar] [CrossRef][Green Version]
- Roberts, C.J.; Raymond, C.K.; Yamashiro, C.T.; Stevens, T.H. Methods for studying the yeast vacuole. Methods Enzym. 1991, 194, 644–661. [Google Scholar]
- Boeke, J.D.; Trueheart, J.; Natsoulis, G.; Fink, G.R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzym. 1987, 154, 164–175. [Google Scholar]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Tomiyasu, A.; Nakamura, M.; Ichiba, M.; Ueno, S.; Saiki, S.; Morimoto, M.; Kobal, J.; Kageyama, Y.; Inui, T.; Wakabayashi, K.; et al. Novel pathogenic mutations and copy number variations in the VPS13A gene in patients with chorea-acanthocytosis. Am. J. Med. Genetics. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2011, 156, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Smolders, S.; Philtjens, S.; Crosiers, D.; Sieben, A.; Hens, E.; Heeman, B.; Van Mossevelde, S.; Pals, P.; Asselbergh, B.; Dos Santos Dias, R.; et al. Contribution of rare homozygous and compound heterozygous VPS13C missense mutations to dementia with Lewy bodies and Parkinson’s disease. Acta. Neuropathol. Commun. 2021, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; de los Santos, P.; Neiman, A.M. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 2004, 15, 1802–1815. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, E.M.; Suda, Y.; Nickas, M.; Snydsman, B.; Davis, T.N.; Muller, E.G.; Neiman, A.M. Vesicle docking to the spindle pole body is necessary to recruit the exocyst during membrane formation in Saccharomyces cerevisiae. Mol. Biol. Cell 2010, 21, 3693–3707. [Google Scholar] [CrossRef]
- Lottridge, J.M.; Flannery, A.R.; Vincelli, J.L.; Stevens, T.H. Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA 2006, 103, 6202–6207. [Google Scholar] [CrossRef]
- Velayos-Baeza, A.; Vettori, A.; Copley, R.R.; Dobson-Stone, C.; Monaco, A.P. Analysis of the human VPS13 gene family. Genomics 2004, 84, 536–549. [Google Scholar] [CrossRef]
- van Leeuwen, J.; Pons, C.; Mellor, J.C.; Yamaguchi, T.N.; Friesen, H.; Koschwanez, J.; Usaj, M.M.; Pechlaner, M.; Takar, M.; Usaj, M.; et al. Exploring genetic suppression interactions on a global scale. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Okumura, Y.; Nakamura, T.S.; Tanaka, T.; Inoue, I.; Suda, Y.; Takahashi, T.; Nakanishi, H.; Nakamura, S.; Gao, X.D.; Tachikawa, H. The Dysferlin Domain-Only Protein, Spo73, Is Required for Prospore Membrane Extension in Saccharomyces cerevisiae. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Parodi, E.M.; Roesner, J.M.; Huang, L.S. SPO73 and SPO71 function cooperatively in prospore membrane elongation during sporulation in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0143571. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.D.; Fink, G.R. Methods in Yeast Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1990. [Google Scholar]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Jin, L.; Zhang, K.; Xu, Y.; Sternglanz, R.; Neiman, A.M. Sequestration of mRNAs modulates the timing of translation during meiosis in budding yeast. Mol. Cell. Biol. 2015, 35, 3448–3458. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, D.D.; Lashkari, D.A.; Morris, D.; Mittmann, M.; Davis, R.W. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 1996, 14, 450–456. [Google Scholar] [CrossRef]
- Nakanishi, H.; Suda, Y.; Neiman, A.M. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 2007, 120 Pt 5, 908–916. [Google Scholar] [CrossRef]

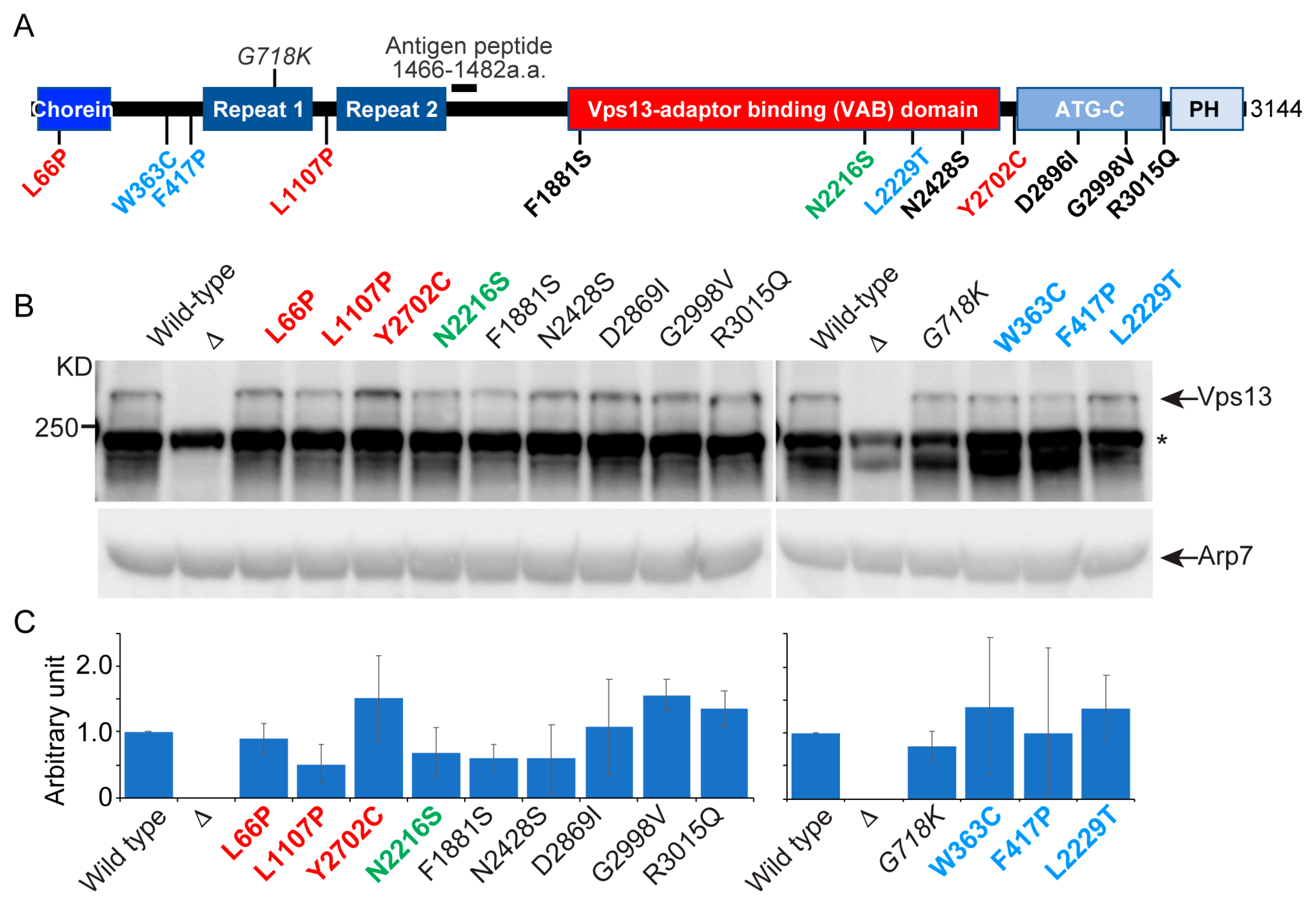

| Human Gene | Mutation | Reference | Cognate Yeast Mutation | Protein Level | CPY Secretion | Growth in mmm1Δ | Sporulation | ER-Phagy |
|---|---|---|---|---|---|---|---|---|
| VPS13 | ++ | - | + | + | + | |||
| vps13Δ | - | +++++ | - | - | - | |||
| VPS13A | L67P | [35] | L66P | ++ | - | - | + | + |
| A1095P | [26] | L1107P | + | ++ | - | + | + | |
| Y2721C | [5] | Y2702C | +++ | - | + | + | + | |
| VPS13B | N2993S | [3] | N2216S | + | - | + | + | + |
| VPS13C | W395C | [36] | W363C | +++ | - | + | + | + |
| A444P | [36] | F417P | ++ | - | + | + | + | |
| I2789T | [36] | L2229T | +++ | - | + | + | + | |
| VPS13D | L2900S | [2] | F1881S | + | - | + | + | ND |
| N3521S | [2] | N2428S | + | ++ | +/− | + | - | |
| N4107I | [6] | D2896I | +++ | - | + | + | ND | |
| A4210V | [6] | G2998V | +++ | - | + | + | ND | |
| R4228Q | [6] | R3015Q | +++ | - | + | + | + |
| Strain | Genotype | Sporulation Efficiency (%) * |
|---|---|---|
| JSYD11 | vps13Δ/VPS13 | 75 ± 5.9 |
| JSYD2 | vps13Δ/vps13Δ | 0 |
| JSYD12 | vps13Δ/vps13-N2428S | 71 ± 4.5 |
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATahis3Δ0 leu2Δ0 met15Δ0 ura3Δ0 ho | [48] |
| KO1 | same as BY4741 except vps13Δ::kanMX6 | [49] |
| KO2 | same as BY4741 except atg40Δ::kanMX6 | [49] |
| KO6 | same as BY4741 except ypt35Δ::kanMX6 | [49] |
| GCY5 | same as BY4741 except RTN1::GFP::SkHIS3MX6 | [47] |
| GCY2 | same as BY4741 except DID2::GFP::SkHIS3MX6 | [47] |
| RP201 | same as BY4741 except vps13-L66P | [13] |
| RP202 | same as BY4741 except vps13-1107P | [13] |
| RP203 | same as BY4741 except VPS13-Y2702C | [13] |
| RP206 | same as BY4741 except VPS13-W363C | this study |
| RP207 | same as BY4741 except VPS13-F417P | this study |
| JSP729 | same as BY4741 except VPS13-L2229T | this study |
| JSP757 | same as BY4741 except VPS13-N2428S | this study |
| JSP758 | same as BY4741 except VPS13-R3015Q | this study |
| JSP770 | same as BY4741 except VPS13-N2216S | this study |
| JSP786 | same as BY4741 except VPS13-F1881S | this study |
| JSP787 | same as BY4741 except VPS13-D2896I | this study |
| JSP788 | same as BY4741 except VPS13-G2998V | this study |
| JSP497 | same as BY4741 except VPS13^GFP | [13] |
| JSP861 | same as BY4741 except VPS13^GFP ypt35Δ::hphMX4 | this study |
| JSP871 | same as BY4741 except VPS13^GFP ypt35Δ::kanMX6 | this study |
| JSP816 | same as BY4741 except vps13-N2428S^GFP | this study |
| JSP857 | same as BY4741 except vps13-N2428S^GFP DID2::GFP::SkHIS3MX6 | this study |
| JSP863 | same as BY4741 except vps13-N2428S^GFP DID2::mRFP | this study |
| JSP832 | same as BY4741 except vps13-G718K N2428S^GFP | this study |
| JSP858 | same as BY4741 except vps13-G718K N2428S^GFP DID2::GFP::SkHIS3MX6 | this study |
| JSP864 | same as BY4741 except vps13-G718K N2428S^GFP DID2::mRFP | this study |
| JSP856 | same as BY4741 except vps13-G718K N2428S^GFP ypt35Δ::hphMX4 | this study |
| JSP845 | same as BY4741 except atg40Δ::kanMX6 RTN1::GFP::SkHIS3MX6 | this study |
| JSP846 | same as BY4741 except vps13Δ::kanMX6 RTN1::GFP::SkHIS3MX6 | this study |
| JSP847 | same as BY4741 except vps13-L66P RTN1::GFP::SkHIS3MX6 | this study |
| JSP848 | same as BY4741 except vps13-L1107P RTN1::GFP::SkHIS3MX6 | this study |
| JSP849 | same as BY4741 except VPS13-Y2702C RTN1::GFP::SkHIS3MX6 | this study |
| JSP851 | same as BY4741 except vps13-N2428S RTN1::GFP::SkHIS3MX6 | this study |
| JSP852 | same as BY4741 except VPS13-R3015Q RTN1::GFP::SkHIS3MX6 | this study |
| JSP854 | same as BY4741 except VPS13-N2216S RTN1::GFP::SkHIS3MX6 | this study |
| JSP883 | same as BY4741 except vps13-G718K RTN1::GFP::SkHIS3MX6 | this study |
| JSP881 | same as BY4741 except vps13-G718K N2428S RTN1::GFP::SkHIS3MX6 | this study |
| JSP441 | MATα mmm1Δ::kanMX6 his3ΔSK leu2 trp1::hisG ura3/pRS316-MMM1 | [13] |
| JSP759 | same as JSP441 except vps13-N2428S | this study |
| JSP443 | MATavps13Δ::SkHIS3MX6 mmm1Δ::kanMX6 leu2 trp1::hisG ura3 his3ΔSK/pRS316-MMM1 | [13] |
| BY4742 | MATα his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 ho | [48] |
| JSP512 | same as BY4742 except VPS13^GFP DID2::mRFP | [13] |
| JSP531 | same as BY4742 except vps13-G718K^GFP | [13] |
| JSP549 | same as BY4742 except vps13-G718K | [13] |
| HI27 | MATα ura3 his3ΔSK trp1::hisG arg4-NspI lys2 hoΔ::LYS2 rme1::LEU2 leu2 vps13Δ::SkHIS3MX6 | [50] |
| JSYD1 | MATa leu2Δ0 his3Δ0 met15Δ0 ura3Δ0 TRP1 ARG4 LYS2 MATα leu2 his3ΔSK MET15 ura3 trp1::hisG arg4-NspI lys2 ho RME1 VPS13 ho::LYS2 rme1::LEU2 VPS13 | [13] |
| JSYD2 | same as JSYD1 only vps13Δ::kanMX6/vps13Δ::SkHIS3MX6 | [13] |
| JSYD4 | same as JSYD1 only VPS13^GFP/vps13Δ::SkHIS3MX6 | [13] |
| JSYD11 | same as JSYD1 only VPS13/vps13Δ::SkHIS3MX6 | this study |
| JSYD12 | same as JSYD1 only vps13-N2428S/vps13Δ::SkHIS3MX6 | this study |
| JSYD13 | same as JSYD1 only vps13-N2428S^GFP/vps13Δ::SkHIS3MX6 | this study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.-S.; Hollingsworth, N.M.; Neiman, A.M. Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs. Int. J. Mol. Sci. 2021, 22, 6200. https://doi.org/10.3390/ijms22126200
Park J-S, Hollingsworth NM, Neiman AM. Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs. International Journal of Molecular Sciences. 2021; 22(12):6200. https://doi.org/10.3390/ijms22126200
Chicago/Turabian StylePark, Jae-Sook, Nancy M. Hollingsworth, and Aaron M. Neiman. 2021. "Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs" International Journal of Molecular Sciences 22, no. 12: 6200. https://doi.org/10.3390/ijms22126200
APA StylePark, J.-S., Hollingsworth, N. M., & Neiman, A. M. (2021). Genetic Dissection of Vps13 Regulation in Yeast Using Disease Mutations from Human Orthologs. International Journal of Molecular Sciences, 22(12), 6200. https://doi.org/10.3390/ijms22126200







